Abstract
The ubiquitin-specific protease HAUSP is a critical component of the p53–Mdm2 pathway by acting as a specific deubiquitinase for both p53 and Mdm2. Recent structural studies have indicated that p53 and Mdm2 bind to the N-terminal TRAF-like domain of HAUSP in a mutually exclusive manner. To understand the mechanism of HAUSP-mediated effects, we have created a p53 mutant that lacks HAUSP binding based on the crystal structure analysis. Indeed, this mutant p53 protein can be degraded byMdm2 but fails to interact with HAUSP both in vitro and in vivo. Surprisingly, however, we have found that direct interaction between HAUSP and p53 is not absolutely required for it to antagonize efficiently Mdm2- mediated ubiquitination of p53 and that HAUSP is capable of enzymatically functioning in trans on p53 by using Mdm2 as a bridge. Further, we show that a trimeric protein complex containing p53, Mdm2 and HAUSP can exist in vivo, despite mutually exclusive binding, with Mdm2 serving as a binding mediator for p53 and HAUSP. These findings reveal the complication of HAUSP-mediated effects in the p53–Mdm2 interplay. It also has important implications for the development of novel chemotherapeutic compounds aimed at blocking this protein–protein interaction.
Keywords: HAUSP, p53, deubiquitination, Mdm2 degradation
The p53signaling pathway is a highly complex, multicomponent signaling network that is of fundamental importance in tumor biology. The function of p53as a sequence-specific transcription factor is critical for the control of cell-cycle arrest, apoptosis and cellular senescence (Brooks and Gu, 2006). Without p53, cells lose pivotal signaling responses to DNA damage events, hypoxia and aberrant oncogene expression, as seen in a large percentage of human tumors that have lost p53 expression.
p53 is predominantly regulated through the ubiquitin– proteasome pathway by the E3 ubiquit in ligase Mdm2 and maintained at low protein levels during normal homeostasis. During times of stress, p53is quickly activated through several mechanisms that block the Mdm2–p53interacti on. Inhibiting this interaction is therefore a critical step for the efficient stabilization and activation of p53a s a transcription factor. When p53is not needed during times of cellular homeostasis, the protein is continuously ubiquitinated and degraded by Mdm2 (Marine et al., 2006).
The deubiquitinase HAUSP was first identified as a Herpes virus-associated cellular factor and subsequently shown to deubiquitinate and stabilize p53(Li et al., 2002). HAUSP is a member of the Ub-specific processing protease (UBP) subfamily of the deubiquitination enzymes (DUBs), a well conserved family of cysteine proteases (Nijman et al., 2005). In addition to p53, HAUSP can also specifically deubiquitinate Mdm2 (Cummins et al., 2004; Li et al., 2004). It also has been recently shown that mitochondrial HAUSP is important for the apoptosis response (Marchenko et al., 2007). These three proteins, p53, Mdm2 and HAUSP have a delicate, intimate balance that maintains p53protein s levels and is critical for an appropriate DNA damage response to occur. Because of the strong p53ind uction in the absence of HAUSP, it represents a potential chemotherapeutic target for the stabilization and activation of p53in cells that retain a wild-type (wt) copy of the gene.
Recently, structural and biochemical studies of the p53–HAUSP and Mdm2–HAUSP interactions have elucidated more defined and precise binding domains within these proteins (Hu et al., 2006; Sheng et al., 2006). p53and Mdm2 both bind within the N-terminal TRAF-like domain of HAUSP in a mutually exclusive manner. Importantly, it was shown that Mdm2 makes more conserved and extensive interactions with HAUSP than p53does, and the minimal binding domain on p53 for HAUSP was narrowly defined to the C-terminal residues 357–367.
