Abstract
Identifying the effects of maternal risk factors during pregnancy on infant and child health is an area of tremendous research interest. However, of interest to policy makers is unraveling the causal effects of prenatal risk factors, not their associations with child health, which may be confounded by several unobserved factors. In this paper, we evaluate the utility of genetic variants in three genes that have unequivocal evidence of being related to three major risk factors – CHRNA3 for smoking, ADH1B for alcohol use, and FTO for obesity – as instrumental variables for identifying the causal effects of such factors during pregnancy. Using two independent datasets, we find that these variants are overall predictive of the risk factors and are not systematically related to observed confounders, suggesting that they may be useful instruments. We also find some suggestive evidence that genetic effects are stronger during than before pregnancy. We provide an empirical example illustrating the use of these genetic variants as instruments to evaluate the effects of risk factors on birth weight. Finally, we offer suggestions for researchers contemplating the use of these variants as instruments.
1. Introduction
The influence of maternal risk factors during pregnancy on fetal and child health has been widely studied in several disciplines. A large body of literature documents negative effects of maternal smoking during pregnancy on birth weight (Rosenzweig and Schultz 1983; Wehby et al. 2011; Wüst 2010) and child neurological development (Wehby et al. 2011a). Others report a negative effect of excessive alcohol consumption on fetal development (Sullivan 2011; Lemoine et al. 1968; Jones and Smith 1973). In contrast, studies of the effects of maternal body weight on child health have produced mixed results (Bhattacharya et al. 2007; Khashan and Kenny 2009; Torloni et al. 2009).
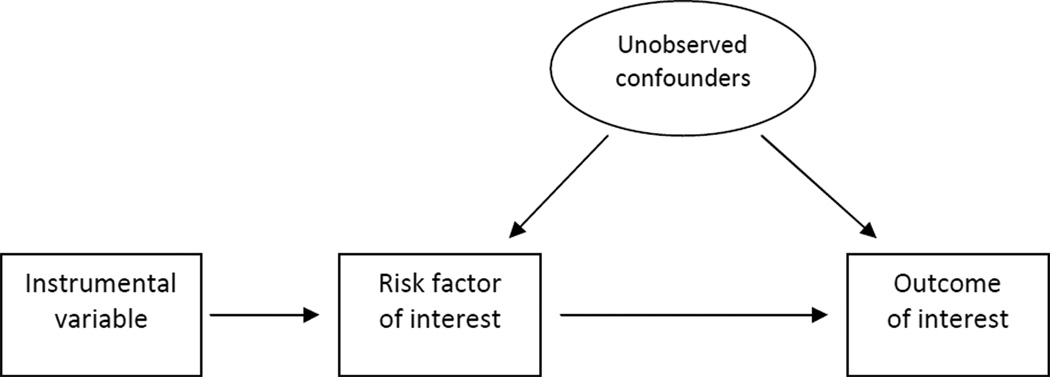
A major challenge in studying the effects of maternal behaviors and risk factors on child health is the role of unobservable confounders. Maternal prenatal behaviors and risk factors are correlated with several typically unobserved or inadequately measured characteristics that are relevant for fetal/child health. If ignored, these may confound the effects of the behaviors and risk factors of interest. Theory suggests that these unobservable confounders include several personality and preference traits, as well as maternal awareness of any risks to fetal/child health.1 Not all such factors are adequately observed. For example, maternal psychosocial (stress and depression/anxiety) and nutritional influences may be inadequately captured. Similarly, maternal or family history of health problems, which may reflect certain health risks for future children and modify maternal behaviors, may not be well measured. Therefore, the correlations between the many maternal behaviors and risk factors, several of which are unobservable, make it virtually impossible to separate the effect of one factor from the other in classical regression models that only adjust for observable confounders. Such models will likely result in biased estimation of the effects of maternal behaviors and risk factors of interest on child health outcomes since it is very unlikely – if not impossible – that one can directly adjust for all relevant confounders. This can be shown graphically in a Directed Acyclic Graph (DAG), such as Figure 1. We are interested in the relationship between the risk factor and the outcome, but there may be several unobserved characteristics that affect both, leading to biased estimates.
Figure 1.
Directed Acyclic Graph for IV Approach
An additional complication of these unobservable confounders is that it is impossible in most cases to a-priori determine the direction of the bias based on theory (Wehby et al. 2009). For example, women who care more about their child’s health than others are more likely to avoid risk factors and to engage in healthy behaviors (such as adequate and appropriate nutrition, exercise, stress control, prenatal care, etc.). Such self-selection would result in overestimating (in absolute value) the negative effects of the risk factors of interest on infant health. In contrast, women who perceive greater risks for fetal/child health are more likely to engage in healthy behaviors and to avoid risk factors during pregnancy. For example, women who expect fetal growth complications due to previous personal or family history of such conditions may be more likely to avoid smoking or alcohol use during pregnancy and more likely to adopt healthy behaviors such as prenatal care or appropriate diet. This type of self-selection may result in underestimating the positive effects of healthy behaviors (such as prenatal care) and understating the negative effects of risk factors (such as smoking). Therefore, the net bias is a function of these opposite biases. Given that it is generally impossible to know which bias dominates, it is challenging to a priori determine the direction of the net bias due to unobservable confounders. This emphasizes the need for empirical investigation to evaluate the sign and magnitude of the bias.
Instrumental variable (IV) methods may be employed to account for such bias resulting from unobservable confounders. The premise is to employ variables (called instruments) that are strong predictors of the risk factors of interest (such as maternal smoking during pregnancy), but otherwise are unrelated to the outcome of interest (such as child health) except through their effects on these risk factors. In other words, these instruments should not affect the outcome directly, nor should they relate to the outcome through unobservable confounders. This is presented graphically in Figure 1, where the instrumental variable is associated with the risk factor of interest, but there is no direct link between the instrument and the outcome of interest, nor with the unobserved confounders. Under these conditions, the only variation in the risk factors used to estimate their effects on the outcome is that predicted by the instrument(s). This variation is unrelated to unobservable confounders under the IV assumptions described above. Therefore, an IV model using instruments that satisfy these IV assumptions allows for estimating the causal effects of risk factors on the outcome.
Until now, studies of the causal effects of prenatal health behaviors and risk factors have generally relied on policy instruments, such as changes in cigarette taxes/prices or smoking policies. While useful, these instruments have several limitations, such as ignoring variation in the etiology of behaviors/risk factors for individuals living in the same geographic area at which the instrument is measured. Also, policies may be endogenous to population behavior since they may be motivated by concerns about population health (Wehby, et al. 2011b).
Genes are implicated in many aspects of human behavior. Several recent studies have used genetic variants such as SNPs as instruments for studying causal behavior effects, also known as Mendelian randomization (Wehby et al. 2008); see e.g. (Chen et al. 2008; Timpson et al. 2009; Kivimaki et al. 2008; von Hinke Kessler Scholder et al. 2010; Lawlor et al. 2008; Smith et al. 2007; Smith and Ebrahim 2004; Davey Smith and Ebrahim 2003b; Fletcher 2009; Fletcher and Lehrer 2011; Fletcher 2011; Wehby et al. 2011a; Wehby et al. 2011b; Wehby et al. 2012). The strength originates from the fact that genetic variants are randomly assigned within a family at conception and are not reversely modified by one’s behaviors or environment. However, there are several potential threats to the validity of genetic instruments, with the main ones being pleiotropy (multifunctionality of genes), potential correlations between different genetic variants relevant for behavior/risk factors and outcomes due to linkage disequilibrium (LD) or co-inheritance of variants, and population stratification, leading to biased genetic effects (Davey Smith and Ebrahim 2003a; Lawlor et al. 2008; Didelez, Meng. S., and Sheehan 2010; von Hinke Kessler Scholder et al. 2011; Sheehan et al. 2008; Colhoun, McKeigue, and Davey Smith 2003). Population stratification occurs when genetic variants differ in their allelic distribution (i.e. the frequency of the minor allele) between subpopulations. The most common example of population stratification is that by ancestry, due to its correlation with social constructs such as race and ethnicity. For example, if the allelic distribution and outcome of interest differ systematically between these subgroups, the estimate of interest will be confounded. As such, population stratification may be particularly important to consider when studying diverse ancestral populations like those in the United States. We discuss this issue in further detail below.
In this study, we evaluate the use of genetic variants in three genes, CHRNA3, ADH1B, and FTO as instrumental variables to study the causal effects of maternal smoking and alcohol consumption during pregnancy, and obesity/body weight prior to pregnancy, respectively, on infant and child health. We argue it is important to evaluate the fit of a genetic variant as an instrument for a particular research question of interest on a case-by-case basis, assessing the extent to which the selected variants are:
Predictors of these maternal risk factors;
Unrelated to other variables that may also affect child health outcomes.
The first evaluation examines the relationship between the genetic variants and the associated risk factor during pregnancy. Most of the evidence on the importance of these genes is based on samples that are very much underrepresented in women of childbearing age and pregnant women. Indeed, less than a handful of studies have specifically evaluated the associations of these genes with maternal prenatal risk factors (Zuccolo et al. 2009; Freathy et al. 2009; Lawlor et al. 2011). However, the effects of genes on behavioral and health phenotypes may differ across the lifecycle. Before and during pregnancy, women go through many biological, hormonal, or behavioral changes, which may modify genetic influences on behavior and risk factors. For instance, many women quit smoking and drinking and modify their diet before and/or during their pregnancy. Given that the effects of certain genetic variants may vary before and during pregnancy compared to a more ordinary time, it is important to assess the effects of these variants specifically during the pregnancy period in order to evaluate their validity as instruments for studying the effects of prenatal risk factors on child health.
The second evaluation aims at indirectly testing whether there are any patterns suggesting that the instruments affect child outcomes other than through affecting the maternal risk factors of interest. Such evidence would violate the IV condition requiring the instruments to only affect the outcome through these factors.
Several genes have been implicated in smoking, alcohol use, and obesity. However, replicability of the reported associations is crucial, as we now know that many initial genetic associations fail to replicate (Colhoun et al. 2003). Indeed, there are only a few genes that have unequivocal evidence of being related to these risk factors in the literature. These include CHRNA3 for smoking, ADH1B for alcohol use, and FTO for obesity and body weight.
CHRNA3 is thought to influence smoking through coding for a receptor of nicotinic acetylchoine, a metabolic form of nicotine (OMIM 2011). CHRNA3 variants have been found to be significantly associated with nicotine dependence measures, including cigarettes per day and smoking intensity (Saccone et al. 2007; Saccone et al. 2009; Berrettini et al. 2008; Thorgeirsson et al. 2008; Weiss et al. 2008; Stevens et al. 2008; Sherva et al. 2008; Baker et al. 2009; Keskitalo et al. 2009; Liu et al. 2010; The Tobacco and Genetics Consortium 2010).
ADH1B is involved in alcohol metabolism by coding for a subunit of alcohol dehydrogenase (ADH) which catalyzes the rate-limiting step for ethanol metabolism (OMIM 2011). The risk allele in the functional variant rs1229984 increases the production of acetaldehyde and consequently alcohol toxicity symptoms believed to lead to lower alcohol consumption. ADH1B has been linked with a variety of alcohol consumption and dependence measures in multiple populations (Reich et al. 1998; Whitfield et al. 1998; Saccone et al. 2000; Loew et al. 2003; Wall et al. 2005; Saccone et al. 2005; Duranceaux et al. 2006; Zintzaras et al. 2006; Luo et al. 2006; Zhang et al. 2007; Tolstrup et al. 2008; Zuccolo et al. 2009; Birley et al. 2009; Macgregor et al. 2009; Sherva et al. 2009; Liu et al. 2011; Ghosh et al. 2008).
