Summary
The transcription factor IRF4 regulates immunoglobulin class switch recombination and plasma cell differentiation. Its differing concentrations appear to regulate mutually antagonistic programs of B and plasma cell gene expression. We show IRF4 to be also required for generation of germinal center (GC) B cells. Its transient expression in vivo induced the expression of key GC genes including Bcl6 and Aicda. In contrast, sustained and higher concentrations of IRF4 promoted the generation of plasma cells while antagonizing the GC fate. IRF4 co-bound with the transcription factors PU.1 or BATF to Ets or AP-1 composite motifs, associated with genes involved in B cell activation and the GC response. At higher concentrations IRF4 binding shifted to interferon sequence response motifs; these enriched for genes involved in plasma cell differentiation. Our results support a model of “kinetic control” in which signaling induced dynamics of IRF4 in activated B cells control their cell fate outcomes.
Introduction
Germinal Center (GC) B cells and plasma cells (PC) develop following the activation of naïve B cells with cognate antigen in combination with signals from T helper cells and dendritic cells (Goodnow et al., 2010). These distinct cellular states, GC and PC, perform key roles in humoral immunity against microbes by enabling generation of high affinity antibodies and their robust expression and secretion, respectively. Considerable progress has been achieved in the analysis of transcription factors that are required for the generation of GC B cells and their plasma cell counterparts. However the molecular mechanisms by which such regulators orchestrate these alternative cellular states and the transition from the GC to the PC differentiation programs are incompletely understood.
The identity and function of plasma cells is dependent on the transcription factors Blimp1, Xbp1 and IRF4 (Nutt et al., 2011). In contrast, GC B cell development requires the transcription factors Bcl6, Pax5, Bach2 and Obf1 (Nutt et al., 2011). Blimp1 and Bcl6 function to counter regulate each other's expression. This reciprocal negative feedback is considered to play a major role in stabilizing the alternate programs of gene expression.
We have proposed that the transcription factor IRF4 is a pivotal regulator of B cell fate dynamics upon antigen encounter (Sciammas et al., 2011; Sciammas et al., 2006). This is based on our findings that IRF4 is required for class switch recombination (CSR) and plasma cell differentiation. It does so by upregulating AID and Blimp1 expression, respectively. We have demonstrated, using a variety of approaches, that differing IRF4 concentrations underlie the generation of these alternative cell states. These experimental analyses have led to the formulation of a “kinetic control” model for the regulation of B cell fate dynamics spanning the CSR and plasma cell states (Sciammas et al., 2011) (see also (Muto et al., 2010). According to this model, the rate of accumulation of IRF4 induced by the BCR determines the duration for which such a cell expresses AID and therefore can undergo CSR and also somatic hypermutation (SHM). Increased expression of IRF4 beyond a critical threshold results in IRF4 activation of the Prdm1 (encoding Blimp1) locus and terminal differentiation into a plasma cell. This is accompanied by repression of Aicda (encoding AID) expression and CSR as well as SHM. However, it remains to be determined if this regulatory model is applicable to T-dependent B cell responses in vivo. It has been suggested that IRF4 is dispensable for the GC response in vivo (Klein et al., 2006). However, this conclusion was based on the use of a conditional allele of IRF4 whose deletion is initiated after antigen encounter raising the possibility that IRF4 protein was not sufficiently depleted in precursors of GC B cells in these mice. Therefore, we sought to address the role of IRF4 in regulating generation of GC B cells using alternative genetic strategies.
IRF4 is a member of the IRF superfamily of transcription factors most highly related to IRF8 (Eisenbeis et al., 1995). Although IRF8 is expressed in activated and GC B cells, it has been shown to be dispensable for antigen-dependent B cell responses (Feng et al., 2011). IRF4 and -8 bind with much lower affinity to the GAAA motif contained within the canonical interferon sequence response element (ISRE). Instead they are recruited to high affinity Ets-IRF composite motifs (EICE) through their interaction with the transcription factors PU.1 or SpiB (Brass et al., 1999; Eisenbeis et al., 1995). The latter are related Ets family members that play key roles in B cell activation and GC B cell function (Garrett-Sinha et al., 2001; Su et al., 1997). Recently IRF4 and IRF8 have shown to cooperatively assemble with BATF containing AP-1 complexes on composite AP-1-IRF (AICE) motifs (Glasmacher et al., 2012). Intriguingly, IRF4 appears to activate the Prdm1 (Blimp1) locus by binding to a site within a conserved intronic sequence that does not contain an EICE motif nor is associated with PU.1 co-binding (Sciammas et al., 2006). These results raised the possibility that alternate modes of IRF4 genome targeting i.e. PU.1 or SpiB dependent and Ets factor independent may be important in regulating distinct states of gene expression, GC vs. PC, within activated B cells.
Herein, using distinct genetic strategies we demonstrate that IRF4 regulates the generation of GC B cells. It does so by controlling the expression of the Bcl6 and Obf1 genes. Furthermore whereas transient induction of IRF4 in vivo was sufficient to induce GC B cells, sustained and higher concentrations of IRF4 promoted the generation of plasma cells while antagonizing the GC fate. To delineate IRF4 target genes and its modes of genomic interaction that are reflective of the GC or plasma cell programs we performed ChIPseq analysis using an antigen specific B cell culture system. Kinetic analysis of IRF4 binding to genomic sites, with or without its DNA partner PU.1, was correlated with changes in gene expression. Interestingly, IRF4 co-targeting with PU.1 at EICE motifs was associated with genes involved with B cell activation and the GC response. During these early stages of B cell activation, IRF4 targeting was also associated with AICE motifs. In striking contrast at a later stage, reflective of plasma cells, IRF4 targeting shifted to lower affinity ISRE motifs that enriched for genes involved in plasma cell differentiation. These results provide molecular insight into the concentration dependent modes of IRF4 action in regulating the GC and PC programs of gene expression. Furthermore, they provide in vivo support for our model of “kinetic control” which posits that the dynamics of accumulation of IRF4 in activated B cells regulates cell fate outcomes during a humoral immune response.
