Abstract
Metabolism is essential to understand human health. To characterize human metabolism, a high-resolution read-out of the metabolic status under various physiological conditions, either in health or disease, is needed. Metabolomics offers an unprecedented approach for generating system-specific biochemical definitions of a human phenotype through the capture of a variety of metabolites in a single measurement. The emergence of large cohorts in clinical studies increases the demand of technologies able to analyze a large number of measurements, in an automated fashion, in the most robust way. NMR is an established metabolomics tool for obtaining metabolic phenotypes. Here, we describe the analysis of NMR-based urinary profiles for metabolic studies, challenged to a large human study (3007 samples). This method includes the acquisition of nuclear Overhauser effect spectroscopy one-dimensional and J-resolved two-dimensional (J-Res-2D) 1H NMR spectra obtained on a 600 MHz spectrometer, equipped with a 120 μL flow probe, coupled to a flow-injection analysis system, in full automation under the control of a sampler manager. Samples were acquired at a throughput of ∼20 (or 40 when J-Res-2D is included) min/sample. The associated technical analysis error over the full series of analysis is 12%, which demonstrates the robustness of the method. With the aim to describe an overall metabolomics workflow, the quantification of 36 metabolites, mainly related to central carbon metabolism and gut microbial host cometabolism, was obtained, as well as multivariate data analysis of the full spectral profiles. The metabolic read-outs generated using our analytical workflow can therefore be considered for further pathway modeling and/or biological interpretation.
The oldest known form of diagnostics in humans using body fluid analysis was done on urine. As early as 400 BC, urine was poured on the ground, and in the event of attracting insects, patients were diagnosed with boils.1 Nowadays, urine analysis is conventionally employed in standard clinical research to identify, among others, kidney and urogenital disorders. Products of cellular metabolism as metabolites, proteins, and salts are excreted in urine. Urine contains many low molecular weight compounds, in particular nitrogen-rich molecules, such as urea, creatinine, and amino acids but also various organic acids and sugars, as well as xenobiotics, and detoxified and nonmetabolized compounds. Probably the most comprehensive list of metabolites identified in urine so far is presented in the Human Metabolome Database, containing over 3000 chemical species obtained by a variety of analytical techniques.2 Urine analysis provides novel insights into the metabolism taking place, in which certain metabolites may associate with the subject’s physiological condition, under the influence of genotypic and environmental settings. According to the type of sample collection (spot or 24 h urine), urinary metabolic profiles give an approximate time-averaged representation of the recent homeostatic metabolic changes of the individual, in addition to carry indirect information on dietary exposure, as well as certain gut microbial metabolic activities. The latter has been recently associated with various conditions, such as cardiovascular disease,3 type 2 diabetes,4 obesity,5 nonalcoholic fatty liver disease,5 central nervous system diseases,6 and cancer.7 The application of metabolomics to urine samples has been largely promoted due to the noninvasiveness of the sample collection, well suited in epidemiological or clinical studies, and yet providing the required proficiency in detecting metabolic changes.
To capture the multitude of metabolites present in urine, analytical methods such as mass spectrometry (MS) and NMR spectroscopy are often used in metabolomics analyses. Particularly when dealing with large population studies, rigorous analytical set-ups and data processing pipelines are required to provide and ensure the following: (1) unequivocal labeling of samples, (2) metabolite-rich information, (3) metabolite identification, (4) metabolite quantitation, (5) selectivity to detect biological differences, (6) high-throughput, (7) robustness, (8) reproducibility, (9) stability over time, and (10) minimal technical errors, so that relevant metabolic changes can be investigated through various data mining strategies. NMR gathers most of these properties and is an expert tool to generate metabolic profiles from biofluids such as urine.
The opportunity to perform “tubeless” NMR, without compromising data quality, seems both attractive (e.g., suppression of tubes altogether) and practical in the analysis of large-scale studies, in particular for high-throughput applications. In the early 1980s, the development of dedicated flow NMR probes has opened the field to on-flow NMR applications, such as liquid chromatography (LC)- and flow injection analysis (FIA)-NMR.8 While LC-NMR methodologies9 were extensively developed with the purpose of compound identification strategies,8,10 FIA-NMR was applied for screening and high-throughput applications.11,12 FIA-NMR was first introduced in 1997, before being extended to a few other metabolomics studies.13,14
NMR is a quantitative technique that therefore offers the opportunity of quantifying metabolites directly from 1H NMR metabolite profiles, avoiding additional sample preparation or analyses, with the possibility of quantifying unexpected metabolites (unlike MS-based quantitative methods). A 1H NMR spectrum of a biofluid, such as urine, can be viewed as the sum of its individual metabolites and if acquired under quantitative conditions can be used for both metabolite identification and quantitation. The NMR signal intensity is essentially related to the concentration (actually, to the number of nuclei responsible for the signal), although influenced by many experimental and sample dependent parameters.15−17 Many quantitation methods have been developed for NMR measurements, leading to both relative and absolute quantitation,15 such as internal or external quantitation references, electronic reference signals,18 and library-based quantitation tools (e.g., the software provided by Chenomx, ACD/Laboratories, Mestrelab, and Bruker AMIX).
Here, we describe an automated high-throughput NMR-based metabolomics method for urine analysis. All analyses are controlled by a sample manager that tracks samples along the full metabolomics pipeline (labeling, FIA-NMR analysis, spectra preprocessing, and data storage). The samples are barcode-identified, prepared for analysis by a robot handling 96-well plates, and submitted to the FIA-NMR setup. High-resolution 1H NMR spectra are obtained, preprocessed, and transferred in an automated way for further data handling. The identification and quantitation of metabolites directly from urine profiles is achieved by using a standard compounds knowledge-base approach. We believe this method will be of relevance for undergoing large-scale urine metabolomics analyses in population studies.