Although its function is not fully understood, FTO has been associated with obesity related traits both in adults and children (Scuteri et al. 2007; Scherag et al. 2010; Frayling et al. 2007; Melen et al. 2010; Meyre et al. 2009; Thorleifsson et al. 2009; Cheung et al. 2011; Willer et al. 2009; Dina et al. 2007; Tung and Yeo 2011). The increase in obesity is thought to occur through an increase in food and energy intake (Cecil et al. 2008; Haupt et al. 2009; Jonsson and Franks 2009; Speakman, Rance, and Johnstone 2008; Timpson et al. 2008) and a decrease in satiety (Wardle et al. 2008; den Hoed et al. 2009; Rutters et al. 2010).2
Using two independent datasets, we find that the genetic variants in these three genes are likely to be appropriate and useful instruments for these prenatal risk factors as well as some suggestive evidence of intensified genetic effects during pregnancy. We also provide an empirical example of using these genetic variants to evaluate the effects of maternal smoking, alcohol consumption, and body weight on child birth weight.
2. Methods and Data
2.1 Evaluating if the instruments fit the two IV conditions
We assess the fit of the selected genetic variants as instruments by evaluating the extent to which they satisfy the two IV conditions:
They are a predictor of the maternal risk factor of interest;
They are unrelated to other variables that may also affect child health.
We investigate the first condition by evaluating the strength of the association between each genetic variant and its related risk factor. This is done by regressing the risk factor on the genetic instrument using an ordinary least squares (OLS) regression and calculating the F statistic for the instrument effect on the risk factor. A common rule of thumb for a non-weak instrument is an F-statistic of at least 10 (Staiger and Stock 1997).3 Lower F-statistics indicate weaker instruments, which are more likely to lead to bias in the IV estimates and may require additional inference approaches. We use a linear probability model for binary risk factors to follow a standard interpretation of the instrument effects across all models and to evaluate the instruments’ strength using a common metric based on the F-statistic threshold. We repeat the estimation for binary risk factors using logit and find similar results to OLS (results available from the authors upon request). Linear probability models provide a good approximation of the average effect in large samples.4
Each of the evaluated SNPs has two alleles. SNPs may be coded in different ways under different assumptions. Under a dominant model, an indicator for the presence of the risk allele in either homozygote or heterozygote forms would be used. Under a recessive model, an indicator for the homozygote genotype of the risk allele would be used. Under an additive model, a variable representing the number of risk allele copies (0, 1, 2) would be appropriate. We specify the FTO and CHRNA3 to have three values assuming an additive model, which is consistent with the literature and is supported in our data. Given that the majority of individuals of European ancestry are homozygous for the common allele of rs1229984 in ADH1B, we use a binary indicator for the risk allele assuming a dominant model, which is also the model supported in our data.
Evaluating the second IV assumption is less straightforward. In fact, it is impossible to fully test this assumption due to the role of unobservables. Therefore, the foremost validation should be based on the current knowledge about the functions of the gene (and genetic variant) and whether they may relate to child outcomes other than through the maternal risk factor of interest. We evaluate this by reviewing the literature on the functions and expression of these genes. In addition, an ad-hoc check of the second IV assumption is to evaluate the association between the genetic variants and observed characteristics that may confound the relationship between the risk factors of interest and child health, but which are not affected by the risk factors (i.e. not on their causal pathway to child health). The premise is that if a genetic variant randomizes unobserved confounders, it would also be expected to randomize observed ones. While this does not provide a complete validation, it would reveal if there are systematic associations between the genetic variants and other characteristics, which would reduce the evidence for the fit of these variants as instruments. We evaluate the association between the genetic variants and such observable characteristics including maternal demographic, socioeconomic, and health indicators, pregnancy wantedness, and others by regressing the instrument on a vector of these characteristics. The two datasets that we employ vary in the available background measures, but they both capture the main maternal demographic, socioeconomic and health characteristics.
2.2 Datasets
2.2a. Danish National Birth Cohort (DNBC) dataset
The first dataset we employ in this study is based on a subsample from the Danish National Birth Cohort (DNBC). The DNBC is a cohort study of more than 100,000 pregnant women and their babies in Denmark in 1996–2003 (Olsen et al. 2001). Pregnant women were identified and recruited during their first prenatal visit, which generally occurred in the first trimester. The women were interviewed for their health and socioeconomic backgrounds, and provided DNA samples. The women were interviewed again around the beginning of the third trimester and after delivery.
The specific dataset that we employ in our study comes from the Prematurity Genome Wide Association Study (GWAS), which included a subsample from the DNBC of 1,000 preterm birth infants and 1,000 randomly selected term infants and their mothers. The vast majority of the sample mothers are thought to be of European ancestry. The GWAS genotyped the maternal DNA samples for 550K SNPs using a high density panel (Illumina Human660W-Quadv1_A platform) at the Center for Inherited Disease Research (CIDR). The covered genes included CHRNA3, FTO, and ADH1B. The SNPs in these genes reported in the literature to have the largest effects on the risk factors of interest were not covered in the GWAS panel. However, there are other covered SNPs in strong or perfect LD with these SNPs in CHRNA3 and FTO. LD, the correlation between alleles of two genetic variants (SNPs), is evaluated by measures of association such as a Pearson correlation coefficient or its squared term, the r-squared (r2).5 We report the results for one SNP per gene that is in high or perfect LD with the SNP in the literature with the largest effect on the risk factor of interest and that has the most explanatory power for the risk factor in this sample.6 There are no genotyped SNPs in the DNBC GWAS panel in strong LD with rs1229984 in ADH1B. Therefore, we only evaluate this SNP in the second dataset we use as described below.
We evaluate rs12914385 in CHRNA3 for its validity as an instrument for smoking measures in the DNBC sample. This SNP is in strong LD (r2 of 0.836) with rs1051730 which has been consistently found to have one of the strongest effects on smoking behavior. We also evaluate rs8050136 in FTO as an instrument for body mass index (BMI) and obesity. This SNP is in perfect LD (r2 of 1.0) with rs9939609, which is the SNP with the most convergent evidence in the literature for having the strongest effects on BMI/obesity (Frayling et al. 2007; Cauchi et al. 2009; Cornes et al. 2009; Hubacek et al. 2008).
The measures on smoking and obesity are obtained from the interviews conducted with the mothers during pregnancy. Our analytical sample ranges from 1811 to 1898 observations depending on the number of women with complete data for each analysis that we employ. CHRNA3 has been mainly reported in previous studies to be related to the number of cigarettes. Therefore, we evaluate the effects of the CHRNA3 variant on the average number of cigarettes per day during pregnancy (including 0 for non-smokers) and on number of cigarettes among smokers. Also, we evaluate its effect on any smoking during pregnancy. When possible, evaluating the genetic effects separately for before and during pregnancy is important to identify if these are modified by pregnancy. However, smoking before pregnancy is not measured in the DNBC dataset but is measured in the second dataset described below.7 We evaluate the effects of the FTO variant on obesity (BMI>30) and body mass index (BMI) before pregnancy. Table 1 describes the measures of risk factors and genetic instruments used in the DNBC analysis.8
Table 1.
Descriptive Statistics for Risk Factors, Birth Weight, and Genetic Instruments in DNBC
| Variable | Description | Mean | Std. Dev. |
N |
|---|---|---|---|---|
| Smoking | 0/1 indicator for any smoking during pregnancy |
0.280 | 0.449 | 1900 |
| Cigarettes | Average number of cigarettes smoked per day |
1.541 | 3.757 | 1813 |
| BMI | Maternal body mass index | 23.527 | 4.308 | 1811 |
| Obese | 0/1 indicator for obese mothers (BMI >30) |
0.087 | 0.282 | 1811 |
| Birth weight | Child birth weight in grams | 3095.077 | 871.277 | 1914 |
| rs12914385 | Number of T alleles of maternal rs12914385 in CHRNA3 |
0.749 | 0.685 | 1934 |
| rs8050136 | Number of A alleles of maternal rs8050136 in FTO |
0.799 | 0.692 | 1937 |
2.2b. Avon Longitudinal Study of Parents and Children (ALSPAC)
The second dataset is a cohort of children born in one geographic area (Avon) in England. Women eligible for enrolment in the population-based ALSPAC study had an expected delivery date between 1 April 1991 and 31 December 1992. Approximately 85% of eligible mothers enrolled, leading to about 14,000 pregnancies. The Avon area has approximately 1 million inhabitants and is broadly representative of the UK as a whole, although slightly more affluent than the general population (Golding, Pembrey, and Jones 2001); see www.bristol.ac.uk/alspac for more a detailed description of the data).
Although the cohort focused on the children and their development, ALSPAC also includes regular data collection on the mothers. For all analyses, we exclude mothers of non-white ethnic origin to reduce the risk of population stratification. All genotyping was performed by KBioscience (http://www.kbioscience.co.uk). SNPs were genotyped using the KASPar chemistry, which is a competitive allele-specific PCR SNP genotyping system using FRET quencher cassette oligos (http://www.kbioscience.co.uk/genotyping/genotyping-chemistry.htm).
We use the SNPs in the (maternal) genes CHRNA3, ADH1B and FTO that have been reported in the literature to have the strongest effects on the risk factors of interest (rs1051730, rs1229984 and rs9939609, respectively).
We observe maternal smoking and alcohol consumption at three points in time during pregnancy: at 8 weeks, 18 weeks and 32 weeks gestation. We refer to these as the first, second, and third trimester respectively.9 We use dummy variables indicating any smoking and alcohol consumption at these three time points, as well as counts indicating the number of cigarettes and units of alcohol.10 We also observe whether the mother binged, measured only in the second trimester and defined as drinking the equivalent of two pints of beer, four glasses of wine, or four pub measures of spirit. Similar to the DNBC analysis, we estimate the gene effects on maternal risk factors before pregnancy including for smoking (any smoking and number of cigarettes per day) and any alcohol consumption. Finally, we evaluate the FTO effects on BMI and obesity, which are obtained based on self-reported measures of pre-pregnancy height and weight. Table 2 describes the variables for the ALSPAC sample.11
Table 2.
Descriptive Statistics for Risk Factors, Birth Weight, and Genetic Instruments in ALSPAC
| Variable | Description | Mean | Std. Dev. |
N |
|---|---|---|---|---|
| Mother smokes pre-pregnancy |
0/1 indicator for any smoking during pregnancy |
0.318 | 0.466 | 10521 |
| Mother smokes in Trimester 1 |
0/1 indicator for any smoking in first trimester |
0.194 | 0.396 | 9864 |
| Mother smokes in Trimester 2 |
0/1 indicator for any smoking in second trimester |
0.193 | 0.395 | 10521 |
| Mother smokes in Trimester 3 |
0/1 indicator for any smoking in third trimester |
0.199 | 0.399 | 9177 |
| No. of cigarettes in Trimester 1 |
Number of cigarettes a day in first trimester |
2.090 | 5.114 | 9864 |
| No. of cigarettes in Trimester 3 |
Number of cigarettes a day in third trimester |
2.148 | 5.249 | 9177 |
| Mother drinks pre- pregnancy |
0/1 indicator for any alcohol during pregnancy |
0.928 | 0.258 | 10449 |
| Mother drinks in Trimester 1 |
0/1 indicator for any alcohol in first trimester |
0.310 | 0.463 | 9500 |
| Mother drinks in Trimester 2 |
0/1 indicator for any alcohol in second trimester |
0.291 | 0.454 | 10414 |
| Mother drinks in Trimester 3 |
0/1 indicator for any alcohol in third trimester |
0.331 | 0.471 | 5849 |
| No. of alcoholic units pre-pregnancy |
Categorical variable indicating the number of units of alcohol per week pre-pregnancy |
1.594 | 0.785 | 10449 |
| No. of alcoholic units in T1 |
No. of units of alcohol a week in first trimester |
1.699 | 4.285 | 9500 |
| No. of alcoholic units in T2 |
No. of units of alcohol a week in second trimester |
1.521 | 3.854 | 10414 |
| No. of alcoholic units in T3 |
No. of units of alcohol a week in third trimester |
1.730 | 3.702 | 5849 |
| Bingeing | Mother binged in second trimester | 0.307 | 0.890 | 10400 |
| BMI | Maternal Body Mass Index | 22.95 | 3.822 | 9403 |
| Obese | 0/1 indicator for mother being obese (BMI>30) |
0.055 | 0.229 | 9403 |
| Birth weight | Child birth weight in grams | 3407.10 | 559.437 | 10869 |
| rs1051730 | Number of T alleles of maternal rs1051730 (CHRNA3) |
0.667 | 0.671 | 7830 |
| rs1229984 | 0/1 indicator for one or two copies of the A allele of maternal rs1229984 (ADH1B) |
0.050 | 0.222 | 7818 |
| rs9939609 | Number of T alleles of maternal rs9939609 (FTO) |
0.799 | 0.690 | 7832 |
3. Results
We first show the results from evaluating the extent to which the instruments satisfy the first IV condition. We describe the effects of each instrument on the related risk factor in separate sections for each gene. Next, we show the results of the indirect tests of the second IV condition that the instruments are unrelated to confounders. In that section, we also describe the correlations between the risk factors and observable confounders to further make the case for the need of IV analysis. Finally, we provide an applied example for using these instruments to evaluate the effects of selective prenatal risk factors on birth weight.