Results
IRF4 regulates GC B cell differentiation
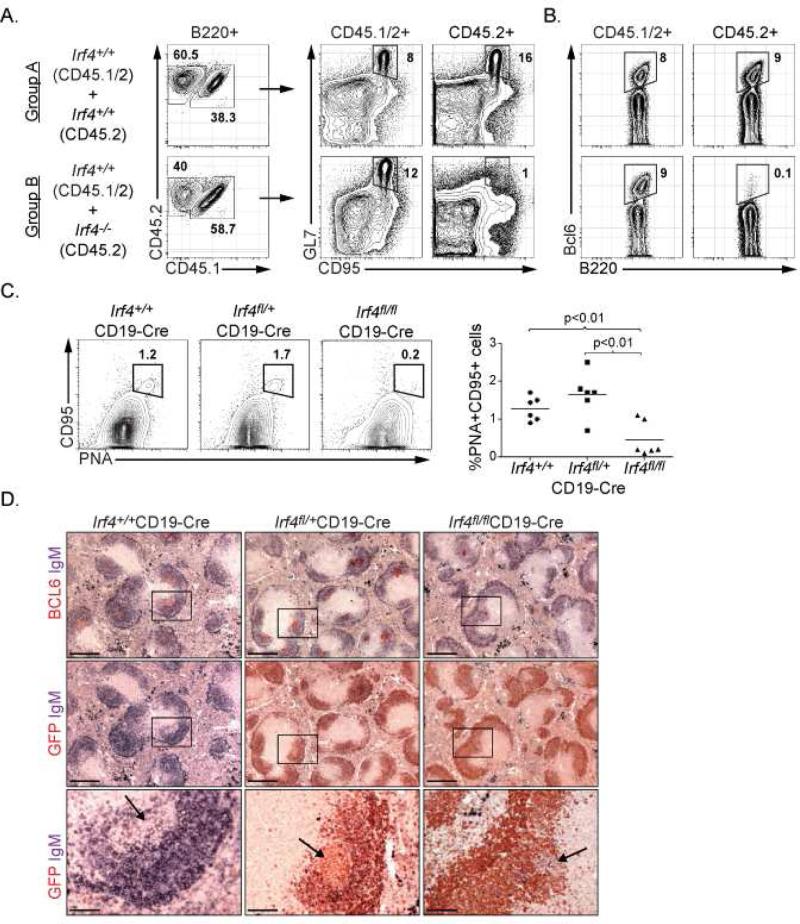
To analyze requirement of IRF4 in GC B cell responses, we generated mixed bone marrow chimeras with Irf4+/+ and Irf4-/- progenitors (Fig. S1A). Following hematopoietic reconstitution, the animals were immunized with sheep red blood cells (SRBC) to elicit a T-dependent GC B cell response. While the wild type (CD45.1+CD45.2+) B220+ compartment contained CD95+GL7+ GC B cells, the Irf4-/- (CD45.2+) B220+ compartment lacked such cells (Fig. 1A). Accordingly, Bcl6 expressing cells were not generated within the Irf4-/- population (Fig. 1B). The defect in GC B cell formation must be intrinsic to Irf4-/- B cells as the hematopoietic compartment in these chimeric animals contains wild type T and dendritic cells. It has been suggested that B cells from Irf4-/- mice are developmentally immature based on expression of CD23 and IgM (Mittrucker et al., 1997). To exclude the possibility that the severe block in GC B cell differentiation was simply due to a developmental arrest at an immature stage, we analyzed Irf4-/- B cells for expression of CD93, a marker of immature and transitional B cells (Allman et al., 2001). CD93 expression on splenic B cells from Irf4+/- and Irf4-/- mice was indistinguishable (Fig. S1B). Moreover, this analysis revealed the basis for the skewed distributions of CD23 and IgM expression in Irf4-/- mice to be likely due to an increase in the proportions of marginal zone B cells (Fig. S1C and D). Thus the defect in GC B cell differentiation caused by loss of IRF4 is not due to a developmental arrest at an immature B cell stage.
Figure 1.
see also Figure S1. IRF4 regulates GC B cell differentiation. (A, B) 1:1 mixed bone marrow chimeras were generated such that the CD45.1 expressing compartments in Groups A and B were reconstituted with Irf4+/+ hematopoietic progenitors whereas the CD45.2 expressing compartments were reconstituted with Irf4+/+ and Irf4-/- hematopoietic progenitors, respectively. Reconstituted mice were immunized with SRBC and GC B cells were analyzed on Day 7 based on expression of GL7 and CD95 or intracellular Bcl6 expression after gating on CD45 polymorphic alleles and the B cell lineage marker B220 as indicated. Data are representative of two independent experiments using 5 mice per group. (C) Conditional deletion of Irf4 in B cells using CD19-Cre. Indicated mice were immunized with SRBC and splenic GC B cells were analyzed on Day 14 based on expression of PNA and CD95 after gating on B220. Each point in the right panel represents the numbers of GC B cells from individual mice. (D) Immunohistochemical analysis of GCs in mice described in (C). Splenic sections were stained for Bcl6, IgM and GFP as indicated.