Experimental Section
Chemicals
Heavy water (99.8 atom % D) used for NMR analysis was obtained from Cortecnet (Paris, France). Sucrose (BioXtra, purity ≥99.5%), d-(+)-glucose (purity 99.5%), hippuric acid (purity 98%), succinic acid (purity ≥99%), d-alanine (purity ≥99%), sodium azide (BioXtra), sodium phosphate dibasic (purity ≥99.0%), and nitric acid (purity ≥65%) were obtained from Sigma–Aldrich (Buchs, Switzerland). Sodium dihydrogen phosphate monohydrate, (purity ≥99.0%) and methanol-d4 (≥99.8% for NMR spectroscopy MagniSolv) were purchased from Merck KGaA (Darmstadt, Germany), and 3-(trimethylsilyl)-2,2′,3,3′-tetradeuteropropionic acid sodium salt (TSP) was obtained from Dr. Glaser AG (Basel, Switzerland). The standard line shape test solution (chloroform in acetone-d6) used for NMR flow injection was provided by Bruker (Rheinstetten, Germany). Ultrapure water was obtained from a Direct-Q 5 ultrapure water system, Millipore (Zug, Switzerland).
Biological Samples
In this multicenter study, 3007 urine samples of healthy men and women (ages 35–74, mean average 58 years old) were collected as morning-spots. All urine samples were obtained within the Mark-Age project (www.mark-age.eu). Appropriate ethical approval from the research ethics committee was acquired to allow the collection of human urine samples for metabolomics purposes. The collection, storage, and associated sample logistics were coordinated to ensure sample integrity: samples were kept cold until being sent in cryogenic containers filled with dry ice. After arrival to the laboratory, the frozen samples were stored at −80 °C before analysis.
Solutions and Eluents
Sample Extraction Buffer
A 0.6 M phosphate-buffered saline pH 7.40 with 0.2 g/L TSP and 0.5 g/L NaN3 was used as sample extraction buffer.
Quality Control (QC) Sample
A 20 mM sucrose solution, 0.5 mM TSP, 2 mM NaN3 in 90% H2O/10% D2O (v/v) was used for line-shape assessment, system equilibration, and quality control.
FIA Push-Eluent and Sample Train Eluent
A 90% H2O/10% D2O solution (v/v) was prepared in a hermetic bottle to be used as eluent in the FIA system.
FIA Cleaning Solution
For each cleaning event, 1 mL of 25% (v/v) 65% HNO3 in 90% H2O/10% D2O solution (v/v) was freshly prepared.
Sample Preparation
Urine samples were randomized before proceeding with sample preparation. All samples (urine and QC) were prepared using the MICROLAB STAR liquid handling workstation (Hamilton) in a 1:2 proportion (buffer/sample). Samples were taken from −80 °C freezers, thawed in batches of 96 (85 urine samples and 11 QC samples), and a sample table was elaborated (as the samples were not provided with barcodes from the collection centers) for each plate. The samples were then prepared plate by plate, progressively, just before NMR acquisition. By using the implemented sample preparation routine, each plate was prepared in less than 15 min. This procedure was done using tailor-made routines under the control of Hamilton method manager software, version 5 (Hamilton) (see the Supporting Information).
FIA
As a FIA device, a Bruker efficient sample transfer (BEST) system was used. A Peltier cooling controller was employed to cool a sample rack able to hold simultaneously two barcode-labeled sample plates. The infusion method was optimized for aqueous solutions (D2O and H2O), and the transfer volumes and transfer speeds parameters were set using the BEST-NMR administration toolbox within TopSpin (Bruker BioSpin) (see the Supporting Information).
NMR
NMR spectral acquisition was performed using a Bruker Avance III NMR spectrometer equipped with 600 MHz magnet Ultrashield Plus (spectrometer frequency: 600.0299446 MHz), a BCU-05 cooling unit, and a 4 mm flow probe FISEI 1H-13C/D Z-GRD NMR with 120 μL active volume. The temperature was fixed to 300 K for all NMR measurements, and temperature calibration was checked every week, after nitrogen fill. The line-shape sensitivity and water suppression were tested to assess probe performance (see the Supporting Information).
All samples were submitted to the following sequential NMR measurements: flow cell profile, 1H one-dimensional version of nuclear Overhauser effect spectroscopy (NOESY-1D, noesygppr1d) and for a subset of spectra, homonuclear 1H J-Resolved two-dimensional correlation (J-Res-2D, jresgpprqf) (see the Supporting Information). All NMR spectral acquisition and preprocessing were done under the control of TopSpin 2.1, patch level 6 (Bruker BioSpin), and the automated submission of a sequence of samples was performed using ICON-NMR, 4.2.6 build 9 for TopSpin 2.1 (Bruker BioSpin).
Automation
Samples were managed by the sample manager software SampleTrack 2.60.01 (Bruker BioSpin) that allows the printing of unique linear barcodes, necessary for identifying samples, according to their container type. It allows importing sample tables (XLS file); it deals with complex sample orders, involving the synchronization of various instruments (FIA and NMR); it submits sequential analytical methods and steps; it coordinates data acquisition and storage, and it detains a universal sample database. In terms of configuration, SampleTrack takes control via a peer-to-peer network the NMR acquisition.
Metabolite Identification
Metabolite identification was achieved by comparing the obtained urine spectra with spectral databases containing spectra of standard compounds, including the database B-BIOREFCODE (Bruker BioSpin) and additional in-house reference compounds. Further information relevant for identification was complemented by consulting HMDB,2 Madison Metabolomics Consortium Database (MMCD),19 Biological Magnetic Resonance Bank (BioMagResBank),20 Birmingham Metabolite Library (BML-NMR),21 NMRShiftDB,22 Platform for RIKEN Metabolomics (PRIMe),23 and literature.2,24−28 The identification of metabolites was further verified with homonuclear and heteronuclear 2D NMR experiments, such as 1H–1H COSY, 1H–1H TOCSY, 1H–13C HMBC, and 1H–13C HSQC.
Metabolite Quantitation
Standard solutions were added to urine in 1:3 dilutions (n = 8): hippuric acid (δH = 7.750 ± 0.050 ppm), succinic acid (δH = 2.335 ± 0.035 ppm), alanine (δH = 1.400 ± 0.050 ppm), and (±)-glucose (alpha-, δH = 5.150 ± 0.050 ppm; beta-, δH = 4.563 ± 0.037 ppm). Spectra were acquired by FIA-NMR, and quantitation of these four compounds was obtained by addition of the standard compounds. In parallel, metabolite quantitation was achieved using a library-based method, by building a knowledge-base using our internal reference database within AMIX (Bruker), taking 1H NMR NOESY and 1H J-Res spectra of reference compounds acquired at pH 7.0. For each metabolite chemical shift intervals, stoichiometry and multiplicity of resonances were described. Creatinine was used as an internal reference, and the obtained quantitative values were expressed as millimoles of metabolite per mole of creatinine.