3.1 Evaluating if instruments are significantly related to risk factors of interest
3.1a CHRNA3
Table 3 lists the coefficients of the regressions of smoking measures on CHRNA3 variants and the F statistics for variant effects (panel A for the Danish sample and panel B for the ALSPAC sample). In the DNBC sample, the CHRNA3 variant has significant effects on the total number of cigarettes smoked, and on the number of cigarettes among smokers, with F statistics of 9 and 10.6, respectively, suggesting overall a reasonably non-weak instrument. Each copy of the risk allele increases the total number of cigarettes by about 0.39 cigarettes per day and the number of cigarettes among smokers by about 1.2 cigarettes per day. In contrast, CHRNA3 has an insignificant effect on the probability of smoking during pregnancy.
Table 3.
Effects of Genetic Instruments in CHRNA3 on Several Maternal Smoking Measures
| Panel A. DNBC | |||
|---|---|---|---|
| Number of cigarettes per day during pregnancy |
Number of cigarettes per day during pregnancy among smokers |
Any smoking during pregnancy |
|
| rs12914385 | 0.389*** | 1.197*** | 0.012 |
| (0.142) | (0.366) | (0.015) | |
| F statistic | 9.05 | 10.662 | 0.621 |
| R2 | 0.005 | 0.024 | 0.0003 |
| N | 1811 | 445 | 1898 |
| Panel B. ALSPAC | ||||
| Any smoking prior to pregnancy |
Any smoking, Trimester 1 |
Any smoking, Trimester 2 |
Any smoking, Trimester 3 |
|
|---|---|---|---|---|
| rs1051730 | 0.005 | 0.021*** | 0.017** | 0.015** |
| (0.008) | (0.007) | (0.007) | (0.007) | |
| F statistic | 0.322 | 8.296 | 6.291 | 4.339 |
| R2 | 0.000 | 0.001 | 0.001 | 0.001 |
| N | 7405 | 7023 | 7405 | 6485 |
| Number of cigarettes a day, trimester 1 |
Number of cigarettes a day among smokers, trimester 1 |
Number of cigarettes a day, trimester 3 |
Number of cigarettes a day among smokers, trimester 3 |
|
|---|---|---|---|---|
| rs1051730 | 0.407*** | 0.923*** | 0.264*** | 0.476* |
| (0.094) | (0.259) | (0.101) | (0.288) | |
| F statistic | 18.534 | 12.664 | 6.875 | 2.735 |
| R2 | 0.003 | 0.010 | 0.001 | 0.002 |
| N | 7023 | 1356 | 6485 | 1288 |
Notes: The Table reports the instrument effects (OLS regression coefficients and their standard errors in parentheses) on several smoking measures separately for the DNBC and ASLPAC datasets.
p < 0.10,
p < 0.05,
p < 0.01.
In the ALSPAC data, the CHRNA3 variant has significant effects on the binary indicators of smoking during pregnancy with F-statistics between 4 and 8 but has an insignificant effect on smoking prior to pregnancy. The CHRNA3 variant has also significant effects on the number of cigarettes smoked per day during the first and third pregnancy trimesters, with an F statistic of 18 and 7, respectively, and on the number of cigarettes smoked among smokers, with an F statistic of 13 and 3 for the first and third trimesters, respectively. Recall that we do not observe the count of the number of cigarettes during the second trimester. The analyses show that each copy of the risk allele increases the total number of cigarettes per day by 0.26–0.41 cigarettes and the number of cigarettes among smokers by 0.5–1 cigarette per day.
3.1b ADH1B
Table 4 lists the effects of the variants in ADH1B on the various alcohol measures and the F statistics in the ALSPAC sample.12 The ADH1B rare allele variant decreases the probability of consuming alcohol mainly during pregnancy. Women who carry at least one copy of the rare allele are between 7 and 10 percentage points less likely to drink during pregnancy. The F-statistics for the ADH1B effects during pregnancy are between 9 (for third trimester) and 20 (for first trimester), suggesting a relatively strong instrument. There is a smaller and marginally significant effect on alcohol consumption prior to pregnancy; women with the rare variant are about 2.9 percentage points less likely to drink (F-statistic of 3.4). When we examine the number of alcoholic units consumed per week during pregnancy, we also find strong genetic effects, with those carrying the rare allele of the ADH1B variant drinking between 0.6 and 0.9 units per week less than those homozygous for the common allele; F-statistics range from 9.2 to 55, with the largest effects on the first trimester. The ADH1B variant also shows significant effects on binge drinking during pregnancy, with those carrying at least one rare allele being on average 13 percentage points less likely to binge.
Table 4.
Effects of the Genetic Instrument in ADH1B on Several Maternal Alcohol Measures in ALSPAC
| Any alcohol Trimester 1 |
Any alcohol Trimester 2 |
Any alcohol Trimester 3 |
|
|---|---|---|---|
| rs1229984 | −.100*** | −0.072*** | −0.086*** |
| (0.022) | (0.022) | (0.029) | |
| F statistic | 20.529 | 10.429 | 9.011 |
| R2 | 0.002 | 0.001 | 0.002 |
| N | 6754 | 7340 | 4553 |
| Number of alcohol units, Trimester 1 |
Number of alcohol units, Trimester 2 |
Number of alcohol units, Trimester 3 |
Binge drinking, Trimester 2 |
|
|---|---|---|---|---|
| rs1229984 | −0.888*** | −0.610*** | −0.580*** | −0.129*** |
| (0.120) | (0.132) | (0.191) | (0.034) | |
| F statistic | 55.242 | 21.379 | 9.193 | 14.355 |
| R2 | 0.002 | 0.001 | 0.001 | 0.001 |
| N | 6754 | 7340 | 4553 | 7316 |
Notes: The Table reports the instrument effects (OLS regression coefficients and their standard errors in parentheses) on several alcohol consumption measures in the ALSPAC dataset.
p < 0.10,
p < 0.01
3.1c FTO
Table 5 shows the FTO variant effects on obesity and BMI before pregnancy (panel A for DNBC; panel B for ALSPAC). In both datasets, the FTO variant has large and significant effects on obesity status and BMI, with F-statistics of 20.9 and 16, respectively, in the DNBC dataset, and 16.5 and 35.5, respectively, in ALSPAC. In the DNBC sample, each copy of the risk allele of rs8050136 increases the probability of obesity by 4 percentage points and increases mean BMI by 0.6 points, respectively. In ALSPAC, each copy of the risk allele of rs9939609 increases the probability of obesity by 2 percentage points and BMI by 0.4 points. .
Table 5.
Effects of Genetic Instruments in FTO on Maternal Obesity and BMI before Pregnancy
| Obesity | BMI | |
|---|---|---|
| Panel A. DNBC | ||
| rs8050136 | 0.044*** | 0.587*** |
| (0.01) | (0.146) | |
| F statistic | 20.87 | 16.119 |
| R2 | 0.011 | 0.009 |
| N | 1811 | 1811 |
| Panel B. ALSPAC | ||
|---|---|---|
| rs9939609 | 0.018*** | 0.427*** |
| (0.004) | (0.072) | |
| F statistic | 16.524 | 35.486 |
| R2 | 0.003 | 0.006 |
| N | 6634 | 6634 |
Notes: The Table reports the instrument effects (OLS regression coefficients and their standard errors in parentheses) on obesity and BMI for the DNBC and ALSPAC datasets.
p < 0.01.
3.2 Evaluating if instruments are unrelated to confounders
We first check the associations between the study factors and several background demographics, socioeconomic status, and maternal health characteristics. The goal is to evaluate self-selection into each of these risk factors and the extent to which they may relate to child health outcomes through unobserved confounders. Consistent patterns of association between the risk factors and observable background characteristics suggest there may also be unobservable confounders. The associations are obtained from an OLS regression of the risk factor of interest on these background characteristics. After that, we show the associations between these same background characteristics and the SNPs to see if these characteristics are randomized by the genetic variants. Consistent patterns of correlations between the genetic variants and these background characteristics would suggest that the variants may be related to other unobservable confounders.
Here we summarize the main associations between the risk factors and the background characteristics (detailed results are in Tables A3 and A4 in the Appendix for the DNBC and ALSPAC datasets, respectively). In the DNBC sample, individuals who smoke more are less likely to be married, have a stable relationship, or plan their pregnancy. Furthermore, they have more previous live births but are younger at menarche (marginally significant). Also, individuals who smoke more are more likely to have mental health problems and to be employed. Furthermore, any smoking is associated with worse general health. Similar to the DNBC sample, smoking is generally characterized by an inverse social gradient in ALSPAC: smokers are younger, less educated, of lower social class, and more likely to be a lone parent than non-smokers. The findings are similar when examining the number of cigarettes.
Several significant associations are also found between alcohol consumption and background characteristics in the ALSPAC sample.13 Mothers who consume alcohol either before or during pregnancy are older, higher educated, and more likely to be employed. Similar results are obtained when examining the number of alcoholic units (available upon request).
Obesity and BMI are also associated with several background characteristics. In DNBC, obesity is associated with an increase in the number of previous miscarriages, a decrease in general health, younger age at menarche, and increase in employment. Similar associations are found for BMI (except for employment which is negative but insignificantly related). In ALSPAC, heavier mothers are less educated, and of lower social class. BMI is positively related to being employed.
These results show strong patterns of associations between maternal risk factors and a large set of background characteristics. As datasets do not provide measures of all relevant determinants of child health outcomes that are correlated with the maternal risk factors of interest, it is impossible to directly control for all such confounders.
Next, we summarize the associations between the SNPs and background characteristics obtained from OLS regressions of the SNPs on these characteristics (detailed results are in Tables A5 and A6 in the Appendix). In stark contrast to the study risk factors, we find no systematic differences in these characteristics by the variants of each SNP. In the DNBC sample, there are no significant associations between the SNPs and any of these characteristics, except for a marginally significant negative association between the CHRNA3 risk variant and being married or in a stable relationship and a marginally significant positive association between the FTO risk variant and previous number of miscarriages, though this effect may go via the effect of FTO on BMI, as higher BMI is associated with increased miscarriage risk (Metwally et al. 2008). Similar to the DNBC, the ALSPAC sample only shows few marginally significant associations. Carrying the risk allele of FTO is positively related to the probability that the mother is a lone parent. However, we do not find any systematic associations with socioeconomic position, social class, or other evaluated characteristics, as none of these other covariates are associated with the SNPs. Hence, these findings indicate that the various genotypes of these SNPs effectively randomize observable background characteristics that are relevant to the study risk factors, suggesting that they are expected to also randomize unobservable confounders.