Previously we suggested that IRF4 was dispensable for GC B cell differentiation based on analysis of Irf4fl/fl mice using a Cγ1-Cre driver that deletes after antigen stimulation of B cells (Klein et al., 2006). The analysis did not rule out the possibility that in this mouse model, the timing of Irf4 deletion and/or stability of the residual IRF4 protein may have obscured its role in GC B cell differentiation. To test this possibility, we crossed Irf4fl/fl mice with CD19-Cre mice so that deletion of the Irf4 gene occurred prior to antigen encounter. Importantly, earlier deletion of Irf4 in B cells resulted in an impaired GC response (Fig. 1C and D). The conditional Irf4 allele activates GFP expression concomitant with CRE-mediated deletion (Klein et al., 2006). Staining of splenic tissue sections revealed that the few GCs developing in Irf4fl/fl x CD19-Cre mice were GFP-negative, in contrast with their controls, demonstrating that these residual GCs were formed with B cells in which the Irf4 allele had not been deleted (Fig. 1D). Thus, IRF4 plays an essential and cell autonomous role in instructing GC B cell differentiation.
IRF4 regulates Bcl6 and Pou2af1 during a GC B cell response
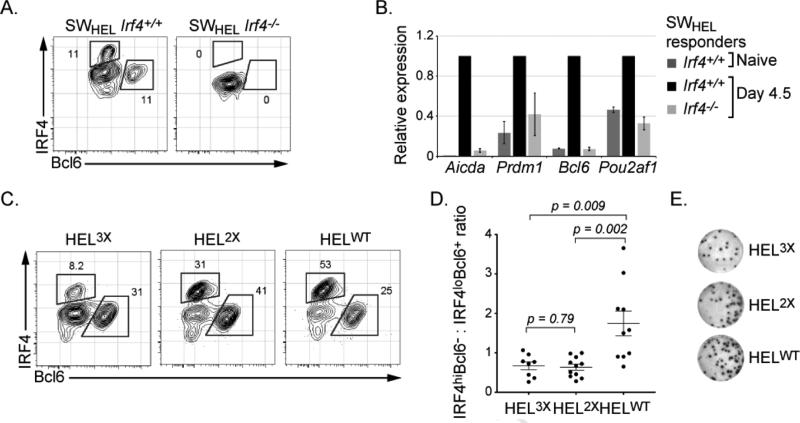
To determine if IRF4 regulates Bcl6 and Obf1 expression within antigen responding B cells in vivo, we bred the Irf4-/- mouse to the SWHEL mouse in which the B cells are specific for the Hen Egg Lysozyme (HEL) antigen (Phan et al., 2005). SWHEL B cells from wild type and Irf4-/- mice were adoptively transferred into CD45.1 mice, which were then immunized with the intermediate affinity HEL2X coupled to SRBC (Fig. S2A). Analysis of IRF4 and Bcl6 expression in wild type SWHEL responder cells, 4.5 days later, revealed two subsets: IRF4loBcl6+ and IRF4hiBcl6- (Fig. 2A and S2B). As IRF4 is highly expressed in plasma cells, we confirmed that IRF4hiBcl6- population represented plasma cells by their expression of cytoplasmic anti-HEL Ig and reduced B220 (Fig. S2C). In contrast, the IRF4loBcl6+ population represented GC B cells based on high expression of Bcl6 and B220. Importantly, Irf4-/- SWHEL cells did not generate either Bcl6 expressing cells or plasmablasts. Notably, Irf4-/- SWHEL B cells responded appropriately to antigen and engaged T cell help as they underwent multiple cell divisions, albeit with reduced efficiency (Fig. S2D) (Phan et al., 2005). Thus, IRF4 plays an essential role in the generation of Bcl6 expressing cells in the context of antigen signaling and cognate T cell interactions in vivo. Furthermore, these results demonstrate that antigen encounter leads to the generation of distinct B cell states that can be discriminated on the basis of IRF4 and Bcl6 expression in vivo, which reflect mutual antagonism between the GC B cell and plasma cell programs of gene expression.
Figure 2.
see also Figure S2. IRF4 regulates GC B cell differentiation via the activation of the Bcl6 and Pou2af1 genes. Irf4+/+ or Irf4-/- SWHEL donor B cells were transplanted into CD45.1 hosts and immunized with HEL2XSRBC. (A) 4.5 days after immunization, donor derived antigen specific cells were identified based on B220+CD45.2+CD45.1- phenotype and binding to HEL antigen. Expression of IRF4 and Bcl6 expression was then analyzed by intracellular staining. (B) Cells described in (A) were sorted and RNA was analyzed by qRT-PCR. Indicated transcripts were normalized to those from Oct1 gene, the data represents the average ±SEM of three independent experiments with two mice per group. (C) Wild type SWHEL donor B cells were adoptively transferred into CD45.1 hosts and immunized with indicated HEL variants conjugated to SRBC. 4.5 days after immunization, donor derived antigen specific cells were identified and analyzed as in (A). (D) Quantitative analysis of experiments described in (C). The ratio of HEL-specific IRF4hiBcl6- to IRF4loBcl6+ expressing cells for individual mice is plotted from 3 independent experiments. (E) ELISpot analysis of HEL-specific IgG secreting PC cells from experiments in (C), representative results are shown, see Fig. S2 for quantitation.
We then tested if IRF4 was required for transcriptional activation of the Bcl6 and Pou2af1 genes. SWHEL responder cells were isolated 4.5 days following immunization (Fig. S2D) and their transcripts analyzed by qRT-PCR. Importantly expression of Bcl6 and Pou2af1 (encoding Obf1) were severely compromised in Irf4-/- SWHEL B cells compared to their wild type counterparts (Fig. 2B). As expected, expression of the Aicda and Prdm1 genes were also impaired (Klein et al., 2006; Sciammas et al., 2006). We note that Pax5 transcripts were comparable between Irf4-/- SWHEL B cells and their wild type counterparts (Fig. S2E). Thus, the Bcl6 and Pou2af1 genes, which regulate GC B cell differentiation, are dependent on IRF4 for their induced expression in B cells upon antigen encounter in vivo.