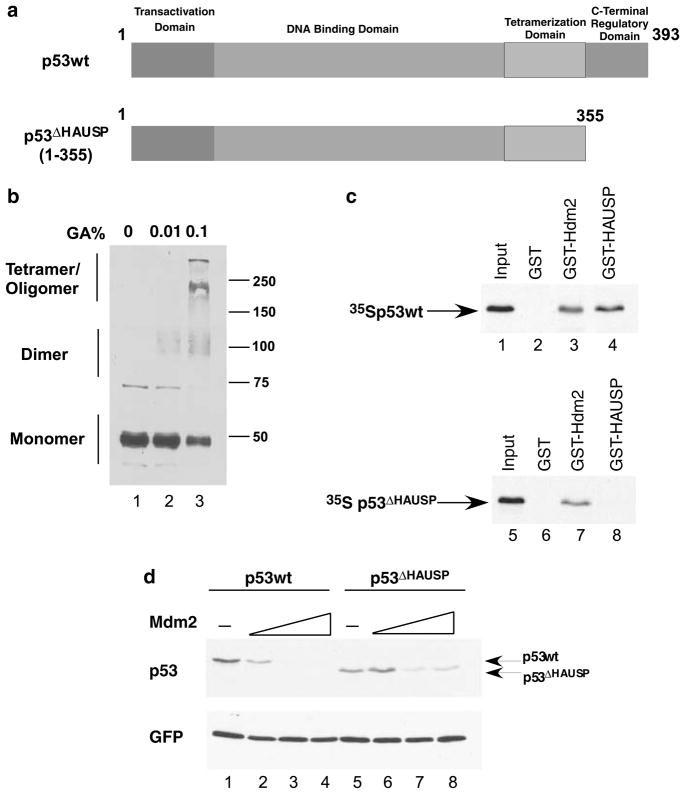
We wished to further characterize the interaction of these three proteins and the effect it had on p53function based on previous structural analysis. Since the minimal binding region for the tumor necrosis factor-receptor associated factor (TRAF)-like domain of HAUSP on p53was mapped to the C-terminal region of amino acids 357–367, and deletion of this region completely abrogates the p53–HAUSP interaction, we hypothesized that deletion of this region would render p53unab le to bind to HAUSP allowing for us to assess the Mdm2–HAUSP interaction in isolation. To confirm this, we first tested whether deletion of these C-terminal amino acids of p53 would inhibit binding to HAUSP. A deletion mutant of p53consis ting of amino acids 1–355 was subcloned into the pCIN4 vector (p53ΔHAUSP) (Figure 1a). An in vitro oligomerization assay was conducted to confirm that this mutant could still tetramerize similar to p53wt (Figure 1b, lanes 1–3). We then used the labeled protein in a glutathione-S-transferase (GST) pulldown assay (Figure 1c). As expected, p53wt interacted with GST– Hdm2 and GST–HAUSP (Figure 1c, lanes 1–4), but p53ΔHAUSP failed to bind to GST–HAUSP (Figure 1c, lanes 5–8). Notably, the deletion of the last 27 amino acids of p53did not disrupt its interaction with GST– Hdm2. This result confirmed previous structural findings and validated the approach described above.
Figure 1.
The TRAF-like domain of HAUSP binds to a 20 amino-acid specific region of p53. (a) A schematic representation of the constructs used in the following assays. (b) In vitro oligomerization assay. H1299 cells were transiently transfected with p53(1–355) and lysed 24 h post-transfection with Flag lysis buffer (50mM Tris–HCl, 137mM NaCl, 10mM NaF, 1mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100 and fresh protease inhibitors). Equal protein amounts were then incubated with the indicated amounts of glutaraldehyde on ice for 30 min and resolved on 8% SDS–PAGE and blotted with DO-1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). (c) In vitro GST pulldown assay. The p53wt and p53ΔHAUSP vectors were 35S-labeled using the trinitrotoulene coupled reticulocyte lysate system (Promega Corporation, Madison, WI, USA). For the in vitro GST pulldown assay, 35S-labeled p53wt (lanes 1–4) or p53ΔHAUSP (lanes 5–8) was incubated with either GST, GST-Hdm2 or GST-HAUSP at 4°C, washed four times with BC200 (20mM Tris–HCl, pH 7.3, 10% glycerol, 0.1mM EDTA and 200mM NaCl) and eluted with 20mM glutathione followed by SDS–PAGE and autoradiography. (d) In vivo degradation assay of p53wt and p53ΔHAUSP. H1299 cells were transiently transfected with green fluorescent protein (GFP), p53wt (lanes 1–4) or p53ΔHAUSP (lanes 5–8) and increasing amounts of Mdm2. Cells were then lysed 24 h post-transfection with Flag lysis buffer followed by western blot analysis with polyclonal α-Mdm2, DO-1 (Santa Cruz) and α-GFP (Clontech, Mountain View, CA, USA). GFP, green fluorescent protein; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; wt, wild type.