We further evaluate the second IV assumption by reviewing the literature and current knowledge about gene expression and functions. CHRNA3 is expressed in the central and peripheral nervous systems where it is involved in detecting chemical signals through synapses as well as in other body parts including tongue, stomach, and others (OMIM, 2011). Other than nicotine dependence, CHRNA3 has been linked to lung cancer (Tournier and Birembaut 2011; Timofeeva et al. 2011) and bladder cancer (Kaur-Knudsen et al. 2011). Nicotinic acetylcholine receptors may also play a role in other health related issues such as chronic obstructive pulmonary disease (COPD) (Saccone et al. 2010; Nakamura 2011; Kaur-Knudsen et al. 2011; Kim et al. 2011). Moreover, a meta-analysis found that CHRNA3 may be related to peripheral arterial disease (PAD) (Zintzaras and Zdoukopoulos 2009). All of these conditions and diseases however, are highly correlated with smoking, where smoking is believed to be the causal factor. Hence, these associations are not likely to invalidate the use of the CHRNA3 variant as an instrument, but are simply another causal effect of smoking.
ADH1B is mainly expressed in the liver, as well as in the central and peripheral nervous system, the stomach, nose, renal system, and reproductive system. ADH1B has been associated with several cancers including upper aerodigestive tract (McKay et al. 2011), esophageal (Zhang et al. 2010a; Zhang et al. 2010b; Yang et al. 2010), oral and laryngeal cancers (Marichalar-Mendia et al. 2011), with potential variation between populations. As ADH1B is expected to affect these conditions through its effects on alcohol consumption, these effects do not compromise using the ADH1B variant as an instrument.
FTO is expressed in the central nervous system, placenta, hypothalamus, and pancreatic islets. While its function is not yet fully understood, some studies have found associations between FTO and diabetes independent of the effect on body weight (Hertel et al. 2011; Rees et al. 2011), but many other large GWA studies find that the association pertains to obesity and not diabetes (Frayling et al. 2007). Variants in FTO have also been related to age at menarche (Elks et al. 2010; Mumby et al. 2011), although there is no significant association between the evaluated FTO variant and age at menarche in our DNBC sample. However, age at menarche itself is known to be strongly associated with BMI (e.g. Wattigney et al. 1999). To the extent that FTO affects age at menarche and diabetes through its effect on maternal body weight, this does not violate the IV assumptions. If maternal FTO variants directly affect infant and child health however, this would invalidate their use instruments, potentially biasing the IV estimates. However, the overall lack of significant associations between the observed characteristics and FTO variants provides some support for using these variants as instruments. Nonetheless, as with all instruments, caution should be used when using genetic variants as instrumental variables for studying causal effects.
3.3 Empirical Example
As an empirical illustration of using the variants in these genes as instruments for studying the effects of prenatal risk factors on infant health, we apply these variants as instruments to study the effects of maternal smoking, alcohol use, and BMI on birth weight, which is measured in the DNBC and ALSPAC datasets (descriptive statistics for birth weight are in Tables 1 and 2).14 We evaluate the effect of each risk factor using 2SLS estimation for the IV model and OLS as a reference model.15
We first discuss the results from the DNBC sample shown in Table 6. Cigarettes are negatively and significantly associated with birth weight under OLS. Each cigarette is associated with about 30 gram decrease in birth weight on average under OLS. Under the IV model, cigarettes have larger (in absolute value) negative effects on birth weight by at least two times than OLS but the effects are overall statistically insignificant and not statistically different from OLS estimates based on a Hausman-type test (Hausman 1978). The estimated IV effect of about 55 gram decrease per cigarette in the total sample combining smokers and non-smokers is very close to a previous estimate using other genetic estimates in a Norwegian sample (Wehby et al, 2011a). In the IV estimation limited to smokers, birth weight decreases by about 90 grams with each cigarette (marginally significant); however, the difference between the OLS and 2SLS estimates is still not significant (p=0.16). These results suggest that the adverse effects of prenatal smoking on birth weight may be underestimated in a classical OLS model, consistent with several previous studies that have suggested that maternal smoking effects on birth weight and other child health outcomes may be underestimated when unobservable confounders are ignored (e.g. Permutt and Hebel 1989; Lien 2005; Wehby et al. 2011a; Wehby et al. 2011b; Wehby et al. 2011c).
Table 6.
Effects of Risk Factors on Birth Weight under OLS and IV (2SLS) Models with Genetic Instruments in DNBC
| Prenatal risk factor | OLS | IV (2SLS) |
|---|---|---|
| Birth Weight (grams) |
Birth Weight (grams) |
|
| Cigarettes | −28.411*** | −55.434 |
| (5.239) [1792] |
(73.937) [1792] {0.712} |
|
| Cigarettes among smokers |
−29.551*** | −93.318** |
| (6.963) [441] |
(50.100) [441] {0.161} |
|
| BMI | 9.657** | −111.637* |
| (4.773) [1791] |
(58.860) [1791] {0.016} |
|
Note: The table reports the coefficients from the regressions of birth weight on prenatal risk factors; standard errors are in parentheses; sample sizes are in brackets; p values for significance of the difference between OLS and 2SLS estimates are in curly brackets. A separate regression is estimated for prenatal factor.
p < 0.1,
p < 0.05,
p < 0.01
The lack of significance of the smoking effects under the IV model is in part due to limited power given that only part of the variation in cigarettes that is predicted by the instrument is used in the estimation, which is expressed in very large standard errors. This highlights the particular need for large samples in IV estimations. Similarly, the lack of significance of differences between the OLS and 2SLS estimates is in part driven by this limited variation and large variances of IV estimates.
Unlike cigarettes, BMI is positively associated with birth weight under OLS. However, the BMI effect on birth weight switches to negative and is large and marginally significant under the IV model. Each unit increase in BMI leads to a 111 gram decrease in birth weight; this is significantly different from the OLS estimate (p=0.016).
Table 7 reports the results from a similar application using the ALSPAC dataset in addition to evaluating the effects of alcohol consumption on birth weight. The results for cigarette number and BMI are overall consistent with those using the DNBC data. OLS shows a negative association between maternal smoking during pregnancy and the child’s birth weight, with each cigarette on average being associated with a reduction in birth weight of 14 grams. Using the variant in CHRNA3 to instrument for the number of cigarettes doubles this estimate. We find similar negative results when conditioning the analysis on those mothers who smoke at least one cigarette, and the difference between the IV and OLS estimates is marginally significant. Although the actual point estimates differ between the DNBC and ALSPAC sample, both show that smoking reduces birth weight and that the IV estimates are larger than OLS. The large standard errors, however, preclude us overall from rejecting the OLS estimate.
Table 7.
Effects of Risk Factors on Birth Weight under OLS and IV (2SLS) Models with Genetic Instruments in ALSPAC
| Prenatal risk factor | OLS | IV (2SLS) |
|---|---|---|
| Number of cigarettes (first trimester) |
−14.035*** (1.280) [6947] |
−28.270 (23.554) [6947] { 0.542} |
| Number of cigarettes among (first trimester) smokers |
−3.422 (2.201) [1341] |
−42.571 (25.323) [1341] {0.087} |
| Any alcohol use (firsttrimester) |
32.877* (14.769) [6683] |
355.315 (332.896) [6683] {0.327} |
| Number of units of alcohol (first trimester) |
1.285 (1.572) [6697] |
−187.651 (301.782) [6697] {0.272} |
| BMI | 19.991*** (1.969) [6566] |
−112.017 (196.182) [6566] {0.363} |
Note: The table reports the coefficients from the regressions of birth weight on prenatal risk factors; standard errors are in parentheses; sample sizes are in brackets; p values for significance of the difference between OLS and 2SLS estimates are in curly brackets. A separate regression is estimated for prenatal factor.
p < 0.1,
p < 0.01
Similar to the DNBC results, we find that maternal BMI is positively related to child birth weight in the OLS analysis, which changes sign in the IV. In addition, the ALSPAC and DNBC analysis report very similar findings: a one unit increase in maternal BMI reduces the child’s birth weight by about 112 grams.
Finally, the association between alcohol consumption and birth weight in ALSPAC is positive in both the OLS and IV analysis, with the IV point estimate being considerably larger than that in the OLS, although the difference is not statistically significant. However, once we examine the number of drinks rather than the binary indicator, we find a clear negative effect of alcohol exposure on child birth weight.
Despite the IV estimates being relatively imprecise, with large standard errors, meaning we cannot always reject the OLS based on the IV estimates, the analyses show that a simple OLS regression may lead to different conclusions from the IV model. Assuming the IV conditions hold, these differences are likely to be due to unobservable factors that affect both the outcome and risk factors of interest. As we show in this illustrative example, not accounting for such unobservables may lead to bias in the estimates.
4. Discussion
This paper evaluates the utility of variants in three genes, CHRNA3, ADH1B, and FTO to be employed as instruments for studying the causal effects of maternal smoking and alcohol intake during pregnancy and obesity/BMI before pregnancy on child health outcomes. Using two independent datasets, we find these genetic variants to have overall relatively strong effects on these measures from a statistical perspective, with F-statistics generally close to or above 10. Furthermore, we find no patterns of significant correlations between these instruments and several maternal demographic, socioeconomic, and health characteristics that are correlated with the study risk factors, supporting the hypothesis that these genetic variants randomize confounders. These findings are consistent with the lack of evidence in the literature to date for strong effects of these three genes on other important confounders.
The study provides support for using these variants as instruments for maternal risk factors during pregnancy. However, as is the case for every (genetic as well as non-genetic) instrument, investigators should always validate the fit of their instruments in each dataset and for the specific outcomes they are studying. We note however, that the lack of evidence to date about other functions of the evaluated variants of interest that could confound their use as instruments does not mean that they do not have such functions. As shown above though, unlike the genetic instruments, the risk factors are highly correlated with several observable characteristics. This suggests that these factors are also likely to be correlated with unobservable confounders. Therefore, employing genetic instrumental variables to study the effects of these risk factors on child health could significantly reduce the bias in classical estimates of these effects.
The motivation for studying the fit of these variants specifically for risk factors during and around pregnancy is the premise that genetic effects may be modified by pregnancy. Effects from previous studies which focused on general and older samples may not be generalizable to risk factors during pregnancy. In this study, similar to Zuccolo et al. (2009) and Freathy et al. (2008) we find some suggestive evidence that genetic effects are intensified during compared to before pregnancy. The stronger effects during pregnancy could suggest an interaction between the variant and the environment; the latter being pregnancy. The alcohol-related symptoms among those with the ADH1B rare allele for example, may be perceived as more unfavorable during than before pregnancy, or these women may find it easier to quit as a result of these symptoms.
Although we aim to evaluate the fit of these variants as instruments for maternal prenatal risk factors, one may argue that the genes are more general, as their effects are not necessarily specific to this period. In other words, mothers who carry the rare allele variant of CHRNA3 and/or ADH1B are likely to smoke more and/or drink less anytime throughout life including after pregnancy, and those with the risk allele of FTO are likely to be slightly heavier throughout life. Hence, any estimated effects of these risk factors on child health outcomes may not necessarily be solely due to prenatal effects, but may partly reflect risk factors after pregnancy. The extent of this analytical challenge will depend on the period at which child outcomes are measured. While this is unlikely to be of relevance to neonatal outcomes, it may be an issue for longer-term child outcomes that may be affected by maternal risk factors after pregnancy. In these cases, one can instrument for both prenatal and postnatal behaviors risk factors if there are enough instruments. If not, one may compare the prenatal behaviors effects between models that alternatively control for or exclude postnatal risk factors (if these cannot be instrumented for due to the lack of instruments).