Increased antigen affinity favors generation of IRF4hi plasma cells
Previously we have shown that increased antigen affinity augments BCR signaling mediated expression of IRF4 and thus favors the generation of plasma cells at the expense of cells undergoing CSR in vitro (Sciammas et al., 2011). Given ability to analyze B cell dynamics in vivo on the basis of IRF4 and Bcl6 expression we tested if varying antigen affinity in vivo had the predicted consequences on GC and plasma cell states. After adoptive transfer of SWHEL cells we immunized with a series of HEL variants that exhibit a 10,000 fold range in affinity for the HyHEL10 BCR (Paus et al., 2006). The highest affinity antigen led to an increased proportion of IRF4hiBcl6- plasma cells (Fig. 2C, 2D, and S2F). The enhancement in plasma cell generation was confirmed by HEL-specific ELISpot analysis (Fig. 2E, S2G). Thus both in vitro as well as in vivo increasing the intensity of signaling through the BCR leads to greater expression of IRF4 and favors the generation of plasma cells.
Transient expression of IRF4 induces generation of Bcl6 expressing GC B cells
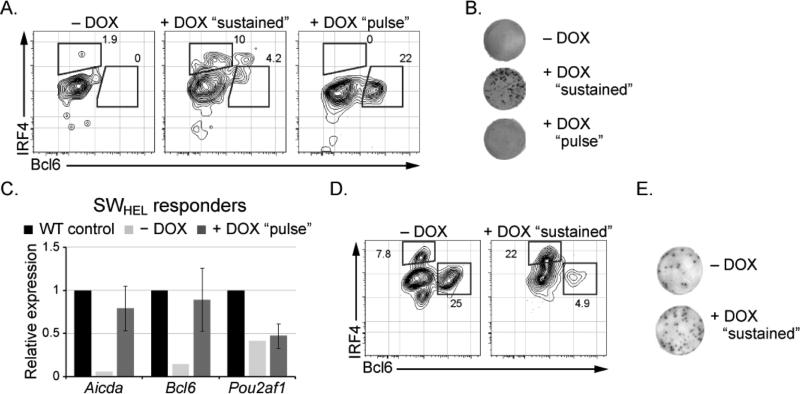
To directly test consequences of manipulating IRF4 concentration on B cell fate dynamics in vivo we utilized a tet-inducible allele (Sciammas et al., 2011) with the SWHEL transgenic system. This transgene is engineered to express IRF4 in a tet-responsive manner via the transcriptional activator (M2rtTA). The tet-inducible Irf4 and SWHEL transgenes were crossed onto the Irf4-/- background so that the former functioned as the sole source of IRF4 protein in vivo (Fig. 3A). For the experiments using Irf4-inducible SWHEL B cells, the CD45.1 host mice were crossed with the M2rtTA allele to prevent rejection of transplanted cells due to the neo-antigen effects of the bacterial transactivator protein. Following adoptive transfer of B cells into CD45.1+Rosa+/M2rtTA congenic hosts and immunization with intermediate affinity HEL2X, the mice were administered doxycycline (DOX) either continuously (“sustained”) or just during the first two days after immunization (“pulsed”). Sustained induction of the tet-inducible Irf4 transgene led to the generation of IRF4hiBcl6- and Ig secreting plasma cells (Figs. 3A, 3B and S3D). We note that rescued B cells secreted HEL-specific IgG demonstrating that sustained expression of IRF4 restored both CSR and secretory function (Fig. 3B and S3D). Importantly, transgenic expression of IRF4 also rescued the generation of Bcl6+ GC B cells (Fig. 3A, S3B and S3C). Strikingly, “pulsed” induction of IRF4 led only to the emergence of Bcl6 expressing GC B cells (Fig. 3A and S3B, S3C) that also expressed Aicda but not Pou2af1 transcripts (Fig. 3C). Importantly, plasma cells did not develop under these conditions (Fig. 3A, 3B and S3D). We note that the HEL-specific IgM spots observed with Irf4-/- cells most likely emanate from host derived B cells as they are also seen in mice immunized with mock-conjugated SRBC (data not shown). Thus, following antigen encounter, a transient burst of IRF4 expression appears sufficient to enable the generation of a stable population of GC B cells that express Bcl6 and AID.
Figure 3.
see also Figure S3. Inducible expression of Irf4 regulates B cell fate dynamics (A) Irf4-inducible (Irf4-/-) SWHEL donor B cells transplanted into CD45.1RosaM2rtTA/+ hosts and immunized with HEL2XSRBC. Mice were administered water lacking DOX (- DOX), containing DOX throughout the 5 day experiment (+ DOX “sustained”) or containing DOX for the first two days only (+ DOX “pulse”). 5 days after immunization, donor derived antigen specific cells were identified and analyzed as in Fig. 2A. (B) Representative HEL-specific IgG ELISpot analysis from experiments in (A). (C) Cells described in (A) were sorted and RNA was analyzed as in Fig. 2B, the data represents the average ±SEM of three individual mice. (D) Irf4-inducible (Irf4+/+) HEL-specific SWHEL donor B cells transplanted into CD45.1, RosaM2rtTA/+ hosts and immunized with HEL3XSRBC. Mice were administered DOX as indicated in (A). 5 days after immunization, donor derived antigen specific cells were identified and analyzed as in Fig. 2A. (E) Representative HEL-specific IgG ELISpot analysis from experiments in (D). Representative results from two independent experiments are shown, see Fig. S3 for quantitation.