Data Analysis
Profiling data were examined using TopSpin and AMIX-Viewer, version 3.9.9 (Bruker BioSpin) by overlaying spectra to detect inconsistencies. Flow cell profiles were inspected as a quality gate; thus, samples that failed to have a correct positioning inside the flow cell were discarded. As a second step, all nonconforming 1H NMR NOESY-1D spectra due to poor water suppression, poor shimming, or intractable baseline correction were also discarded. Extraction of a data matrix from 1H NMR NOESY-1D spectra was done using the Statistics toolbox within AMIX: bin size was selected according to data analysis purpose, ranging from 0.04 to 0.0005 (larger bin sizes depreciate chemical shift variation, at the expense of lower spectral resolution); the water signal was excluded (4.95–4.70 ppm); a spectral width of 12–0.05 ppm was chosen; TSP signal intensity was used for technical normalization (0.05 to −0.05 ppm interval); and baseline correction was performed by noise estimation using the signal-free interval 11–9.5 ppm. A Pareto-scaled principal component analysis was used to model the urine spectra.
Results and Discussion
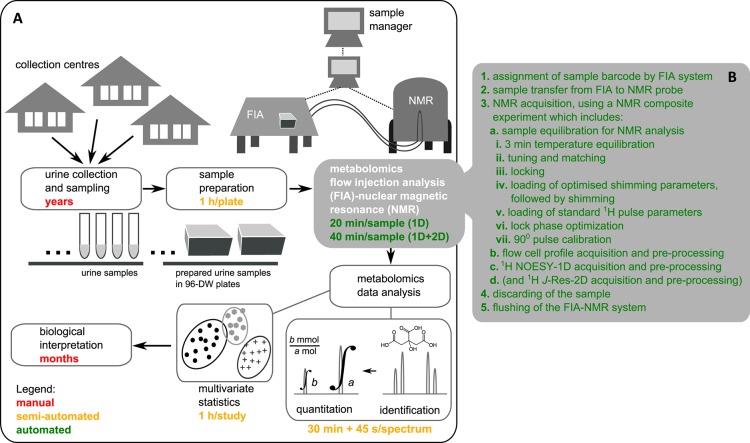
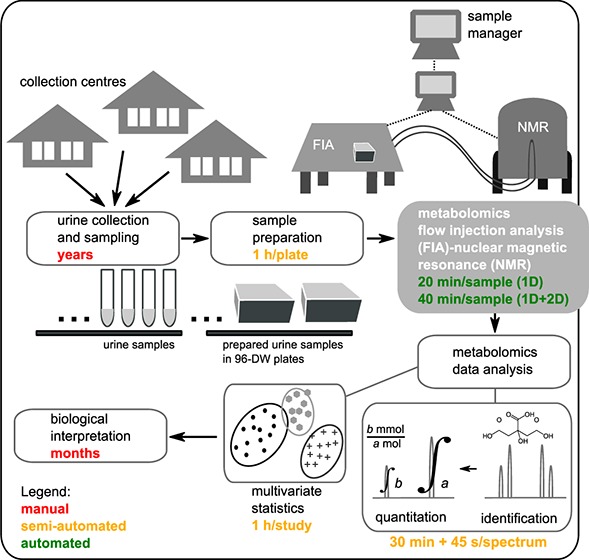
Being able to analyze samples at a high throughput opens the application of metabolomics to a systems level, where metabolomics technologies are comparable to genomics or proteomics in providing large-scale metabolic information about various processes in living organisms. The primary aim of this study was to establish a robust high-throughput metabolomics method, providing an untargeted and quantitative overview of endogenous urinary metabolites, which would be implementable in future epidemiological studies. Given the large number of samples dealt in this study, particular emphasis was given on automating the whole metabolomics pipeline (sample preparation, sample management, sample injection, NMR acquisition, and data extraction and analysis, Figure 1A). The methodology aims for a minimum of manual intervention while ensuring data quality, reproducibility, quantifiability, and traceability.
Figure 1.
(A) Pipeline of the quantitative metabolomics analysis of urine by FIA-NMR, including an approximate timing for each step (method implementation time is not included). (B) Step-by-step description of the automated FIA-NMR setup.
Planning and timing
Large human population studies require years to plan, approve, and implement; therefore, special attention was given to sample storage and integrity. According to Lauridsen et al.,29 urine samples are stable below −25 °C. We have stored our samples at −80 °C prior to sample preparation, and to ensure sample integrity, an antimicrobial agent was added. Sample preparation required manual intervention in the preparation of sample lists (as urine samples were provided with hand-written labels), after which the transfer and buffer dilution were performed automatically using a sample preparation robot (Figure 1A). Samples were kept at 10 °C in a cooled tray prior to FIA-NMR analysis.
The FIA-NMR method was fully automated in terms of NMR data acquisition (Figure 1B). The sample manager attributed a unique sample identifier to each sample and tracked the samples by submitting analytical orders and controlling all analysis steps. Samples were transferred into the NMR probe using an implemented transfer method by FIA. Once inside the magnet, automatic programs were set so that all required parameter adjustments for NMR analysis were automated, followed by data acquisition and preprocessing. After NMR acquisition, samples were discarded, the probe was rinsed, and the following sample was transferred. Sample orders could be submitted, plate by plate, while acquired data was archived and stored in a server, without having to halt the acquisition. The total FIA-NMR analysis time per sample was 20 min (and 40 min when J-Res-2D was included). Because J-Res-2D was recorded for the first 15 plates only, the total analysis time was reduced by roughly half. This study required 4 months of FIA-NMR analysis, including weekly breaks for nitrogen fill, reconditioning, preventive cleaning, and troubleshooting.
Sample Traceability
To make sure all sample information is recorded and traced, a sample manager associated to a database was used to deal with sample identification and labeling (associated barcode, sample name, and sample history), FIA-NMR method used (date of analysis, transfer method, NMR measurements, and error occurrence), and data management (folder organization, archiving, storage, and access) (Figure 1).