We were able to evaluate the genetic instruments in CHRNA3 and FTO in two independent datasets and find consistent results, which supports the validity of the findings. We could not evaluate the ADH1B instrument (rs1229984) in the DNBC sample since it was not genotyped and there were no other genotyped SNPs in strong LD with this variant. Therefore, it is important to evaluate this instrument in other datasets in the future.
One study caveat is that both of our samples are very homogenous on race/ethnicity. Although this reduces the bias in instrument effects due to population stratification, it may limit the generalizability of the study results to other populations. This highlights the need to study the utility of these genetic variants as instruments in other populations and samples. However, it is important to recognize the potential bias due to population stratification when studying diverse populations such the United States. If the allelic distributions of the genetic instruments vary by ancestry, the IV estimates may be biased. Furthermore, direct adjustment for self-reported measures of race and ethnicity may be inadequate to remove this bias, as these may not effectively account for differences in allelic frequency by ancestry. Although this is not likely to affect our estimation, we report in Table A7 in the Appendix the minor allele frequencies for the three main evaluated SNPs in CHRNA3, ADH1B, and FTO. There is a substantial variation in the allelic frequencies of these SNPs by ancestry. For example, the frequency of the minor allele for rs1051730 in CHRNA3 is about 39%, 10% and 3% in individuals of European, Sub-Saharan African, and Asian (Han Chinese) ancestry, respectively. Therefore, when using genetic instruments that vary in allelic distribution by ancestry and data from populations with diverse ancestries, it is important to appropriately account for population stratification using effective methods such as genome-wide based indicators of ancestry based on principal component analysis when possible (Paschou et al. 2007).
IV models provide an estimate of the local average treatment effect (also known as LATE), which is the effect that applies to individuals whose behavior/risk factor was affected by the instrument, and whose behavior/risk factor would have changed had they been in a different instrument group (Angrist et al. 1996). These individuals are commonly described as the group on the margin (or “marginal” group). However, it is impossible to identify those individuals since none is observed under different instrument groups. Therefore, an important question is the generalizability of the LATE estimate to the total population. Again, this question is not specific to genetic instruments per se, but to all instruments. Two complications for the generalizability of the LATE estimate might arise in the context of genetic instruments: 1- gene-environment interactions and 2- limited explanatory power of instruments. We discuss each of these below in detail.
The effects of genetic variants on behaviors and risk factors may be modified by environmental factors. Although such gene-environment interactions should not bias the IV estimate, ignoring them may mask important differences between subgroups defined by these environmental factors. As such, this may limit the generalizability of the results. While it is impossible to identify the “marginal” group to whom the LATE applies, some subgroups of certain environmental characteristics may include a larger representation of the “marginal” individuals than others. In general, gene-environment interactions are not expected to affect the internal validity of the IV estimate, but rather the external validity and interpretation of the estimates. Therefore, it is important to evaluate any suspected gene-environment interactions in the first stage. If significant interactions are found, the IV model (both first and second stages) should be stratified by the environmental variables with significant interactions, provided that the instrument has sufficiently strong effects on the risk factor in the stratified subgroups (Wehby et al. 2008).
It is important to evaluate the extent of variation in the risk factor that the instrument explains, beyond whether the instrument passes the minimum threshold to be considered non-weak from a statistical perspective (e.g. an F-statistic above 10).16 An instrument may be considered statistically strong, but it may still explain a small percentage of the variation in the risk factor. This does not bias the analysis, but it may lead to an imprecise estimate with potentially limited generalizability as it is based on only little variation in the risk factor. Again, the issue here is whether the LATE estimate that is applicable to the “marginal” group may generalize to the majority of the population for whom the risk factor was not explained by the instrument. This will depend on whether the effects of the risk factors being instrumented for on outcomes may vary between subgroups that have different underlying etiologies for these risk factors. Therefore, it is important that researchers consider the “clinical relevance” of the variation explained by the genetic instruments and implications for generalizability.
In our case, the instruments explain a small percent of the variation in the risk factors (see R2 in Tables 3 through 5). The CHRNA3 instruments explain about 0.2–2.4% of the variation in number of cigarettes among smokers. Similarly, the ADH1B variants explain about 0.1–0.2% of the variation in alcohol consumption during pregnancy. The FTO variants explain about 0.3–1% of the variation in obesity and BMI. The small explanatory power is not surprising given the complex etiology of these risk factors. In fact, most models of socioeconomic and environmental characteristics explain less than 10% of the variation in such risk factors.17 However, the small variation explained by the genetic instruments raises a question about the applicability of the IV estimates to the larger population. In this particular case, there is no consistent knowledge that the effects of these risk factors on infant health outcomes vary significantly between maternal groups with different etiologies for these risk factors. For example, there is no expectation a priori that the effects of smoking on birth weight vary between those who smoke more cigarettes because of lower prices or because of having the risk CHRNA3 variant or both. As the genetic etiologies of these risk factors are further unraveled allowing for the use of additional instruments, it is possible to evaluate the IV estimates of the risk factor effects under different instruments. If the IV estimates are overall insensitive to different instrument (sets), then this would support the generalizability of the results to other groups, and the validity of the instruments and IV approach (Wehby et al. 2008; Davey Smith 2011; Palmer et al. 2011; von Hinke Kessler Scholder et al. 2010).
Acknowledgements
Data analysis of the DNBC dataset was supported by NIH/NIDCR grant 1 R01 DE020895. We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council, the Wellcome Trust and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors, who will serve as guarantors for the contents of this paper and does not reflect the views of the ALSPAC executive. We thank Drs. Mads Melbye and Jeffrey C. Murray for providing access to the DNBC data. We also thank Dr. Bjarke Feenstra for his thoughtful review of the paper.
Appendix
Table A1.
Appendix Descriptive Statistics for Background Characteristics in DNBC
| Variable | Description | Mean | Std. Dev. |
N |
|---|---|---|---|---|
| Maternal age | Maternal age in years | 29.565 | 4.244 | 1930 |
| Child: boy | 0/1 indicator for a male birth | 0.525 | 0.500 | 1937 |
| Married/stable relationshipa |
0/1 indicator for mother being married or a stable relationship |
0.989 | 0.106 | 1839 |
| Previous live births |
Number of previous live births at time of 1st DNBC interview |
0.727 | 0.833 | 1841 |
| Previous miscarriages |
Number of previous miscarriages at time of 1st DNBC interview |
0.258 | 0.608 | 1839 |
| Unplanned pregnancy |
0/1 indicator for not planning the pregnancy |
0.099 | 0.298 | 1842 |
| Average healthb | 0/1 indicator for mother reporting her health as “average” |
0.429 | 0.495 | 1840 |
| Not so good healthb |
0/1 indicator for mother reporting her health as “not so good” |
0.047 | 0.211 | 1840 |
| Mental problems | 0/1 indicator for any previous or current mental health problems |
0.126 | 0.332 | 1840 |
| Menarche | Mother’s age at menarche in years | 13.330 | 1.421 | 1678 |
| Employed | 0/1 indicator for being employed at the time of the 1st DNBC interview |
0.213 | 0.409 | 1842 |
Notes: reference category is single mothers
reference category is good health
Table A2.
Descriptive Statistics for Background Characteristics in ALSPAC
| Variable | Description | Mean | Std. Dev. |
N |
|---|---|---|---|---|
| Mother’s age | Maternal age in years | 28.27 | 4.850 | 10999 |
| Child: girl | 0/1 indicator for female birth | 0.485 | 0.500 | 11428 |
| Lone Parent | 0/1 indicator for mother being a lone parent |
0.070 | 0.255 | 8576 |
| # siblings | Number of siblings of cohort child | 0.871 | 0.958 | 9009 |
| Locus of control | Mother’s locus of control | −0.033 | 0.991 | 8605 |
| CCEI Trimester 2 | Crown-Crisp Experimental Index, Trimester 2 |
13.29 | 7.583 | 9279 |
| EPDS Trimester 2 | Edinburgh Postnatal Depression Score, Trimester 2 |
6.759 | 4.757 | 9717 |
| Mother is happy | Variable ranging from 1–4 with 4 being most happy |
3.260 | 0.805 | 6733 |
| Mother is content | Variable ranging from 1–4 with 4 being most content |
3.100 | 0.908 | 6692 |
| Maternal education | Variable ranging from 1–4; 4 is highest education |
2.297 | 0.926 | 10372 |
| Social class | Variable ranging from 1–6; 6 is lowest social class |
3.021 | 1.320 | 9008 |
| Mother is employed | 0/1 indicator for mother being employed |
0.480 | 0.500 | 8531 |
Notes: Maternal educational includes four indicators: less than ordinary (O) level, O-level only, advanced (A) level that permits higher educational study, and having a university degree. We use the standard (reversed, so that higher values correspond to higher social classes) UK classification of social class based on occupation (professional (I), managerial/technical (II), non-manual skilled (IIInm), manual skilled (IIIm), semi-skilled (IV) and unskilled (V)). EPDS (Edinburgh Postnatal Depression Score) and CCEI (Crown-Crisp Experimental Index) aim to capture aspects of maternal mental health. EPDS indicates to what extent the mother is at risk of perinatal depression; CCEI captures a broader definition of mental health, measuring general anxiety, depression and somaticism. Higher scores mean the mother is more affected.
Table A3.
Coefficients from OLS Regressions of Study Risk Factors on Background Characteristics in DNBC
| Number of cigarettes |
Any smoking during pregnancy |
Obese | Pre- pregnancy BMI |
|
|---|---|---|---|---|
| Maternal age | −0.024 | −0.011*** | −0.000 | 0.029 |
| (0.026) | (0.003) | (0.002) | (0.029) | |
| Child: boy | 0.163 | 0.027 | 0.015 | 0.177 |
| (0.187) | (0.022) | (0.014) | (0.210) | |
| Married/stable relationship |
−4.748*** | −0.292*** | −0.008 | 0.619 |
| (0.858) | (0.099) | (0.064) | (0.980) | |
| Previous live births |
0.295** | 0.022 | −0.002 | 0.106 |
| (0.128) | (0.015) | (0.009) | (0.143) | |
| Previous miscarriages |
0.049 | 0.006 | 0.035*** | 0.351** |
| (0.160) | (0.018) | (0.011) | (0.175) | |
| Unplanned pregnancy |
0.986*** | 0.107*** | −0.019 | −0.476 |
| (0.323) | (0.037) | (0.024) | (0.364) | |
| Average health | 0.199 | 0.036 | 0.072*** | 1.050*** |
| (0.193) | (0.022) | (0.014) | (0.218) | |
| Not so good health |
0.593 | 0.134** | 0.069** | 1.487*** |
| (0.475) | (0.053) | (0.034) | (0.515) | |
| Mental problems |
1.377*** | 0.120*** | −0.038 | −0.560 |
| (0.359) | (0.040) | (0.026) | (0.392) | |
| Menarche | −0.117* | −0.013* | −0.016*** | −0.457*** |
| (0.067) | (0.008) | (0.005) | (0.075) | |
| Employed | 0.497** | 0.054** | 0.040** | −0.135 |
| (0.237) | (0.027) | (0.017) | (0.266) | |
| Constant | 7.834*** | 0.977*** | 0.260* | 27.508*** |
| (1.399) | (0.161) | (0.103) | (1.580) | |
| N | 1607 | 1668 | 1643 | 1643 |
| R2 | 0.057 | 0.048 | 0.035 | 0.047 |
Notes: The Table reports the coefficients (with standard errors in parentheses) from OLS regressions of the study risk factors on several background characteristics in the DNBC dataset. All characteristics are included simultaneously in the regression.
p < 0.10,
p < 0.05,
p < 0.01.
Table A4.