Next, we tested if increased expression of IRF4 in wild type B cells might promote plasma cell differentiation at the expense of GC B cells. To do so, we adoptively transferred Irf4-inducible SWHEL B cells on the Irf4+/+ background into CD45.1+Rosa+/M2rtTA congenic mice, immunized with HEL3X and administered DOX in the drinking water (Fig. S3E). Remarkably, SWHEL responders in the “sustained” DOX group were impaired in their ability to generate Bcl6+ cells (Fig. 3D, S3B and S3C). DOX-mediated induction of the Irf4 transgene led to an increase in IRF4hi expressing cells and was accompanied by a corresponding increase in HEL-specific plasma cells (Fig. 3E and S3H). Interestingly, the increase in plasma cells was predominantly observed in the IgM class of HEL-specific cells (Fig. S3H), as predicted by our model in which high IRF4 concentrations prevent durable AID expression. Thus transient induction of IRF4 is sufficient to induce the GC program. In contrast sustained and higher concentrations of IRF4 terminate the GC program while promoting the generation of plasma cells.
Genomic targeting analysis of IRF4 and PU.1 in an antigen-dependent differentiation system
To gain insight into IRF4 regulation of distinct programs of B cell gene expression we performed ChIPseq analyses in an antigen specific in vitro system that results in CSR and efficient plasma cell differentiation (Fig. S4A and B) (Sciammas et al., 2011). IRF4 expression is induced under these conditions with BCR engagement and exhibits a wide range of cellular concentrations at Day 1 (Fig. S4C). By Day 3, a bimodal pattern of IRF4 expression is observable; the cells expressing low or high concentrations of IRF4, which correspond to those undergoing CSR or differentiating into plasma cells, respectively (Sciammas et al., 2011; Sciammas et al., 2006). We reasoned that kinetic analysis of the genome binding landscape of IRF4 in this cellular system might reveal a relationship between its differing concentrations and the regulation of distinct programs of gene expression. We note that, these conditions do not promote the generation of GC B cells; however, given that AID is expressed and functions to promote SHM in these cells (Sciammas et al., 2011), we hypothesized that some molecular features of GC B cell differentiation would be manifested in cells expressing the lower amounts of IRF4.
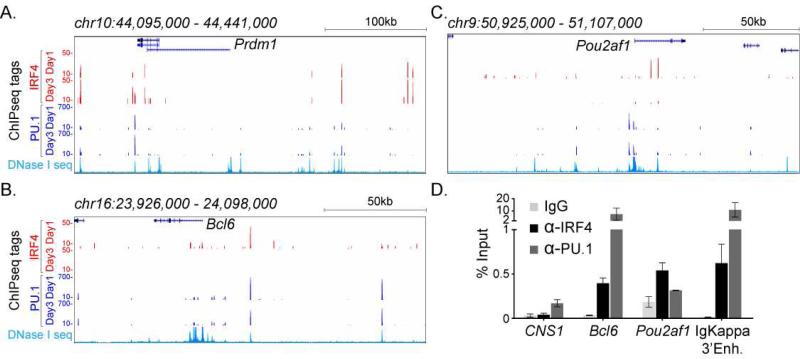
To analyze distinct modes of IRF4 genome targeting, we performed parallel ChIPseq analyses with its major interaction partner, PU.1 (Eisenbeis et al., 1995). This comparison enabled us to identify genomic regions that were targeted by IRF4 in conjunction with or in the absence of PU.1 (Fig. S4A and D). Table S1 reports the details of sample processing with regard to sequencing, alignment and peak calls. The overall distribution of genomic sites of these two transcription factors is shown in Fig. S4E and revealed that the majority of binding events were extragenic and occurred within 100 kbp of the nearest TSS. A small number of binding peaks were randomly chosen and validated by ChIP (Fig. S4F).
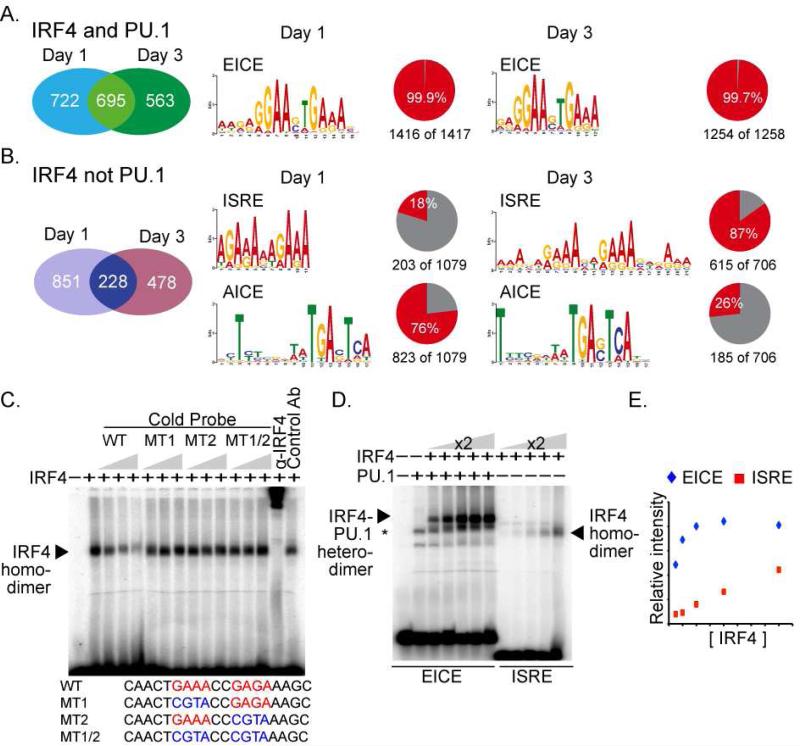
To determine the dynamics of IRF4 and PU.1 binding, the extent of coincident peaks between the Day 1 and Day 3 datasets were compiled (Fig. S4D). Temporally specific binding events were observed for both IRF4 and PU.1, suggesting that a shift in the genome binding landscape is associated with the bimodal expression of IRF4. Next, we determined the extent to which IRF4 targets the genome with or without PU.1 (Fig. 4A and B). IRF4 co-bound with PU.1 at a majority of the genomic sites. A third of IRF4 binding events were not associated with PU.1; this mode of IRF4 genome targeting is denoted IRF4 (not PU.1). Comparison of our data with DNaseI seq analysis in naive B cells (ENCODE data, Fig. S4G) revealed ~90% of IRF4 (and PU.1) co-targeted regions (Day 1 and 3) to be contained within DNaseI hypersensitive sites in naive B cells demonstrating that EICE motifs are located in accessible chromatin. In contrast, IRF4 (not PU.1) regions overlapped with ≤50% of the DNaseI sites present in naive B cells suggesting that this mode of IRF4 genomic targeting involved the de novo establishment of accessible regions.