We next wished to assess the Mdm2-mediated degradation susceptibility of p53ΔHAUSP compared to that of p53wt. Although prominent, C-terminal lysines residues are not the exclusive sites for Mdm2-mediated ubiquitination as C-terminal truncation mutants of p53 are still able to be degraded, albeit less efficiently (Lohrum et al., 2001; Brooks and Gu, data not shown). To assess the degradation capability of p53ΔHAUSP, we performed an in vivo degradation assay in the H1299 cell line. A fixed amount of p53wt or p53ΔHAUSP was transfected into these cells with increasing amounts of Mdm2 and assessed by western blot 24 h post-transfection (Figure 1d).Wt p53was readily degraded byMdm2 in a dose-dependent manner (Figure 1d, lanes 1–4), while p53ΔHAUSP was degraded by Mdm2 with less efficiency (Figure 1d, lanes 5–8). Alhough p53ΔHAUSP was more stable than p53wt due to a loss of its major ubiquitination sites, it was still capable of being degraded and provided evidence that Mdm2-mediated ubiquitination and degradation could occur with this mutant of p53.
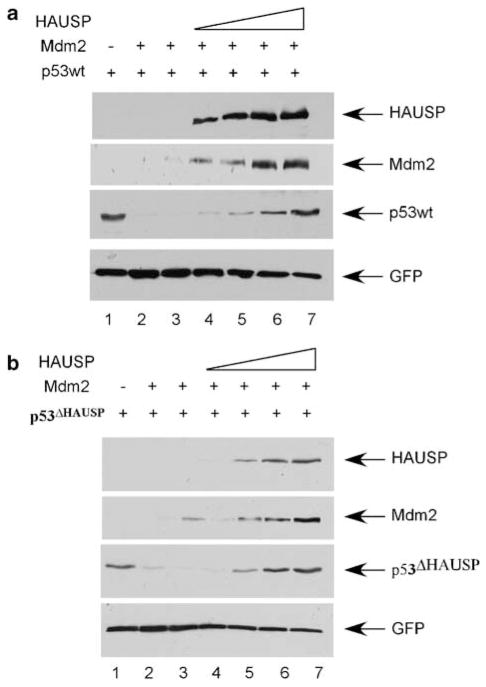
We next wanted to assess the ability of HAUSP to antagonize Mdm2-mediated degradation of a mutant of p53that could not bind HAUSP (p53ΔHAUSP) compared to p53wt. We hypothesized that HAUSP would not be able to rescue Mdm2-mediated degradation of p53ΔHAUSP due to a loss of direct binding. H1299 cells were first transfected with p53and two different amounts of Mdm2 and assessed by western blot (Figure 2a). We observed complete Mdm2-mediated degradation of p53 compared to control (Figure 2a, compare lanes 1 and 2 with 3). We then added increasing amounts of HAUSP on top of this. As shown in Figure 2a, HAUSP was capable of completely rescuing Mdm2-dependent p53 degradation in a dose-dependent manner (Figure 2a, lanes 4–7). We next performed the same assay using p53ΔHAUSP (Figure 2b). As expected, p53ΔHAUSP was degraded by Mdm2 at levels comparable to p53wt (Figure 2b, lanes 1–3). Surprisingly, the addition of HAUSP on top of this lead to the dose-dependent rescue of Mdm2-mediated p53degradat ion in a similar fashion to that of p53wt (Figure 2b, compare lanes 4–7). This result indicated that HAUSP retained its deubiquitinase function on p53despite losing binding ability and suggested that these proteins may form a three-protein complex in vivo with HAUSP functioning enzymatically in trans.
Figure 2.
HAUSP antagonizes Mdm2-dependent p53degradation through direct and indirect mechanisms. The H1299 cell line was transiently transfected with GFP, p53wt or p53ΔHAUSP, Mdm2 and increasing amounts of Flag-HAUSP. Western blot analysis was then carried out with α-Flag, α-Mdm2, DO-1 and α-GFP. (a) Complete Mdm2-mediated degradation of p53(lane 2 vs lane 1) is rescued by HAUSP in a dose-dependent manner (lanes 4–7). (b) HAUSP is capable of antagonizing Mdm2-dependent p53degradation in trans. HAUSP efficiently recovers p53ΔHAUSP from degradation by Mdm2 is a dose-dependent manner (lanes 4–7). GFP, green fluorescent protein; wt, wild type.