FIA-NMR Method Performance
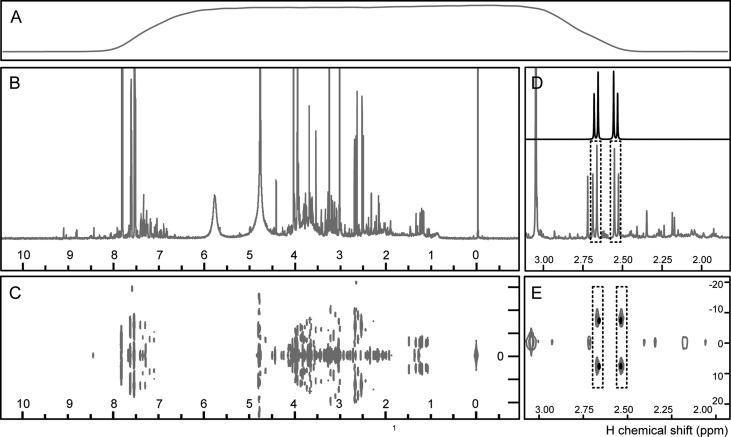
A prerequisite to implement a FIA-NMR methodology is the quality of acquired spectra that should be comparable to conventional “tube”. In flow-NMR, the correct placing of the sample in the active detection cell is a prerequisite for spectra acquisition and should be optimized beforehand for proper alignment. Once the optimization is done for a specific solvent, these parameters (transfer volumes and speeds) are constant if no modification is applied. A simple and fast test to confirm the fullness of the active detection cell is done by acquiring a flow cell profile (Figure 2A) that is routinely implemented for every injected sample by FIA-NMR.
Figure 2.
1H NMR metabolomics profiles of urine, acquired as a routine analysis by FIA-NMR: (A) flow cell profile; (B) 1H NMR NOESY-1D; (C) 1H J-Res-2D; citric acid signal in 1H NMR NOESY-1D and 1H J-Res-2D (E) spectra of (gray) urine and (black) standard compound.
To assess the NMR performance of the flow probe (in comparison to “tube NMR” probes), the suite of standard NMR tests (line shape, sensitivity, and water suppression) was performed. Standard performance tests use nonaqueous solvents (1% chloroform in acetone-d6 for line shape; 0.1% ethylbenzene in cloroform-d1 for sensitivity) that are impractical when used in FI systems: the solutions would have to be freshly prepared before each analysis, making it difficult to standardize the concentration over time, and it would have to be injected directly into the probe, given the incompatibility of the solvents with the FI system. Therefore, we attempted to assess performance using a QC sample. The line-shape test using the conventional standard sample (1% chloroform in acetone-d6) gave the following results: hump 6.9/11.9 Hz with a 0.45 Hz resolution; thereafter, it was assessed by calculating the resolution of the anomeric proton of sucrose in the QC sample (Figure 3A) that was optimized to as low as 25%. The sensitivity was tested using the QC sample, in which the signal-to-noise was calculated for the anomeric proton of sucrose, yielding the ratio 1186.26:1. In terms of water suppression, the ratio 8.8/23.4 Hz was obtained for the water signal of the QC sample. Thus, all these tests indicate that the flow probe performs well using defined solutions, as QCs. When analyzing biological samples, such as urine, the probe is also performing well, as seen by the obtained resolution and water suppression (Figure 2B,C).
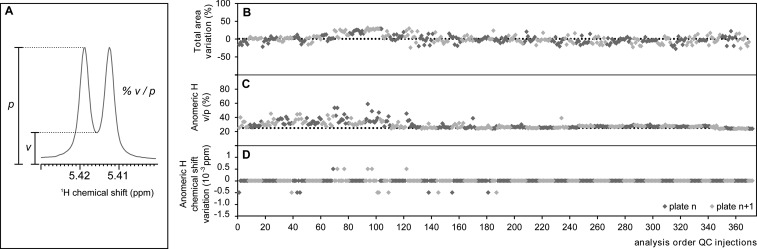
Figure 3.
(A) Anomeric proton of sucrose, appearing as a doublet at δH = 5.416 ppm, indicating the valley (v)-to-peak (p) ratio (%) as a measurement to quantify the shimming quality of a 1H NMR spectrum. (B–D) Stability of the FIA-NMR during 4 months of analyses. The quality controls acquired in-between analyses are depicted, after data extraction using a bin width of 0.0005 (n = 371). (B) Signal intensity variation, expressed as variation of total area of 1H NMR NOESY-1D to average ratio (%). (C) Shimming quality, expressed as % v/p of anomeric proton of sucrose. (D) Chemical shift stability of anomeric proton of sucrose, expressed as a difference to average (ppm).
In terms of FIA-NMR performance, the sample carry-over was estimated by comparing the signal-to-noise of the anomeric proton of a QC sample after a blank sample (9:1, H2O/D2O) injection, in automated mode. A ratio of 3.3% was obtained with the implemented in-between analysis cleaning procedures, which was considered acceptable, and it is lower than reported elsewhere.13
In sum, FIA-NMR allows the acquisition of NMR spectra in a proficient manner, suppressing the usage of NMR tubes altogether, by using cheaper and easier to trace plates, avoiding the tedious and rigorous manipulation of filling and handling glass tubes.
FIA-NMR Method Stability
The technical robustness of a method is a required parameter to consistently provide high quality data usable for biological insight. Because the total analysis time took place over months, we implemented the analysis of QC samples not only for system equilibration and optimization but also as a stability check: for every 10 biological samples, a QC sample was injected, along the full duration of the analyses. Biological QC samples are often used, in particular in LC/MS analysis, but given the large volume that would be needed for this study, a sucrose standard solution was used instead. The advantages are the large volume of QC sample that can be prepared prior to this study, so that the same solution (at exactly the same concentration) could be used for comparison. Thus, the stability of QC samples over time (and plate) was assessed (Figure 3); in particular, the following parameters were closely monitored: stability of the signal intensity, spectral resolution, and chemical shift variation.
The NMR signal is influenced by many experimental and sample dependent parameters, including analytical variants as sample preparation, FIA parameters, and NMR experimental parameters. To check the stability of the signal intensity, the total 1H NMR NOESY-1D spectral intensities of 377 QC samples acquired along the full length of the study were compared, yielding an associated error of 12% (with a spread of ±30%), Figure 3B.