Figure 4.
see also Figure S4. Analysis of IRF4 cistrome in antigen activated B cells reveals three distinct motifs and binding modes. B cells from B1-8i anti-NP knock-in mice were stimulated with NP-Ficoll, CD40L, and IL-2,4 and 5. On Days 1 and 3, chromatin was crosslinked and processed for IRF4 and PU.1 specific ChIP coupled to massively parallel sequencing. Left panels display the union analysis of the number of IRF4 (and PU.1) or IRF4 (not PU.1) binding peaks, respectively, at Day 1 or Day 3 after B cell stimulation. Right panels display over represented sequence motifs revealed by MEME in Logo format. The associated pie charts indicate the number of regions analyzed and the frequency with which the motif is found. EICE, ISRE, and AICE represent the Ets-IRF composite element, Interferon Stimulated Response Element, and AP-1-IRF composite element, respectively. (C) IRF4 binds with lower affinity to ISRE motifs than to EICE motifs. EMSA with the Blimp1 CNS9 ISRE sequence as probe. All binding reactions contained a wild type probe and nuclear extract from IRF4 expressing 293T cells. Increasing amounts of competitor DNAs, wild type or mutant ISREs, were included as indicated. Anti-IRF4 or control antibodies were used in supershift assays to confirm identity of the IRF4 complex. (D) Binding saturation curves of IRF4 to EICE or ISRE motifs. Binding reactions using the EICE DNA probe derived from the Ig Kappa 3’ enhancer, were carried out in the presence of a constant amount of PU.1 protein. IRF4 protein was increased in 2-fold increments as indicated. The ISRE probe and binding reactions were conducted as in panel (C). Positions of relevant protein-DNA complexes are indicated by arrows. (E) Densitometry analysis of (D). Data in panels C, D is representative of three independent experiments.
Distinct DNA motifs comprise the IRF4 cistrome
We searched for over-represented sequence motifs using the MEME pattern finding algorithm in the IRF4 cistrome (Fig. 4A and 4B). Within the IRF4 (and PU.1) bound regions, the EICE motif occurred with an incidence approaching 100%. This finding demonstrated the fundamental importance of the EICE motif in recruitment of IRF4 by PU.1 to genomic sites in differentiating B cells.
In contrast, within IRF4 (not PU.1) bound regions, two distinct DNA motifs were enriched (Fig. 4B). The first represented the Interferon Stimulated Response Element (ISRE), which is composed of two IRF motifs (GAAA) separated by two basepairs. The second motif was a canonical AP-1 motif that was often found near the peak's summit (Fig. S4I). Inspection of the surrounding sequences identified an IRF motif (GAAA) either abutting or separated by 4 nucleotides from the AP-1 motif (Fig. 4B and S4H) suggesting the presence of an AP-1-IRF composite element (AICE) (Glasmacher et al., 2012). Accordingly, we found that BATF and IRF4 co-bound to a sampling of these AICE motifs in B cells (Fig. S4K). Thus, the IRF4 cistrome in B cells comprises three distinct DNA binding modes characterized by EICE, AICE and ISRE motifs.
Within the IRF4 (not PU.1) peaks, the incidence of AICE and ISRE motifs was inverted between the Day 1 and Day 3 time points; the ISRE predominated at Day 3 (Fig. 4B). This demonstrates that the nature of the IRF4 binding landscape shifts during the process of B cell differentiation and the higher concentration of IRF4 is accompanied by increased occupancy of ISRE motifs.
IRF4 binds the ISRE as a dimer with lower affinity
Given the above finding we analyzed the relative affinity of IRF4 for the ISRE and EICE motifs. An ISRE motif from Prdm1 (Sciammas et al., 2006) was used in electrophoretic mobility shift assays (EMSAs) (Fig. 4C). IRF4 generated a protein-DNA complex (Fig. 4C) that was competed by a wild type oligonucleotide but not ones in which one or both of the IRF sites were mutated. This suggested that IRF4 bound the ISRE as a dimer which was confirmed by analyzing the migration of the protein-DNA complexes formed by mixing two different carboxy-terminal truncations of IRF4 (Fig. S4J). Thus IRF4 binds the ISRE as a homodimer in contrast with its binding to an EICE as a heterodimer with PU.1 or to an AICE as a heterotrimer with a BATF containing AP-1 complex.
To determine the relative affinity of IRF4-PU.1 heterodimeric or IRF4 homodimeric complexes for the EICE vs. ISRE motifs, respectively, we analyzed binding over a wide range of IRF4 concentrations (Fig. 4D, E). Whereas, increasing IRF4 concentration in the presence of PU.1 resulted in saturation of IRF4 binding to the EICE, saturation was not observed for IRF4 binding (in the absence of PU.1) to the ISRE within the concentration range that was tested. These data demonstrated that IRF4 binds with higher affinity to EICE motifs as a hetrodimer with PU.1 than to ISRE motifs as a homodimer and suggest that IRF4 is able to occupy EICE motifs at a lower concentration in vivo. Thus, higher IRF4 concentrations would be needed to drive binding onto ISRE motifs and this is consistent with their increased utilization in differentiating IRF4hi B cells at day 3 (Fig. 4B and S4C).