Coefficients from OLS Regressions of Study Risk Factors on Background Characteristics in ALSPAC
| Any smoking before pregnancy |
Any smoking during pregnancy |
Number of cigarettes (first trimester) |
Any alcohol before pregnancy |
Any Alcohol during 1st pregnancy trimester |
Obese | Pre- pregnancy BMI |
|
|---|---|---|---|---|---|---|---|
| Mother’s age | −0.008*** | −0.003* | −0.024 | 0.004*** | 0.011*** | −0.000 | 0.028 |
| (0.002) | (0.001) | (0.018) | (0.001) | (0.002) | (0.001) | (0.016) | |
| Child: girl | −0.011 | −0.013 | −0.060 | 0.005 | −0.012 | −0.009 | −0.184 |
| (0.013) | (0.010) | (0.123) | (0.007) | (0.014) | (0.007) | (0.113) | |
| Lone parent | 0.128** | 0.102** | 1.202* | −0.025 | 0.032 | −0.032 | −0.832** |
| (0.042) | (0.038) | (0.544) | (0.024) | (0.041) | (0.017) | (0.313) | |
| # siblings | −0.010 | 0.008 | 0.262** | −0.024*** | 0.005 | 0.009* | 0.144 |
| (0.007) | (0.006) | (0.084) | (0.005) | (0.009) | (0.004) | (0.074) | |
| Locus of control |
0.062*** | 0.046*** | 0.451*** | −0.005 | 0.026** | 0.009* | 0.202** |
| (0.008) | (0.006) | (0.085) | (0.004) | (0.009) | (0.004) | (0.073) | |
| CCEI Trimester 2 |
0.003 | 0.002 | 0.031* | 0.000 | 0.001 | 0.001 | 0.050*** |
| (0.001) | (0.001) | (0.013) | (0.001) | (0.002) | (0.001) | (0.012) | |
| EPDS Trimester 2 |
0.004 | 0.002 | 0.019 | 0.001 | 0.001 | −0.002 | −0.068*** |
| (0.002) | (0.002) | (0.022) | (0.001) | (0.002) | (0.001) | (0.020) | |
| Mother is happy |
−0.023 | −0.011 | −0.129 | −0.009 | −0.029 | −0.005 | −0.008 |
| (0.013) | (0.011) | (0.127) | (0.007) | (0.015) | (0.007) | (0.128) | |
| Mother is content |
0.004 | −0.003 | −0.062 | 0.007 | 0.028* | 0.007 | 0.050 |
| (0.012) | (0.010) | (0.108) | (0.006) | (0.013) | (0.007) | (0.112) | |
| Mother’s education |
−0.035*** | −0.031*** | −0.433*** | 0.012** | 0.035*** | −0.013** | −0.368*** |
| (0.008) | (0.007) | (0.081) | (0.005) | (0.010) | (0.004) | (0.072) | |
| Father’s social class |
−0.025*** | −0.019*** | −0.212*** | −0.000 | 0.015* | −0.014*** | −0.293*** |
| (0.006) | (0.004) | (0.056) | (0.003) | (0.006) | (0.003) | (0.053) | |
| Mother is employed |
−0.000 | −0.007 | −0.141 | 0.014* | 0.036* | 0.007 | 0.393*** |
| (0.013) | (0.010) | (0.123) | (0.007) | (0.015) | (0.007) | (0.115) | |
| Constant | 0.686*** | 0.402*** | 4.021*** | 0.812*** | −0.185** | 0.134*** | 23.740*** |
| (0.064) | (0.053) | (0.659) | (0.036) | (0.069) | (0.038) | (0.633) | |
| N | 4234 | 4220 | 4220 | 4223 | 4065 | 4015 | 4015 |
| R2 | 0.087 | 0.076 | 0.071 | 0.023 | 0.026 | 0.022 | 0.044 |
Notes: The Table reports the coefficients (with standard errors in parentheses) from OLS regressions of the study risk factors on several background characteristics in the ALSPAC dataset. All characteristics are included simultaneously in the regression.
p < 0.10,
p < 0.05,
p < 0.01.
Table A5.
Coefficients from OLS Regressions of Genetic Instruments on Background Characteristics in DNBC
| rs12914385 (CHRNA3) |
rs8050136 (FTO) |
|
|---|---|---|
| Maternal age | −0.003 | −0.005 |
| (0.005) | (0.005) | |
| Child: boy | −0.021 | −0.019 |
| (0.034) | (0.034) | |
| Married/stable relationship |
−0.282* | −0.065 |
| (0.154) | (0.154) | |
| Previous live births | 0.002 | 0.009 |
| (0.023) | (0.023) | |
| Previous miscarriages |
0.014 | 0.047* |
| (0.028) | (0.028) | |
| Unplanned pregnancy | 0.058 | −0.009 |
| (0.058) | (0.058) | |
| Average health | −0.020 | 0.018 |
| (0.035) | (0.035) | |
| Not so good health | −0.025 | 0.135 |
| (0.082) | (0.082) | |
| Mental problems | 0.010 | 0.056 |
| (0.063) | (0.063) | |
| Menarche | 0.003 | 0.007 |
| (0.012) | (0.012) | |
| Employed | −0.011 | 0.010 |
| (0.043) | (0.043) | |
| Constant | 1.077*** | 0.872*** |
| (0.253) | (0.253) | |
| N | 1667 | 1668 |
| R2 | 0.004 | 0.005 |
Notes: The Table reports the coefficients (with standard errors in parentheses) from OLS regressions of the study SNPs on several background characteristics in the DNBC dataset. All characteristics are included simultaneously in the regression.
p < 0.10,
p < 0.01.
Table A6.
Coefficients from OLS Regressions of Genetic Instruments on Background Characteristics in ALSPAC
| rs1051730 (CHRNA3) |
rs1229984 (ADH1B) |
rs9939609 (FTO) |
|
|---|---|---|---|
| Mother’s age | 0.000 | −0.001 | 0.002 |
| (0.003) | (0.001) | (0.003) | |
| Child: girl | −0.007 | −0.001 | −0.044 |
| (0.025) | (0.008) | (0.025) | |
| Lone parent | 0.063 | 0.047 | 0.183* |
| (0.071) | (0.031) | (0.072) | |
| # siblings | 0.012 | 0.005 | −0.015 |
| (0.015) | (0.005) | (0.016) | |
| Locus of control |
0.006 | −0.008 | 0.004 |
| (0.015) | (0.005) | (0.015) | |
| CCEI Trimester 2 |
0.000 | 0.000 | 0.001 |
| (0.003) | (0.001) | (0.003) | |
| EPDS Trimester 2 |
0.001 | 0.000 | −0.001 |
| (0.004) | (0.002) | (0.004) | |
| Mother is happy |
−0.002 | 0.010 | −0.000 |
| (0.026) | (0.010) | (0.026) | |
| Mother is content |
0.009 | −0.009 | 0.004 |
| (0.023) | (0.009) | (0.023) | |
| Mother’s education |
0.008 | 0.001 | 0.006 |
| (0.017) | (0.006) | (0.016) | |
| Father’s social class |
−0.010 | −0.002 | −0.006 |
| (0.011) | (0.004) | (0.011) | |
| Mother is employed |
−0.027 | −0.013 | 0.044 |
| (0.025) | (0.009) | (0.026) | |
| Constant | 0.662*** | 0.075* | 0.747*** |
| (0.117) | (0.036) | (0.122) | |
| N | 2943 | 2932 | 2958 |
| R2 | 0.002 | 0.004 | 0.005 |
Notes: The Table reports the coefficients (with standard errors in parentheses) from OLS regressions of the study SNPs on several background characteristics in the ALSPAC dataset. All characteristics are included simultaneously in the regression.
p < 0.10,
p < 0.01.
Table A7.
Minor Allele Frequencies for the Genetic Instruments by Ancestry
| Gene/SNP (minor allele) | Minor Allele Frequency (%) | ||
|---|---|---|---|
| European | Sub-Saharan African |
Asian (Han Chinese) |
|
| CHRNA3/rs1051730 (C) | 38.5 | 9.7 | 3.5 |
| ADH1B/rs1229984 (A) | <1% | ∼0% | 75.6 |
| FTO/rs9939609 (A) | 46.0 | 11.6 | 50.9 |
Note: The table reports the minor allele frequencies for three SNP instruments in three ancestral populations based on HapMap allele frequencies (obtained from NCBI dbSNP).
Footnotes
Examples of such personality/preference traits include the extent to which the mother cares about and values her child’s health, whether she is a present- or a future-oriented individual, and whether she takes risks or not. The other type of unobservable factors include maternal perceptions of the presence and extent of risks for fetal/child health, which determine her expectations about fetal/child health outcomes and may modify her behaviors. Mothers may themselves or through their healthcare providers identify specific risks for fetal health, such as maternal illnesses, family history of disease, and indicators for fetal problems that cause them to modify their behaviors or risk factors. Therefore, such risks are confounders since they affect both maternal behaviors/risk factors as well as fetal/child health outcomes.
Some studies focused on food/energy intake as the phenotype while others focused on satiety. The two are strongly related but not the same, as an individual could get full with a low calorie diet.
Since the genetic instruments we evaluate are selected based on the literature, there is no need to adjust for multiple testing when evaluating their significance in predicting the risk factors.
Non-linear regressions may result in biased estimates in IV models if the functional form is not accurately specified (Angrist 2001). IV models are commonly estimated using Two Stage Least Squares (2SLS) models where both the first and second stages of the IV model are estimated by OLS.
r2 values of 1 indicate perfect LD or correlation; values less than 1 indicate less than perfect correlation.
Ideally, we would have used the exact SNPs with the strongest association with these risk factors in the literature which we are able to employ in the second data source described below (ALSPAC). However, since these SNPs are not covered in the GWAS panel, using other SNPs that are very strongly correlated with them is the next best approach.
Although it is important to understand the instrument effects on cessation behavior, it is not the main aim of our study. Our main interest is in evaluating instruments to study the effects of smoking (and alcohol consumption) during pregnancy per se and not the effects of cessation behavior of women who engaged in these behaviors before pregnancy. This is consistent with the majority of the literature and research interest which focuses on studying the effects of these maternal risk factors during pregnancy (and not their cessation). However, other studies that use the ALSPAC data have shown the variants in ADH1B and CHRNA3 to be significantly related to the cessation of alcohol and cigarette consumption during pregnancy (Zuccolo et al., 2009; Freathy et al. 2009).
Table A1 in the Appendix provides the descriptive statistics for background characteristics in the DNBC sample.
Note that the first trimester questionnaire was only sent out to mothers who enrolled before 14 weeks gestation; this is almost half of all mothers in our data. Those who enrolled at a later date were sent a different questionnaire, but were asked similar questions about their alcohol consumption. For ease of description and discussion however, we refer to the week 8 questionnaire as the first trimester, though note that this is not strictly confined to this period, but includes some mothers during a later gestation.
For the number of cigarettes, we only observe the count variable for the first and third trimesters. Similarly, we only observe counts of the number of alcohol units during pregnancy (see also the notes to the tables).
Table A2 in the Appendix provides the descriptive statistics for background characteristics used in the ALSPAC analysis.
As mentioned above, rs1229984 was not on the GWAS panel used for the DNBC sample and there were no other genotyped SNPs in ADH1B with strong LD with this SNP.
This is not evaluated in the DNBC sample since we have no instrument for alcohol consumption in that sample to correlate with these background characteristics.
In the DNBC sample, we only evaluate the effects of cigarette number and BMI on birth weight since we do not have an instrument for alcohol consumption as mentioned above.
Given that the instruments are independent of the observable demographic and socioeconomic characteristics, we do not control for these characteristics in the main models (both 2SLS and OLS). The adjusted and unadjusted results, however, are similar (results available from the authors upon request).
The statistical strength of the instrument as measured by the F-statistic increases with sample size. However, it is unlikely that a truly weak instrument will be considered strong in a large sample.