The spectral resolution is assessed by the shimming quality; therefore, this parameter was inspected by comparing the valley-to-peak ratio (%v/p, Figure 3A) of the anomeric proton of sucrose present in the QC sample, Figure 3C. The shimming parameters were optimized at least every week using a QC sample to a starting value of at least 25%. Throughout the study, the average obtained resolution was 29%. After the analysis of the 15th plate, the shimming quality improved to an average %v/p of 27%, with an associated error of 2%, Figure 3C.
The third parameter analyzed for robustness was the stability of the proton chemical shift, δH. It is known that the δH values of a particular molecule depend primarily on the molecule itself but also on the solvent used, concentration, temperature, pH, metal ion concentrations, protein–metabolite and metabolite–metabolite interactions, or other chemical interactions. In aqueous solutions, such as H2O/D2O, the δH highly depends on pH; therefore, the samples are buffered before analysis. In addition, all spectra were calibrated to the TSP signal (δH = 0) in a preprocessing step, undergoing a one-point δH axis calibration. The variation of the anomeric proton δH of sucrose in QC samples was calculated for this study, and the variation obtained was 2 × 10–3%, Figure 3D. Obviously, this effect is likely to be more predominant in matrices such as urine, in which the presence of salt influences the ionic strength, and resonances corresponding to certain molecules can experience larger deviations.
Metabolite Coverage in Urine Profiles by 1H NMR
More than 3000 samples, including biological and QC samples, were analyzed by FIA-NMR analysis (Figure 1A–C). The identification of compounds from complex mixtures, as in metabolomics, is an intricate endeavor, as metabolite signals overlap and can lead to small chemical shift variations that hinder assignments. In this study, spectral databases containing spectra of standard compounds were used to proceed with the identification of urine metabolites using NOESY-1D profiles and J-Res-2D (Figure 2D,E). J-Res-2D adds increased resolution for 1D-NMR assignments, as multiplicities are put in evidence, facilitating identification. A list of 85 known urine metabolites was compiled from literature and HMDB. Except for glycerophosphocholine, found by Wishart,25 all metabolites have been found in urine according to HMDB. Of these, 81 were present in our internal spectral database. For 45 metabolites, their detection was not feasible either due to low signal-to-noise or overlap. Thus, 36 endogenous metabolites were found in urine, belonging to central metabolism, as glycolysis/gluconeogenesis and Krebs cycle, pyrimidine metabolism, amino acid metabolism, creatinine-related metabolites, and gut microbial–host cometabolites (Table 1). In the identification of metabolites from NMR spectra of mixtures, spectral-driven approaches were the most reliable. The spectra of standard compounds acquired in the same conditions as the mixture spectra (solvent, temperature, and pH) can be overlaid with mixture spectra, for better assessment of chemical shift, resonance shape, and stoichiometry. From all the databases consulted, urine-specific literature and urine-specific databases, such as HMDB, were the most helpful in providing additional confirmation for the resonances observed (biological occurrence and complementary evidence, such as MS or 2D-NMR information).
Table 1. Nonexhaustive List of Urine Metabolites Based on Literature and HMDB and Its Relative Quantitationa Detected by 1H NMR Spectroscopy, Using NOESY-1D and J-Res-2D, in One of the Urine Samples of This Study (Figure 1)b.
| compound | δH (multiplicity, x Hs) | q | compound | δH (multiplicity, x Hs) | q |
|---|---|---|---|---|---|
| glycolysis/gluconeogenesis | creatinine metabolism | ||||
| lactic acid | 1.33 (d, 3) | 15.7 | creatine | 3.04 (s, 3) | 0.8 |
| acetic acid | 1.93 (s, 3) | 14.8 | acetylcarnitine | 3.20 (s, 9) | 14.7 |
| β-glucose | 4.63 (d, 1) | 9.0 | creatinine | 4.07 (s, 2) | 1000 |
| α-glucose | 5.20 (d, 1) | 3.4 | gut microbial–host cometabolism | ||
| pyruvate metabolism | phenylacetic acid | 3.54 (s, 1) | 12.7 | ||
| formic acid | 8.46 (s, 1) | 19.1 | 4-cresol-sulfate | 7.15 (d, 2) | 16.6 |
| pyruvic acid | 2.35 (s, 3) | 24.4 | 4-hydroxyphenylacetic acid | 7.18 (d, 2) | 19.0 |
| citrate cycle (TCA cycle) | indoxyl-sulfate | 7.5 (dt, 1) | 58.2 | ||
| succinic acid | 2.38 (s, 4) | 34.0 | 4-hydroxyhippuric acid | 7.75 (d, 2) | 7.6 |
| citric acid | 2.70 (d, 2) | 110.6 | hippuric acid | 7.83 (d, 2) | 1106.9 |
| fumaric acid | n.d. | benzoic acid | n.d. | ||
| cis-aconitic acid | n.d. | indole-3-acetic acid | 7.250 (t, 1) | 39.88 | |
| 2-oxoglutaric acid | n.d. | 3-(3-hydroxyphenyl)propionic acid | n.d. | ||
| pyrimidine metabolism | phenylacetylglycine | 7.36 (m, 2) | 63.61 | ||
| 3-aminoisobutyric acid | 1.20 (d, 3) | 2.1 | butanoate metabolism | ||
| methylguanidine | 2.83 (s, 3) | 7.5 | butyric acid | n.d. | |
| pseudouridine | 4.30 (t, 1) | 38.6 | 3-hydroxybutyric acid | n.d. | |
| orotic acid | n.d. | acetoacetic acid | n.d. | ||
| uracil | n.d. | acetone | 2.24 (s, 6) | 6.07 | |
| urea | adb | methane metabolism | |||
| malonic acid | a | dimethylamine | 2.73 (d, 6) | 36.0 | |
| amino acid metabolism | trimethylamine | 2.92 (t, 9) | 0.3 | ||
| β-hydroxyisovaleric acid | 1.28 (s, 6) | 10.6 | trimethylamine oxide | 3.27 (s, 9) | 75.4 |
| threonine | 1.35 (d, 3) | 10.6 | methanol | n.d. | |
| alanine | 1.45 (d, 3) | 14.5 | bile metabolism | ||
| N,N-dimethylglycine | 2.93 (s, 6) | 6.6 | taurine | 3.43 (t, 2) | 27.1 |
| glycine | 3.56 (s, 2) | 90.4 | carnitine | n.d. | |
| betaine | 3.89 (s, 2) | 5.7 | choline | n.d. | |
| arginine | n.d. | phosphocholine | adb | ||
| aspartic acid | n.d. | nicotinate and nicotinamide metabolism | |||
| glutamic acid | n.d. | trigonelline | 8.85 (d, 2) | 46.9 | |
| glutamine | n.d. | methylnicotinamide | n.d. | ||
| histidine | n.d. | caffeine metabolism | |||
| isoleucine | n.d. | theobromine | 7.92 (s, 1) | 11.1 | |
| leucine | n.d. | 7-methylxanthine | n.d. | ||
| lysine | n.d. | xanthine | n.d. | ||
| tryptophan | n.d. | hypoxanthine | 8.22 (s, 1) | 4.41 | |
| tyrosine | n.d. | glycerophospholipid metabolism | |||
| valine | n.d. | ethanolamine | n.d. | ||
| serine | n.d. | glycerophosphocholine | n.d. | ||
| phenylalanine | n.d. | others | |||
| 3-methylhistidine | n.d. | adipic acid | n.d. | ||
| anserine | n.d. | allantoin | n.d. | ||
| dimethylglycine | n.d. | ascorbic acid | n.d. | ||
| pyroglutamic acid | n.d. | fucose | a | ||
| 2-hydroxyisobutyric acid | n.d. | glycolic acid | n.d. | ||
| N-acetylglutamic acid | n.d. | methylguanidine | n.d. | ||
| acetylglycine | n.d. | guanidinoacetic acid | n.d. | ||
| mannitol | n.d. | ||||
Only the metabolite resonance used for quantitation purposes is indicated.