IRF4 targeting of the Prdm1 locus
Our previous analysis had suggested that IRF4 directly activates Prdm1 transcription to enable plasma cell differentiation. The ChIPseq analysis confirmed that IRF4 bound to the CNS9 region in Prdm1 (Fig. 5A and S5A). In addition, we identified multiple peaks surrounding the Prdm1 gene that increased in intensity at Day 3 (Fig. S5A). As IRF4 is necessary for promoting Blimp1 expression, we reasoned that IRF4 binding to the Prdm1 locus may be required for the deposition of activating H3K4me1 and H3K27Ac chromatin marks. Analysis of wild type B cells showed that these chromatin marks were present at low levels at some of these regions at the Day 0 and Day 1 time points but sharply increased at Day 3 when Prdm1 was maximally expressed (Fig. S5B). In the absence of IRF4, these marks failed to accumulate not only at IRF4 targeted but also at non-targeted regions that included the Prdm1 promoters (Fig. S5C). Overall, this analysis demonstrates an extensive targeting landscape of IRF4 at the Prdm1 locus that includes both IRF4 (and PU.1) as well as IRF4 (not PU.1) binding modes. Importantly IRF4 binding to multiple sites at the Prdm1 locus appears to be required for the acquisition of an activated chromatin state.
Figure 5.
see also Figure S5. IRF4 targets the Prdm1, Bcl6, and Pou2af1 genes. ChIP-seq tag enrichment (y-axis) is displayed as a histogram for the (A) Prdm1 (Blimp1) (B) Bcl6 and (C) Pou2af1 (Obf1) loci at indicated time points for antigen activated B cells described in Fig. 4. The x-axis indicates the genomic interval (build mm9). The lower most histogram in each panel shows genomic accessibility within naive CD19+ wild type B cells, as assessed by DNaseI-seq (data downloaded from ENCODE). (D) IRF4 binding to the Bcl6 and Pou2af1 loci in purified GC B cells. Enrichment values (% input chromatin) with control IgG, IRF4, and PU.1 antibodies are shown for indicated genomic regions: CNS1 (negative control), Bcl6, Pou2af1, and Igkappa 3’enhancer (positive control). The average enrichment and SEM is from two independent experiments.
IRF4 targeting of Bcl6 and Pou2af1 loci
As Bcl6 and Pou2af1 expression is also dependent on IRF4, we sought to determine if it targeted these genes. IRF4 bound to a region ~24 kbp upstream of the Bcl6 gene (Fig. 5B) and to several sites within the first intron. Notably, at the upstream position, PU.1 was found to co-bind with IRF4 and this region coincided with a DNaseI hypersensitive site (Fig. 5B and S5D). We did not find evidence of IRF4 targeting the Bcl6 promoter as was shown in human B lymphoma cell lines (Saito et al., 2007). There were two prominent IRF4 (not PU.1) peaks within the first intron of Pou2af1 gene (Fig. 5C). IRF4 binding to sites in the Bcl6 and Pou2af1 genes diminished from Day 1 to Day 3 of B cell activation as compared with its occupancy of the Prdm1 locus, which underwent an increase. To confirm targeting of the Bcl6 and Pou2af1 genes by IRF4 in GC B cells we performed ChIP analysis with such cells isolated from immunized mice. We observed binding of IRF4 to the fore-mentioned regions of the Bcl6 and Pou2af1 genes (Fig. 5D). Thus these data, along with those in Fig. 2B, demonstrate that IRF4 directly targets and activates the expression of key regulatory genes that are required for GC B cell differentiation.
Divergent IRF4 binding modes correlate with complementary patterns of gene activity
We next analyzed how the distinct modes of IRF4 targeting correlate with IRF4 dependent gene expression programs that are reflective of the “GC-like” and plasma cell states. Thus we performed genome-wide transcriptome analyses and compared wild type or Irf4-inducible B cells (Sciammas et al., 2011) on the Irf4-/- background.
We reasoned that we could relate distinct patterns of gene expression to divergent modes of IRF4 genome targeting by analyzing the expression changes between cellular conditions in which IRF4 is expressed to varying extents at the different timepoints. To this end, we employed Expectation Maximization of Binding and Expression Profiles (EMBER) (Maienschein-Cline et al., 2011), which uses an unsupervised machine learning algorithm to infer target genes from transcription factor binding and expression data. EMBER scores genes that are likely regulated by a given transcription factor within 100kpb of its binding peaks based on their conforming to an over-represented gene expression pattern (see Supplement for further details).
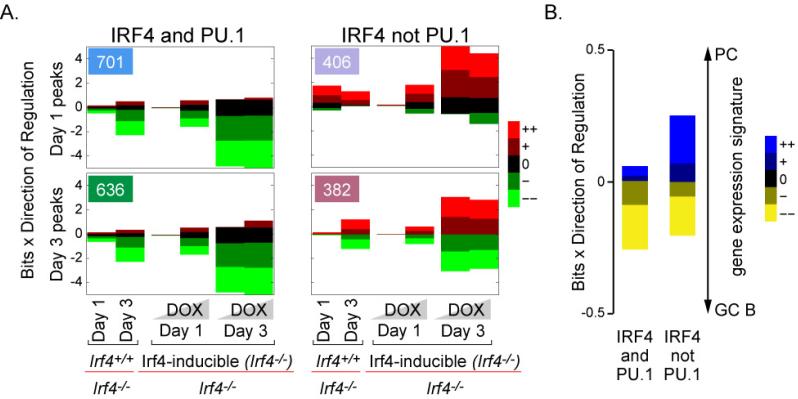
We applied EMBER to the IRF4 (and PU.1) and IRF4 (not PU.1) peaks at the Day 1 and Day 3 time points (Fig. 4A and 4B). The EMBER analysis is shown in Fig. 6A and Table S2. The numbers in the upper left hand boxes of each graph indicate the number of peaks that were assigned at least one target gene (Fig. 6A). Roughly half of all genome targeting events (compare to the number of peaks in Fig. 4A and 4B) scored above a statistical threshold (see Supplement). To simplify analysis of the gene expression data, differential expression between pairwise sample comparisons was binned into five categories: (++) large up regulation, (+) small up regulation, (0) no change, (–) small down regulation, and (– –) large down regulation (see Supplement). Each of the pairwise gene expression comparisons are plotted against the log-likelihood ratio of finding a given differential expression pattern. IRF4 peak-associated trends in differential gene expression are represented as changes in the relative sizes of colored bars corresponding to each of the 5 bins of differential expression. For example, in Fig. 6A, in the IRF4 (and PU.1) Day 1 expression pattern (top left), the large bright green bars in the 2nd, 4th, 5th, and 6th columns indicate that the majority of inferred target genes tend to be strongly down-regulated in the presence of IRF4 (either in wild type cells or in Irf4-/- cells after restoration of IRF4 expression with DOX). Conversely, the much smaller red bars in the same columns indicate that very few of the inferred gene targets are strongly up regulated in the presence of IRF4.