We next wished to deduce whether p53, Mdm2 and HAUSP could indeed form a three-protein complex in vivo. Owing to the mutually exclusive manner in which Mdm2 and p53bind to HAUSP, it was possible that the interaction between p53and Mdm2 was sufficient for bringing HAUSP within a functionally accessible distance through the formation of three-protein complex. To address this, we overexpressed two of the proteins containing epitope tags, namely Flag-tagged HAUSP and HA-tagged p53, and performed a double immunoprecipitation to visualize the presence of endogenous Mdm2 in the three-protein complex. The eluates were resolved by sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDS–PAGE) and western blot using the α-HA, α-Flag and α-Hdm2 antibodies (Figure 3). The protein complex of co-immunoprecipitated HAUSP and p53 contained endogenous Hdm2 protein as well (Figure 3, lane 3). This indicated that the pure p53–HAUSP protein complex also contained Mdm2, and suggested that these three proteins can form a three-protein complex in vivo.
Figure 3.
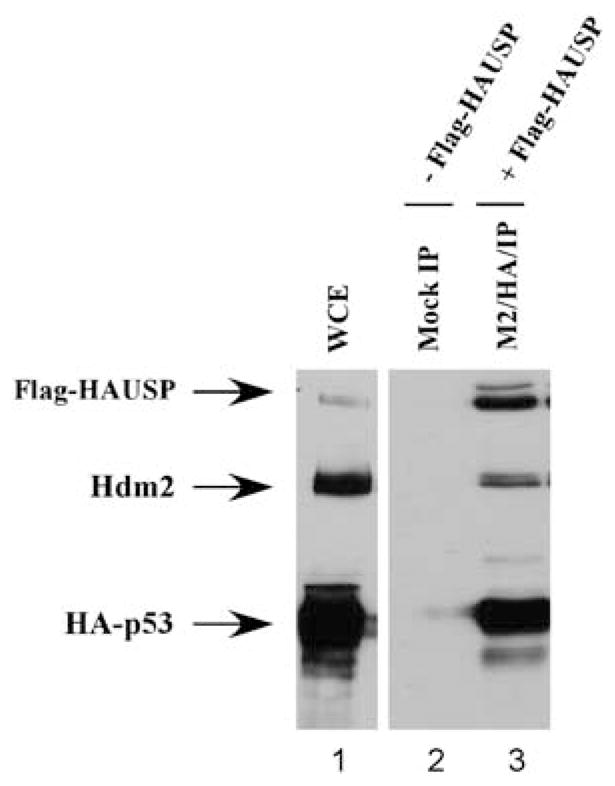
p53–Mdm2–HAUSP form a three-protein complex in vivo. H1299 cells were transfected with Flag-HAUSP and HA-p53and subjected to a two-step immunoprecipitation first with anti-Flag (M2) followed by anti-HA conjugated agarose beads. Western blot analysis and blotted with monoclonal α-FLAG, DO-1 and polyclonal α-Mdm2 to visualize HAUSP, p53and endogenous Mdm2 protein, respectively.