δH, 1H chemical shift used for the quantification, in ppm; multiplicity, multiplicity of the resonance used for the quantification; x Hs, number of protons associated with resonance; q, relative quantification towards creatinine, in mmol/mol of creatinine; s, singlet; d, doublet; t, triplet; dt, double triplet; n.d., not detected; adb, absent from our internal database.
Metabolite Quantitation from 1H NMR Urine Profiles
NMR offers the opportunity of quantifying metabolites directly from 1H NMR metabolite profiles. Metabolite quantifiability was tested for the established FIA-NMR method using a standard addition and a calibration curve method for four chemically different compounds: a sugar ((±)-glucose), an aromatic carboxylic acid (hippuric acid), an organic acid (succinic acid), and an amino acid (alanine). For all compounds, a regression r > 0.99 was obtained using urine as a background, suggesting the adequacy of the FIA-NMR method for quantitation (see Figure 1, Supporting Information). These results are in accordance with previous findings where, in optimum conditions, the linearity response of a regression r > 0.999 was reported for NMR measurements.30 Nevertheless, this quantitation approach has the disadvantage of being labor-intensive, as requiring the preparation and analysis of additional samples proportional to the number of analytes to quantify. Due to the large number of metabolites coexisting in a 1H NMR spectrum profile, the usage of library-based integration tools allows not only the identification but also the quantitation of a series of metabolites directly from spectra. There are different tools that offer this type of approach; however, we have chosen AMIX, as our internal reference database is integrated in this tool. Our quantitation strategy (example of one metabolite, citric acid, in Figure 2D,E) was implemented for 85 previously identified metabolites in urine (Table 1). As an example, this procedure was applied to one spectrum (Figure 2) where metabolite quantitative values were obtained for 36 detected metabolites, Table 1 (taking about half an hour of analysis time, Figure 1A). In the event of obtaining the absolute molarity of creatinine by other means, these relative quantitative values can become absolute. Surely, this strategy evolves initial effort in establishing the knowledge-base, but once implemented, it can be readily applied to specific or a batch of spectra within this study. Extrapolation of this knowledge-base to other urine studies is still to be assessed. Higher metabolite quantitation coverage and enhanced data analysis performance (advanced peak-picking, faster analyses) can be further achieved by including advanced 2D NMR spectra (e.g., 1H–13C at the expense of acquisition time) and computational strategies31−33 or a combination of the two34,35 to face the signal overlap in 1D NMR spectra.
Urine Metabolomics Data Extraction and Analysis
Urine metabolite profiles (1H NOESY-1D spectra) were inspected by spectral overlay for data quality. Spectra with poor water suppression or baseline or failure of acquisition or sample transfer (probe not correctly filled or communication issue) were discarded. Ninety-five percent of the analyzed biological samples were successfully acquired (2879 spectra) and can be used for data modeling and subsequent biological interpretation. In terms of data transformation, normalization to the TSP signal was necessary to correct for slight variations in the sample volume, inherent to FIA-NMR. Multivariate statistics was used for a preliminary inspection of the technical quality of the data, as a whole (taking about 1 h of analysis, Figure 1A). By applying principal component analysis, urine spectra were modeled (Figure 4), and no plate-specific tendencies were identified, putting in evidence the robustness of the method. Some biological outliers can be detected due to the presence of glucose in urine. According to specific hypotheses that might be pertinent to test, dedicated data analysis routines could be implemented using more advanced statistical strategies.
Figure 4.
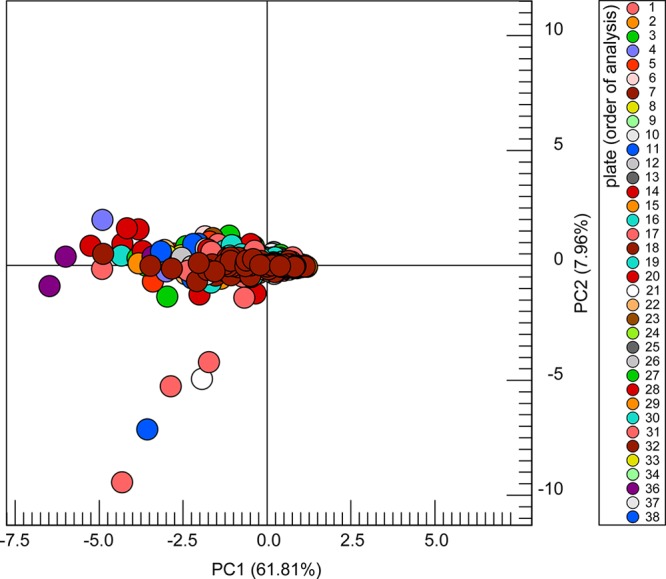
Pareto-scaled scores plot (principal component 1 vs principal component 2) derived from a principal component analysis of 2879 urine spectra (number of rectangular bins used: 287, taken from 12–0.05 ppm), scaled to intensity of the TSP signal, with a confidence level of 99.00%. The five seemingly outliers correspond to samples with a high concentration of glucose, as confirmed by spectral inspection.