See R2 of regressions of these risk factors on background characteristics (Tables A3 and 4 in the Appendix).
Contributor Information
George L. Wehby, Assistant Professor, Department of Health Management and Policy, College of Public Health, University of Iowa, 200 Hawkins Drive, E205 GH, Iowa City, IA 52242, Phone: 1-319-384-5133, Fax: 1-319-384-5125, george-wehby@uiowa.edu.
Stephanie von Hinke Kessler Scholder, Assistant Professor, Department of Economics and Related Studies, University of York, York, YO10 5DD, United Kingdom, Phone: +44 1904 32 3084, Stephanie.Scholder@york.ac.uk.
References
- Angrist Joshua D. Estimations of Limited Dependent Variable Models with Dummy Endogenous Regressors: Simple Strategies for Empirical Practice. Journal of Business and Economic Statistics. 2001;no. 19(1):2–16. [Google Scholar]
- Angrist Joshua D, Imbens Guido W, Rubin Donald B. Identification of Causal Effects Using Instrumental Variables. Journal of the American Statistical Association. 1996;no. 91(434):444–455. [Google Scholar]
- Baker TB, Weiss RB, Bolt D, von Niederhausern A, Fiore MC, Dunn DM, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2009;no. 11(7):785–796. doi: 10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;no. 13(4):368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Campbell DM, Liston WA, Bhattacharya S. Effect of Body Mass Index on pregnancy outcomes in nulliparous women delivering singleton babies. BMC Public Health. 2007;no. 7:168. doi: 10.1186/1471-2458-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Martin NG, et al. ADH single nucleotide polymorphism associations with alcohol metabolism in vivo. Human molecular genetics. 2009;no. 18(8):1533–1542. doi: 10.1093/hmg/ddp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchi S, Stutzmann F, Cavalcanti-Proenca C, Durand E, Pouta A, Hartikainen AL, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. Journal of molecular medicine. 2009;no. 87(5):537–546. doi: 10.1007/s00109-009-0451-6. [DOI] [PubMed] [Google Scholar]
- Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. The New England journal of medicine. 2008;no. 359(24):2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- Chen L, Davey Smith G, Harbord RM, Lewis SJ. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS medicine. 2008;no. 5(3):e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Tso AW, Cheung BM, Xu A, Ong KL, Law LS, et al. Genetic variants associated with persistent central obesity and the metabolic syndrome in a 12-year longitudinal study. European journal of endocrinology / European Federation of Endocrine Societies. 2011;no. 164(3):381–388. doi: 10.1530/EJE-10-0902. [DOI] [PubMed] [Google Scholar]
- Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;no. 361(9360):865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- Cornes BK, Lind PA, Medland SE, Montgomery GW, Nyholt DR, Martin NG. Replication of the association of common rs9939609 variant of FTO with increased BMI in an Australian adult twin population but no evidence for gene by environment (G × E) interaction. International journal of obesity. 2009;no. 33(1):75–79. doi: 10.1038/ijo.2008.223. [DOI] [PubMed] [Google Scholar]
- Davey Smith G. Use of genetic markers and gene-diet interactions for interrogating population-level causal influences of diet on health. Genes and Nutrition. 2011;no. 6:27–43. doi: 10.1007/s12263-010-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? International journal of epidemiology. 2003a;no. 32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- Davey Smith George, Ebrahim Shah. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? International Journal of Epidemiology. 2003b;no. 32(1):1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- den Hoed M, Westerterp-Plantenga MS, Bouwman FG, Mariman EC, Westerterp KR. Postprandial responses in hunger and satiety are associated with the rs9939609 single nucleotide polymorphism in FTO. The American journal of clinical nutrition. 2009;no. 90(5):1426–1432. doi: 10.3945/ajcn.2009.28053. [DOI] [PubMed] [Google Scholar]
- Didelez V, Meng S, Sheehan MA. Assumptions of IV methods for observational epidemiology. Statistical Science. 2010;no. 25(1):22–40. [Google Scholar]
- Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature genetics. 2007;no. 39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- Duranceaux NC, Schuckit MA, Eng MY, Robinson SK, Carr LG, Wall TL. Associations of variations in alcohol dehydrogenase genes with the level of response to alcohol in non-Asians. Alcoholism, clinical and experimental research. 2006;no. 30(9):1470–1478. doi: 10.1111/j.1530-0277.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nature genetics. 2010;no. 42(12):1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JM. The promise and pitfalls of combining genetic and economic research. Health Econ. 2011;no. 20(8):889–892. doi: 10.1002/hec.1745. [DOI] [PubMed] [Google Scholar]
- Fletcher JM, Lehrer SF. Genetic lotteries within families. J Health Econ. 2011;no. 30(4):647–659. doi: 10.1016/j.jhealeco.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Fletcher The effects of adolescent health on educational outcomes: Causal evidence using genetic lotteries between siblings. Forum on Health Economics and Policy. 2009;no. 12(2) [Google Scholar]
- Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;no. 316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freathy RM, Ring SM, Shields B, Galobardes B, Knight B, Weedon MN, et al. A common genetic variant in the 15q24 nicotinic acetylcholine receptor gene cluster (CHRNA5-CHRNA3-CHRNB4) is associated with a reduced ability of women to quit smoking in pregnancy. Human molecular genetics. 2009;no. 18(15):2922–2927. doi: 10.1093/hmg/ddp216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Bierut LJ, Porjesz B, Edenberg HJ, Dick D, Goate A, et al. A novel non-parametric regression reveals linkage on chromosome 4 for the number of externalizing symptoms in sib-pairs. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2008;no. 147B(7):1301–1305. doi: 10.1002/ajmg.b.30735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding J, Pembrey M, Jones R. ALSPAC--the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatric and perinatal epidemiology. 2001;no. 15(1):74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- Haupt A, Thamer C, Staiger H, Tschritter O, Kirchhoff K, Machicao F, et al. Variation in the FTO gene influences food intake but not energy expenditure. Experimental and clinical endocrinology & diabetes : official journal, German Society of Endocrinology [and] German Diabetes Association. 2009;no. 117(4):194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- Hausman JA. Specification Tests in Econometrics. Econometrica. 1978;no. 46(6):1251–1271. [Google Scholar]
- Hertel JK, Johansson S, Sonestedt E, Jonsson A, Lie RT, Platou CG, et al. FTO, type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41,504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes. 2011;no. 60(5):1637–1644. doi: 10.2337/db10-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hinke Kessler Scholder S, Smith GD, Lawlor DA, Propper C, Windmeijer F. Mendelian randomization: the use of genes in instrumental variable analyses. Health economics. 2011;no. 20(8):893–896. doi: 10.1002/hec.1746. [DOI] [PubMed] [Google Scholar]
- von Hinke Kessler Scholder S, Davey Smith G, Lawlor DA, Propper C, Windmeijer F. Child Height, Health and Human Capital: Evidence using Genetic Markers. 2010 doi: 10.1016/j.euroecorev.2012.09.009. CMPO working paper 10/245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubacek JA, Bohuslavova R, Kuthanova L, Kubinova R, Peasey A, Pikhart H, et al. The FTO gene and obesity in a large Eastern European population sample: the HAPIEE study. Obesity. 2008;no. 16(12):2764–2766. doi: 10.1038/oby.2008.421. [DOI] [PubMed] [Google Scholar]
- Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;no. 302(7836):999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Jonsson A, Franks PW. Obesity, FTO gene variant, and energy intake in children. The New England journal of medicine. 2009;no. 360(15):1571–1572. doi: 10.1056/NEJMc090017. author reply 1572. [DOI] [PubMed] [Google Scholar]
- Kaur-Knudsen D, Bojesen SE, Tybjaerg-Hansen A, Nordestgaard BG. Nicotinic acetylcholine receptor polymorphism, smoking behavior, and tobacco-related cancer and lung and cardiovascular diseases: a cohort study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;no. 29(21):2875–2882. doi: 10.1200/JCO.2010.32.9870. [DOI] [PubMed] [Google Scholar]
- Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M, et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Human molecular genetics. 2009;no. 18(20):4007–4012. doi: 10.1093/hmg/ddp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khashan AS, Kenny LC. The effects of maternal body mass index on pregnancy outcome. European journal of epidemiology. 2009;no. 24(11):697–705. doi: 10.1007/s10654-009-9375-2. [DOI] [PubMed] [Google Scholar]
- Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. Plos Medicine. 2011;no. 8(11) doi: 10.1371/journal.pmed.1001116. e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Hersh CP, Washko GR, Hokanson JE, Lynch DA, Newell JD, et al. Epidemiology, radiology, and genetics of nicotine dependence in COPD. Respiratory research. 2011;no. 12:9. doi: 10.1186/1465-9921-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivimaki M, Smith GD, Timpson NJ, Lawlor DA, Batty GD, Kahonen M, et al. Lifetime body mass index and later atherosclerosis risk in young adults: examining causal links using Mendelian randomization in the Cardiovascular Risk in Young Finns study. European heart journal. 2008;no. 29(20):2552–2560. doi: 10.1093/eurheartj/ehn252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Fraser A, Macdonald-Wallis C, Nelson SM, Palmer TM, Davey Smith G, et al. Maternal and offspring adiposity-related genetic variants and gestational weight gain. The American journal of clinical nutrition. 2011;no. 94(1):149–155. doi: 10.3945/ajcn.110.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Statistics in Medicine. 2008;no. 27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- Lemoine P, Harousseau H, Borteyru J-P, Menuet J-C. Les enfants de parents alcooliques. Anomalies Observees. A propos de 127 cas. OQuest-Medical. 1968;no. 21:476–482. [Google Scholar]
- Lien DS, Evans WN. Estimating the impact of large cigarette tax hikes: The case of maternal smoking and infant birth weight. Journal of Human Resources. 2005;no. 40(2):373–392. [Google Scholar]
- Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;no. 42(5):436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhou Z, Hodgkinson CA, Yuan Q, Shen PH, Mulligan CJ, et al. Haplotype-based study of the association of alcohol-metabolizing genes with alcohol dependence in four independent populations. Alcoholism, clinical and experimental research. 2011;no. 35(2):304–316. doi: 10.1111/j.1530-0277.2010.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew M, Boeing H, Sturmer T, Brenner H. Relation among alcohol dehydrogenase 2 polymorphism, alcohol consumption, and levels of gamma-glutamyltransferase. Alcohol. 2003;no. 29(3):131–135. doi: 10.1016/s0741-8329(03)00015-6. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2006;no. 31(5):1085–1095. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, et al. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Human molecular genetics. 2009;no. 18(3):580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marichalar-Mendia X, Acha-Sagredo A, Rodriguez-Tojo MJ, Rey-Barja N, Hernando-Rodriguez M, Aguirregaviria JI, et al. Alcohol-dehydrogenase (ADH1B) Arg48His polymorphism in Basque Country patients with oral and laryngeal cancer: preliminary study. Anticancer research. 2011;no. 31(2):677–680. [PubMed] [Google Scholar]
- McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, Byrnes G, et al. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS genetics. 2011;no. 7(3) doi: 10.1371/journal.pgen.1001333. e1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen E, Himes BE, Brehm JM, Boutaoui N, Klanderman BJ, Sylvia JS, et al. Analyses of shared genetic factors between asthma and obesity in children. The Journal of allergy and clinical immunology. 2010;no. 126(3):631–637. doi: 10.1016/j.jaci.2010.06.030. e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metwally M, Ong KJ, Ledger WL, Li TC. Does high body mass index increase the risk of miscarriage after spontaneous and assisted conception? A meta-analysis of the evidence. Fertil Steril. 2008;no. 90:714–726. doi: 10.1016/j.fertnstert.2007.07.1290. [DOI] [PubMed] [Google Scholar]
- Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nature genetics. 2009;no. 41(2):157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Mumby HS, Elks CE, Li S, Sharp SJ, Khaw KT, Luben RN, et al. Mendelian Randomisation Study of Childhood BMI and Early Menarche. Journal of obesity. 2011 doi: 10.1155/2011/180729. 180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H. Genetics of COPD. Allergology international : official journal of the Japanese Society of Allergology. 2011 doi: 10.2332/allergolint.11-RAI-0326. [DOI] [PubMed] [Google Scholar]
- Olsen Jorn, Melbye Mads, Olsen Sjurdur F, Sorensen Thorkild IA, Peter Aaby, Andersen Anne-Marie Nybo, et al. The Danish National Birth Cohort - its background, structure and aim. Scand J Public Health. 2001;no. 29(4):300–307. doi: 10.1177/14034948010290040201. [DOI] [PubMed] [Google Scholar]
- OMIM. Online Mendelian Inheritance in Man. Baltimore, MD: McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; 2011. [Google Scholar]
- Palmer T, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, et al. Statistical Methods in Medical Research. 2011 doi: 10.1177/0962280210394459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschou P, Ziv E, Burchard EG, Choudhry S, Rodriguez-Cintron W, Mahoney MW, et al. PCA-correlated SNPs for structure identification in worldwide human populations. PLoS Genet. 2007;no. 3(9):1672–1686. doi: 10.1371/journal.pgen.0030160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permutt T, Hebel JR. Simultaneous-equation estimation in a clinical trial of the effect of smoking on birth weight. Biometrics. 1989;no. 45(2):619–622. [PubMed] [Google Scholar]
- Rees SD, Islam M, Hydrie MZ, Chaudhary B, Bellary S, Hashmi S, et al. An FTO variant is associated with Type 2 diabetes in South Asian populations after accounting for body mass index and waist circumference. Diabetic medicine : a journal of the British Diabetic Association. 2011;no. 28(6):673–680. doi: 10.1111/j.1464-5491.2011.03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, et al. Genome-wide search for genes affecting the risk for alcohol dependence. American journal of medical genetics. 1998;no. 81(3):207–215. [PubMed] [Google Scholar]
- Rosenzweig M, Schultz TP. Estimating a household production function: Heterogeneity, the demand for health inputs, and their effects on birth weight. The Journal of Political Economy. 1983;no. 91(5):723–746. [Google Scholar]
- Rutters F, Lemmens SG, Born JM, Bouwman F, Nieuwenhuizen AG, Mariman E, et al. Genetic associations with acute stress-related changes in eating in the absence of hunger. Patient education and counseling. 2010;no. 79(3):367–371. doi: 10.1016/j.pec.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS genetics. 2010;no. 6(8) doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, et al. A genome screen of maximum number of drinks as an alcoholism phenotype. American journal of medical genetics. 2000;no. 96(5):632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;no. 150B(4):453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;no. 16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Saccone NL, Neuman RJ, Rice JP. Genetic analysis of the maximum drinks phenotype. BMC genetics. 2005;no. 6(Suppl 1):S124. doi: 10.1186/1471-2156-6-S1-S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS genetics. 2010;no. 6(4) doi: 10.1371/journal.pgen.1000916. e1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS genetics. 2007;no. 3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS medicine. 2008;no. 5(8):e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Rice JP, Neuman RJ, Rochberg N, Saccone NL, Bierut LJ. Associations and interactions between SNPs in the alcohol metabolizing genes and alcoholism phenotypes in European Americans. Alcoholism, clinical and experimental research. 2009;no. 33(5):848–57. doi: 10.1111/j.1530-0277.2009.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Wilhelmsen K, Pomerleau CS, Chasse SA, Rice JP, Snedecor SM, et al. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with 'pleasurable buzz' during early experimentation with smoking. Addiction. 2008;no. 103(9):1544–1552. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;no. 33(1):30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- Smith GD, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;no. 4(12):e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity. 2008;no. 16(8):1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- Staiger Douglas, Stock James H. Instrumental Variables Regression with Weak Instruments. Econometrica. 1997;no. 65(3):557–586. [Google Scholar]
- Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2008;no. 17(12):3517–3525. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan WC. A note on the influence of maternal inebriety on the offspring. 1899. International journal of epidemiology. 2011;no. 40(2):278–282. doi: 10.1093/ije/dyr006. [DOI] [PubMed] [Google Scholar]
- The Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature genetics. 2010;no. 42(5):441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;no. 452(7187):638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nature genetics. 2009;no. 41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Timofeeva MN, McKay JD, Smith GD, Johansson M, Byrnes GB, Chabrier A, et al. Genetic Polymorphisms in 15q25 and 19q13 Loci, Cotinine Levels, and Risk of Lung Cancer in EPIC. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011 doi: 10.1158/1055-9965.EPI-11-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson NJ, Emmett PM, Frayling TM, Rogers I, Hattersley AT, McCarthy MI, et al. The fat mass- and obesity-associated locus and dietary intake in children. The American journal of clinical nutrition. 2008;no. 88(4):971–978. doi: 10.1093/ajcn/88.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson NJ, Harbord R, Davey Smith G, Zacho J, Tybjaerg-Hansen A, Nordestgaard BG. Does greater adiposity increase blood pressure and hypertension risk?: Mendelian randomization using the FTO/MC4R genotype. Hypertension. 2009;no. 54(1):84–90. doi: 10.1161/HYPERTENSIONAHA.109.130005. [DOI] [PubMed] [Google Scholar]
- Tolstrup JS, Nordestgaard BG, Rasmussen S, Tybjaerg-Hansen A, Gronbaek M. [Genetic variations in alcohol dehydrogenase, drinking habits and alcoholism] Ugeskrift for laeger. 2008;no. 170(35):2672–2675. [PubMed] [Google Scholar]
- Torloni MR, Betran AP, Daher S, Widmer M, Dolan SM, Menon R, et al. Maternal BMI and preterm birth: a systematic review of the literature with meta-analysis. The journal of maternal-fetal & neonatal medicine : the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstetricians. 2009;no. 22(11):957–70. doi: 10.3109/14767050903042561. [DOI] [PubMed] [Google Scholar]
- Tournier JM, Birembaut P. Nicotinic acetylcholine receptors and predisposition to lung cancer. Current opinion in oncology. 2011;no. 23(1):83–87. doi: 10.1097/CCO.0b013e3283412ea1. [DOI] [PubMed] [Google Scholar]
- Tung YC, Yeo GS. From GWAS to biology: lessons from FTO. Annals of the New York Academy of Sciences. 2011;no. 1220:162–171. doi: 10.1111/j.1749-6632.2010.05903.x. [DOI] [PubMed] [Google Scholar]
- Wall TL, Shea SH, Luczak SE, Cook TA, Carr LG. Genetic associations of alcohol dehydrogenase with alcohol use disorders and endophenotypes in white college students. Journal of abnormal psychology. 2005;no. 114(3):456–465. doi: 10.1037/0021-843X.114.3.456. [DOI] [PubMed] [Google Scholar]
- Wardle J, Carnell S, Haworth CM, Farooqi IS, O'Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. The Journal of clinical endocrinology and metabolism. 2008;no. 93(9):3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- Wattigney WA, Srinivasan SR, Chen W, Greenlund KJ, Berenson GS. Secular trend of earlier onset of menarche with increasing obesity in black and white girls: the Bogalusa Heart Study. Ethnicity & disease. 1999;no. 9(2):181–189. [PubMed] [Google Scholar]
- Wehby GL, Fletcher JM, Lehrer SF, Moreno LM, Murray JC, Wilcox A, Lie RT. A genetic instrumental variables analysis of the effects of prenatal smoking on birth weight: evidence from two samples. Biodemography and social biology. 2011a;no. 57(1):3–32. doi: 10.1080/19485565.2011.564468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehby GL, Jugessur A, Moreno LM, Murray JC, Wilcox A, Lie RT. Genes as instruments for studying risk behavior effects: An application to maternal smoking and oraofacial clefts. Health Services and Outcomes Research Methodology. 2011b;no. 11(1–2):54–78. doi: 10.1007/s10742-011-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehby GL, Ohsfeldt RL, Murray JC. 'Mendelian randomization' equals instrumental variable analysis with genetic instruments. Statistics in Medicine. 2008;no. 27(15):2745–2749. doi: 10.1002/sim.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehby GL, Prater KN, McCarthy AM, Castilla EE, Murray JC. The Impact of Maternal Smoking during Pregnancy on Early Child Neurodevelopment. Journal of Human Capital. 2011c;no. 5(2):207–254. doi: 10.1086/660885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehby GL, Murray JC, Wilcox A, Lie RT. Smoking and body weight: Evidence using genetic instruments. Economics and human biology. 2012;no. 10(2):113–126. doi: 10.1016/j.ehb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehby GL, Murray JC, Castilla EE, Lopez-Camelo JS, Ohsfeldt RL. Prenatal care demand and its effects on birth outcomes by birth defect status in Argentina. Economics & Human Biology. 2009;no. 7(1):84–95. doi: 10.1016/j.ehb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS genetics. 2008;no. 4(7) doi: 10.1371/journal.pgen.1000125. e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB, Nightingale BN, Bucholz KK, Madden PA, Heath AC, Martin NG. ADH genotypes and alcohol use and dependence in Europeans. Alcoholism, clinical and experimental research. 1998;no. 22(7):1463–1469. [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature genetics. 2009;no. 41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst M. The effect of cigarette and alcohol consumption on birth outcomes. In Working Paper 10-05: Aarhus School of Business, Department of Economics. 2010 [Google Scholar]
- Yang SJ, Yokoyama A, Yokoyama T, Huang YC, Wu SY, Shao Y, et al. Relationship between genetic polymorphisms of ALDH2 and ADH1B and esophageal cancer risk: a meta-analysis. World journal of gastroenterology : WJG. 2010;no. 16(33):4210–4220. doi: 10.3748/wjg.v16.i33.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FF, Hou L, Terry MB, Lissowska J, Morabia A, Chen J, et al. Genetic polymorphisms in alcohol metabolism, alcohol intake and the risk of stomach cancer in Warsaw, Poland. International journal of cancer. Journal international du cancer. 2007;no. 121(9):2060–2064. doi: 10.1002/ijc.22973. [DOI] [PubMed] [Google Scholar]
- Zhang GH, Mai RQ, Huang B. Meta-analysis of ADH1B and ALDH2 polymorphisms and esophageal cancer risk in China. World journal of gastroenterology : WJG. 2010a;no. 16(47):6020–6025. doi: 10.3748/wjg.v16.i47.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Mai R, Huang B. ADH1B Arg47His polymorphism is associated with esophageal cancer risk in high-incidence Asian population: evidence from a meta-analysis. PloS one. 2010b;no. 5(10) doi: 10.1371/journal.pone.0013679. e13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintzaras E, Stefanidis I, Santos M, Vidal F. Do alcohol-metabolizing enzyme gene polymorphisms increase the risk of alcoholism and alcoholic liver disease? Hepatology. 2006;no. 43(2):352–361. doi: 10.1002/hep.21023. [DOI] [PubMed] [Google Scholar]
- Zintzaras E, Zdoukopoulos N. A field synopsis and meta-analysis of genetic association studies in peripheral arterial disease: The CUMAGAS-PAD database. American journal of epidemiology. 2009;no. 170(1):1–11. doi: 10.1093/aje/kwp094. [DOI] [PubMed] [Google Scholar]
- Zuccolo L, Fitz-Simon N, Gray R, Ring SM, Sayal K, Smith GD, Lewis SJ. A non-synonymous variant in ADH1B is strongly associated with prenatal alcohol use in a European sample of pregnant women. Human molecular genetics. 2009;no. 18(22):4457–4466. doi: 10.1093/hmg/ddp388. [DOI] [PMC free article] [PubMed] [Google Scholar]