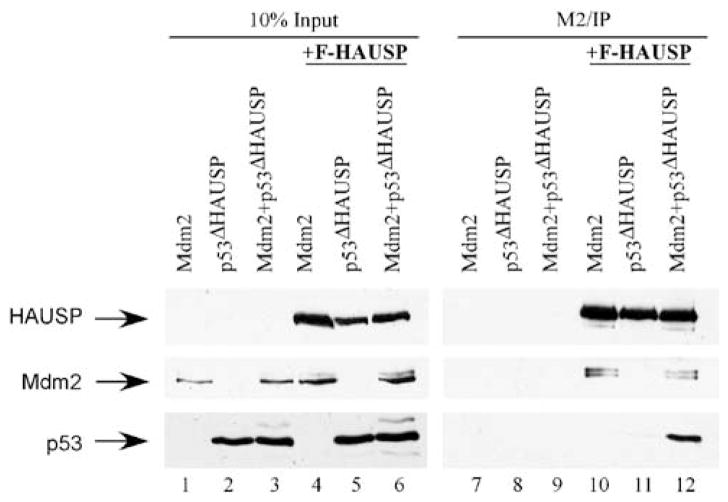
To support further our hypothesis that HAUSP could function in trans as a deubiquitinase for p53through indirect binding with Mdm2, we performed co-immunoprecipitation rescue experiments (Figure 4). We surmised that since HAUSP was not capable of interacting with p53ΔHAUSP, if HAUSP could indeed function indirectly through binding with Mdm2, then the addition of Mdm2 into the HAUSP-p53ΔHAUSP system would restore the three-protein complex in vivo. To assess this, Flag-HAUSP was co-transfected into H1299 cells with either Mdm2, p53ΔHAUSP or both Mdm2 and p53ΔHAUSP together. After immunoprecipitation the eluates were resolved by SDS–PAGE and western blot (Figure 4). As expected, Flag-HAUSP interacted with Mdm2 but failed to interact with p53ΔHAUSP (Figure 4, lanes 10 and 11). However, when Mdm2 and p53ΔHAUSP were transfected together, Flag-HAUSP was able to immunoprecipitate p53ΔHAUSP as a three-protein complex (lane 12). This result indicates that Mdm2 is capable of bridging the interaction between HAUSP and p53ΔHAUSP and that the 3 proteins can exist as a three-protein complex. Collectively, the data also suggest that HAUSP can sufficiently stabilize p53 in trans without direct interaction.
Figure 4.
Mdm2 can act as a binding mediator and rescue the binding deficiency between HAUSP and p53ΔHAUSP. H1299 cells were transiently transfected with Mdm2, p53ΔHAUSP or p53ΔHAUSP and Mdm2 together with or without Flag-HAUSP. Input (lanes 1– 6) and M2/IP (lanes 7–12) were resolved and blotted with α-Flag, α-Mdm2 and DO-1 antibodies.
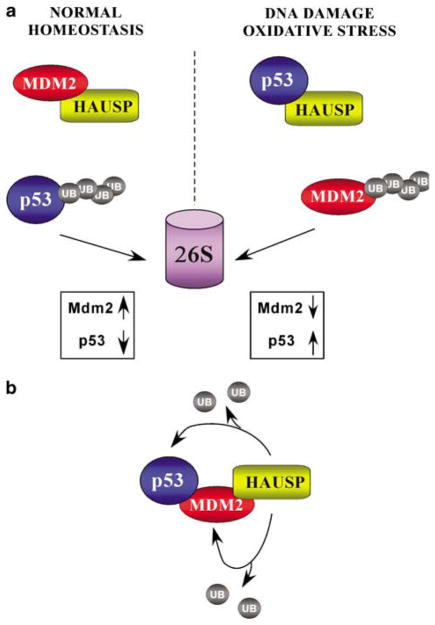
HAUSP has been shown to act as a specific deubiquitinase for several proteins including p53, Mdm2, FOXO4 and MdmX (Li et al., 2002; Meulmeester et al., 2005; Tang et al., 2006; van der Horst et al., 2006). Structural and biochemical analysis of the direct interactions between p53-HAUSP and Mdm2-HAUSP have elucidated defined binding regions and provide mechanistic explanations for their functions in vivo. p53 and Mdm2 bind to the same TRAF-like domain of HAUSP in a mutually exclusive manner with the binding affinity of Mdm2 for HAUSP several fold higher than that of p53. It has been suggested that the true physiologic binding partner for HAUSP under normal, unstressed cellular conditions is Mdm2. The stronger affinity and more extensive interactions Mdm2 makes with HAUSP, combined with the striking reduction of Mdm2 protein level in HAUSP ablated cells suggest preferential binding in vivo. Mechanistically, it is possible that p53–HAUSP becomes the preferential interaction upon times of DNA damage and cell stress. Indeed, treatment of cells with the radiomimetic agent neocarzinostatin (NCS) reduces the amount of Mdm2 protein that co-immunoprecipitates with HAUSP (Meulmeester et al., 2005). However, our data presented here show that p53, Mdm2 and HAUSP can exist as a three-protein complex in vivo and that HAUSP is capable of deubiquitinating p53 in trans through an indirect interaction mediated by Mdm2. In this regard, Mdm2 serves as a binding mediator between p53and HAUSP in addition to its functions as a specific E3ligase for p53. We hypothesize that there are two possible physiologic scenarios occurring in vivo: (a) direct binding between p53–HAUSP or Mdm2–HAUSP where mutually exclusive competition occurs and (b) a three-protein complex where HAUSP is able to function in trans without direct binding (Figure 5). In the first scenario, Mdm2 is the preferred binding partner for HAUSP under normal, unstressed cellular conditions (Figure 5, top panel). Since Mdm2 has inherent self-ubiquitination function, this interaction maintains a sufficient protein level for controlling p53through the ubiquitin–proteasome pathway. However, upon some type of DNA damage event, such as treatment with NCS, the Mdm2–HAUSP binding affinity is reduced through phosphorylation events, creating a pool of free HAUSP molecules that can readily interact with p53. This interaction provides a mechanism