Concluding Remarks
The presented FIA-NMR method is well suited for acquiring high-quality 1H NMR metabolomics data, from a large number of subjects, as proven by its performance, stability, and robustness. This method offers the following advantages: suppression of tubes that not only decreases analysis price but also reduces the sample preparation time; increased throughput, as more samples can be continually analyzed, in comparison to regular tube holders; decreased human intervention, given the integration of a sample manager that coordinated all analytical steps and provided data storage in an orderly manner. We therefore recommend this method in laboratories where there is a high demand in standard urine metabolomics analysis. The implementation might not be trivial for FIA beginners, but once the method is implemented, thousands of samples can be robustly analyzed. Concerning the applicability of this method to other biological matrices, aqueous-based samples of comparable viscosity to urine should provide analogous results; in terms of robustness, however, high viscous matrices as plasma or serum might require stricter cleaning routines and should be carefully tested.
Concerning the direct quantitation of metabolites from 1H NMR spectra using a standard compound knowledge-base, even though more sophisticated methods are available, this remains a simple strategy. It does not require additional NMR acquisition time or elaborate computational algorithms, providing quantitative values for a fair amount of metabolites, with minimum effort.
Acknowledgments
S.M. especially thanks Silke Keller, Frank Roth, Ulrich Braumann, Martin Hofmann, and Hartmut Schaefer for their valuable assistance in setting up the FIA-NMR, especially in troubleshooting. L.D. and S.M. acknowledge the help of Guillaume Gesquiere and Emilie Reginato for their contribution in setting up the automated sample preparation method. L.D., F.P.J.M., S.C., and S.M. thank Serge Rezzi and Sunil Kochhar for managerial support. We thank the European Commission for financial support through the FP7 large-scale integrating project “European Study to Establish Biomarkers of Human Ageing” (MARK-AGE; grant agreement no.: 200880).
Supporting Information Available
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
● Laeticia Da Silva, François-Pierre J. Martin, Sebastiano Collino: Molecular Biomarkers, Nestle Institute of Health Sciences, EPFL campus, Innovation square, H, 1015 Lausanne, Switzerland.
Author Present Address
◇ Tilman Grune: Chair of Food Toxicology, University of Jena, 07743 Jena, Germany.
Author Present Address
⊗ Sofia Moco: Natural Bioactives and High Throughput Screening, Nestle Institute of Health Sciences, EPFL campus, Innovation square, H, 1015 Lausanne, Switzerland.
Author Contributions
S.C., A.B, M.M.-V., J.B., O.T., B.G.-L., E.S.G, E.S., T.G., N.B., C.F., and A.H. provided the urine samples used in this study. Set-up of the automated sample preparation method and sample preparation was done by L.D. NMR spectral acquisition was done by L.D. and S.M. Development of the experimental setup, quality control, FIA-NMR method development, NMR acquisition parameters, cleaning procedures, troubleshooting, data transfer, data preprocessing, and quantification strategies was done by S.M., M.G., and M.S. Writing and critical review of the manuscript was done by F.-P.J.M., M.G., and S.M..
The authors declare no competing financial interests.
Supplementary Material
References
- Berger D. Med. Lab. Obs. 1999, 31, 28–40. [PubMed] [Google Scholar]
- Wishart D. S.; Knox C.; Guo A. C.; Eisner R.; Young N.; Gautam B.; Hau D. D.; Psychogios N.; Dong E.; Bouatra S.; Mandal R.; Sinelnikov I.; Xia J.; Jia L.; Cruz J. A.; Lim E.; Sobsey C. A.; Shrivastava S.; Huang P.; Liu P.; Fang L.; Peng J.; Fradette R.; Cheng D.; Tzur D.; Clements M.; Lewis A.; De Souza A.; Zuniga A.; Dawe M.; Xiong Y.; Clive D.; Greiner R.; Nazyrova A.; Shaykhutdinov R.; Li L.; Vogel H. J.; Forsythe I. Nucleic Acids Res. 2009, 37, D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Klipfell E.; Bennett B. J.; Koeth R.; Levison B. S.; Dugar B.; Feldstein A. E.; Britt E. B.; Fu X.; Chung Y. M.; Wu Y.; Schauer P.; Smith J. D.; Allayee H.; Tang W. H.; DiDonato J. A.; Lusis A. J.; Hazen S. L. Nature 2011, 472, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J.; Li Y.; Cai Z.; Li S.; Zhu J.; Zhang F.; Liang S.; Zhang W.; Guan Y.; Shen D.; Peng Y.; Zhang D.; Jie Z.; Wu W.; Qin Y.; Xue W.; Li J.; Han L.; Lu D.; Wu P.; Dai Y.; Sun X.; Li Z.; Tang A.; Zhong S.; Li X.; Chen W.; Xu R.; Wang M.; Feng Q.; Gong M.; Yu J.; Zhang Y.; Zhang M.; Hansen T.; Sanchez G.; Raes J.; Falony G.; Okuda S.; Almeida M.; LeChatelier E.; Renault P.; Pons N.; Batto J. M.; Zhang Z.; Chen H.; Yang R.; Zheng W.; Yang H.; Wang J.; Ehrlich S. D.; Nielsen R.; Pedersen O.; Kristiansen K. Nature 2012, 490, 55–60. [DOI] [PubMed] [Google Scholar]