for the quick stabilization of p53 and allows unbound Mdm2 to self-ubiquitinate for subsequent degradation. The mutually exclusive manner in which these proteins bind allows for a balance of protein levels and ultimate functional regulation of p53. However, our data suggest that the B scenario could exist for a subpopulation of protein in vivo (Figure 5, bottom panel). This pool of protein exists as a three-protein complex, where HAUSP deubiquitinates p53through an indirect interaction mediated by Mdm2. The close proximity of HAUSP to p53through this indirect interaction is sufficient for the deubiquitination of p53.
Figure 5.
Schematic representation of two possible scenarios for in vivo interactions between p53, Mdm2 and HAUSP. (a) The preferred protein binding partner for HAUSP in vivo under unstressed physiologic conditions is Mdm2. Upon DNA damage or other types of cellular stress, the HAUSP–Mdm2 interaction is abrogated through ATM-mediated phosphorylation creating a pool of free HAUSP molecules available for p53binding and subsequent deubiquitination and stabilization. (b) A subpopulation of a three-protein complex may exist in vivo. HAUSP can function enzymatically in trans and deubiquitinate p53indirectly through its interactions with Mdm2.
Although the direct binding between p53–HAUSP and Mdm2–HAUSP has been well established through defined crystal structures, we have provided evidence that direct p53–HAUSP binding is not required for HAUSP-mediated deubiquitination of p53. This particular point has important implications for strategies in the development of novel chemotherapeutic compounds aimed at the p53 pathway. A standard approach for the chemotherapeutic-mediated stabilization of p53ha s been through the development of small molecule inhibitors that target the protein–protein interaction between Mdm2 and p53. Indeed, the promising potential of the small molecule Mdm2 antagonist Nutlin-3A in stabilizing p53 in a variety of tumor types indicates that this approach has yielded the targeted goal of activating p53through the inhibition of Mdm2 interaction (Vassilev et al., 2004). However, our data indicate that this approach may not be the most appropriate for targeting the protein– protein interactions of HAUSP. Ablation of HAUSP in vivo has a profound stabilizing effect on p53as seen in both transient knockdown and somatic knockout systems. Although HAUSP is a logical target for the development of small compound inhibitors as a mechanism for activating p53, we have shown here that HAUSP can function as a deubiquitinase for p53 in trans, indicating that blocking the protein–protein interaction may not efficiently inhibit its effect on p53. In this case, a more logical approach would be to develop a small molecule inhibitor of the catalytic site, a small region of HAUSP that is highly conserved among the family of DUBs (Nijman et al., 2005). This would inhibit the enzymatic activity of HAUSP on all substrates and would ensure that there would be no trans effect that could result from targeting protein– protein interactions. Future studies will address the feasibility of this approach and will assess the ability of HAUSP to function in trans on other known substrates such as Mdm2 and MdmX.
References
- Brooks CL, Gu W. p53ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. following 486. [DOI] [PubMed] [Google Scholar]
- Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53by HAUSP is an important pathway for p53stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH. C-terminal ubiquitination of p53contributes to nuclear export. Mol Cell Biol. 2001;21:8521–8532. doi: 10.1128/MCB.21.24.8521-8532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J. 2007;26:923–934. doi: 10.1038/sj.emboj.7601560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, et al. Molecular recognition of p53and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. FOXO4 transcriptional activity is regulated by monoubi-quitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]