- Henao-Mejia J.; Elinav E.; Jin C.; Hao L.; Mehal W. Z.; Strowig T.; Thaiss C. A.; Kau A. L.; Eisenbarth S. C.; Jurczak M. J.; Camporez J. P.; Shulman G. I.; Gordon J. I.; Hoffman H. M.; Flavell R. A. Nature 2012, 482, 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J. F.; Dinan T. G. Nat. Rev. Neurosci. 2012, 13, 701–712. [DOI] [PubMed] [Google Scholar]
- Arthur J. C.; Perez-Chanona E.; Muhlbauer M.; Tomkovich S.; Uronis J. M.; Fan T. J.; Campbell B. J.; Abujamel T.; Dogan B.; Rogers A. B.; Rhodes J. M.; Stintzi A.; Simpson K. W.; Hansen J. J.; Keku T. O.; Fodor A. A.; Jobin C. Science 2012, 338, 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exarchou V.; Krucker M.; van Beek T. A.; Vervoort J.; Gerothanassis I. P.; Albert K. Magn. Reson. Chem. 2005, 43, 681–687. [DOI] [PubMed] [Google Scholar]
- Braumann U.; Spraul M. In On-Line LC-NMR And Related Techniques; Albert K., Ed.; John Wiley & Sons, Ltd: Chichester, U.K., 2003; pp 23–43. [Google Scholar]
- Moco S.; Vervoort J. In Plant Metabolomics: Methods and Protocols; Hardy N. W., Hall R. D., Eds.; Springer-Verlag: New York, 2012; pp 287–316. [Google Scholar]
- Keifer P. A.; Smallcombe S. H.; Williams E. H.; Salomon K. E.; Mendez G.; Belletire J. L.; Moore C. D. J. Comb. Chem. 2000, 2, 151–171. [DOI] [PubMed] [Google Scholar]
- Spraul M.; Hofmann M.; Ackermann M.; Shockcor J. P.; C. Lindon J.; Nicholls A. W.; Nicholson J. K.; Damment S. J. P.; Haselden J. N. Anal. Commun. 1997, 34, 339–341. [Google Scholar]
- Teng Q.; Ekman D. R.; Huang W.; Collette T. W. Analyst 2012, 137, 2226–2232. [DOI] [PubMed] [Google Scholar]
- Sukumaran D. K.; Garcia E.; Hua J.; Tabaczynski W.; Odunsi K.; Andrews C.; Szyperski T. Magn. Reson. Chem. 2009, 47, S81–S85. [DOI] [PubMed] [Google Scholar]
- Bharti S. K.; Roy R. TrAC, Trends Anal. Chem. 2012, 35, 5–26. [Google Scholar]
- Barding G. A. Jr.; Salditos R.; Larive C. K. Anal. Bioanal. Chem. 2012, 404, 1165–1179. [DOI] [PubMed] [Google Scholar]
- Lacey M. E.; Subramanian R.; Olson D. L.; Webb A. G.; Sweedler J. V. Chem. Rev. 1999, 99, 3133–3152. [DOI] [PubMed] [Google Scholar]
- Akoka S.; Barantin L.; Trierweiler M. Anal. Chem. 1999, 71, 2554–2557. [DOI] [PubMed] [Google Scholar]
- Cui Q.; Lewis I. A.; Hegeman A. D.; Anderson M. E.; Li J.; Schulte C. F.; Westler W. M.; Eghbalnia H. R.; Sussman M. R.; Markley J. L. Nat. Biotechnol. 2008, 26, 162–164. [DOI] [PubMed] [Google Scholar]
- Ulrich E. L.; Akutsu H.; Doreleijers J. F.; Harano Y.; Ioannidis Y. E.; Lin J.; Livny M.; Mading S.; Maziuk D.; Miller Z.; Nakatani E.; Schulte C. F.; Tolmie D. E.; Kent Wenger R.; Yao H.; Markley J. L. Nucleic Acids Res. 2008, 36, D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C.; Easton J.; Lodi A.; Tiziani S.; Manzoor S.; Southam A.; Byrne J.; Bishop L.; He S.; Arvanitis T.; Günther U.; Viant M. Metabolomics 2012, 8, 8–18. [Google Scholar]
- Steinbeck C.; Krause S.; Kuhn S. J. Chem. Inf. Comput. Sci. 2003, 43, 1733–1739. [DOI] [PubMed] [Google Scholar]
- Akiyama K.; Chikayama E.; Yuasa H.; Shimada Y.; Tohge T.; Shinozaki K.; Hirai M. Y.; Sakurai T.; Kikuchi J.; Saito K. In Silico Biol. 2008, 8, 339–345. [PubMed] [Google Scholar]
- Jacobs D. M.; Spiesser L.; Garnier M.; de Roo N.; van Dorsten F.; Hollebrands B.; van Velzen E.; Draijer R.; van Duynhoven J. Anal. Bioanal. Chem. 2012, 404, 2349–2361. [DOI] [PubMed] [Google Scholar]
- Wishart D. S. TrAC, Trends Anal. Chem. 2008, 27, 228–237. [Google Scholar]
- Bales J. R.; Higham D. P.; Howe I.; Nicholson J. K.; Sadler P. J. Clin. Chem. 1984, 30, 426–432. [PubMed] [Google Scholar]
- Yang W.; Wang Y.; Zhou Q.; Tang H. Sci. China, Ser. B: Chem. 2008, 51, 218–225. [Google Scholar]
- Pathmasiri W.; Pratt K.; Collier D.; Lutes L.; McRitchie S.; Sumner S. C. J. Metabolomics 2012, 8, 1037–1051. [Google Scholar]
- Lauridsen M.; Hansen S. H.; Jaroszewski J. W.; Cornett C. Anal. Chem. 2007, 79, 1181–1186. [DOI] [PubMed] [Google Scholar]
- Malz F. In NMR Spectroscopy in Pharmaceutical Analysis; Holzgrabe U., Wawer I., Diehl B., Eds.; Elsevier: Amsterdam, The Netherlands, 2008; pp 43–62. [Google Scholar]
- Aranibar N.; Borys M.; Mackin N. a.; Ly V.; Abu-Absi N.; Abu-Absi S.; Niemitz M.; Schilling B.; Li Z. J.; Brock B.; Russell R. J.; Tymiak A.; Reily M. D. J. Biomol. NMR 2011, 49, 195–206. [DOI] [PubMed] [Google Scholar]
- Hao J.; Astle W.; De Iorio M.; Ebbels T. M. Bioinformatics 2012, 28, 2088–2090. [DOI] [PubMed] [Google Scholar]
- Zheng C.; Zhang S.; Ragg S.; Raftery D.; Vitek O. Bioinformatics 2011, 27, 1637–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylla R. A.; Hu K.; Ellinger J. J.; Markley J. L. Anal. Chem. 2011, 83, 4871–4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette S. L.; Ajredini R.; Rasheed H.; Zeinomar A.; Schroeder F. C.; Dossey A. T.; Edison A. S. Anal. Chem. 2011, 83, 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.