Figure 6.
see also Figure S6. IRF4 (and PU.1) and IRF4 (not PU.1) genome targeting are associated with distinct patterns of transcriptional regulation. (A) Genome-wide expression data was derived from purified B cells isolated from wild type (Irf4+/+), mutant (Irf4-/-), or Irf4-inducible mice that were stimulated with CD40L and cytokines. Total RNA was isolated on Days 1 and 3 and processed for Affymetrix arrays. B cells from the Irf4-inducible mice were also cultured in the presence of 16ng/mL or 100ng/mL doxycycline (DOX) to induce the expression of the Irf4 transgene, indicated by the shaded gradients on the x-axis. EMBER expression patterns for the four peak combinations (IRF4 at Days 1 and 3 with and without PU.1). The numbers in the upper left hand boxes of each graph indicate the number of peaks that scored above a threshold and were assigned at least one target gene. The x-axis displays pairwise comparisons between expression measurements (specifically wild type cells versus Irf4-/- cells or Irf4-inducible cells (on the Irf4-/- background) with and without DOX). The size of each bar in the y-axis indicates the significance of changes in expression and the color indicates the nature of the change; see (Maienschein-Cline et al., 2011) for further details. (B) Day 3 inferred target gene expression in the context of ex vivo antigen specific GC B cell and plasma cell transcriptomes (from NCBI GEO). Data presented as in (A).
EMBER analysis revealed three dominant patterns of transcriptional control by IRF4 (and PU.1) binding versus IRF4 (not PU.1) binding events (Fig. 6A). In the first pattern, inferred target genes associated with IRF4 (and PU.1) binding tended to be repressed regardless of timepoint (the green bars corresponding to mild and strong repression are large). This unexpected finding is consistent with an IRF4 dependent manner of regulation as (i) these genes were de-repressed in the absence of IRF4 and (ii) the same genes were repressed when IRF4 expression was restored by DOX-mediated induction of the inducible allele of Irf4 in Irf4-/- B cells (Fig. 6A). In the second pattern, genes associated with IRF4 (not PU.1) binding were preferentially activated at Day 1. The third pattern comprises a mixture of up and down regulated targets of IRF4 (not PU.1) on Day 3. We note that the transition in the nature of transcriptional output accompanies both the change in IRF4 concentration (Fig. S4C) as well as the differentiation state of these cells (Fig. S4B). Collectively, this analysis shows that IRF4 (not PU.1) genome targeting, regardless of time, is associated with markedly different behaviors of gene expression compared to those associated with IRF4 (and PU.1) genome targeting, suggesting that each binding mode functions to control distinct developmental programs.
Divergent IRF4 binding modes correlate with distinct developmental programs
To substantiate the hypothesis that IRF4 (and PU.1) and IRF4 (not PU.1) modes of genome targeting regulate different developmental programs, we tested whether the expression patterns of inferred target genes obtained from differentiated cells at the Day 3 time point correlated with plasma cell or GC B cell states. To perform this analysis, we used transcriptome experiments derived from ex vivo sorted antigen specific plasma cells and GC B cells (Luckey et al., 2006). Using these data from GEO, we compared GC B cell and plasma cell transcriptomes and classified differential gene expression using the same discrete binning scheme as above (++, +, 0, –, – –). Then, using inferred target genes from either IRF4 (and PU.1) or IRF4 (not PU.1) Day 3 peaks, we computed the log odds ratio of finding preferential expression of these genes in plasma cells or GC B cells (Fig. 6B, S6A and Table S2). With some exceptions, we found that the majority of IRF4 (and PU.1) inferred target genes were expressed at lower levels in plasma cells compared to GC B cells (large yellow bar). In contrast, inferred target genes associated with IRF4 (not PU.1) binding at Day 3 enriched for a substantial set of genes that were expressed at higher levels in plasma cells compared to GC B cells (large bright blue bar). Similar trends were observed when we analyzed the naive to GC B cell and the naive to plasma cell transitions (Fig. S6B and S6C). These data indicate that, during the transition of a naive or GC B cell to a plasma cell, both IRF4 (and PU.1) as well as IRF4 (not PU.1) binding events are associated with the repression of GC genes. In contrast, IRF4 (not PU.1) binding, particularly to the ISRE motif seems to preferentially function in activation of the PC program of gene expression. The proposed functions of these distinct modes of IRF4 genome targeting in relation to its intracellular concentrations and B cell fate dynamics are depicted in Fig. S7.
To further corroborate the finding that each binding mode is controlling distinct developmental programs, we determined whether the inferred target genes controlled by IRF4 (and PU.1) and IRF4 (not PU.1) genome targeting were associated with different functional classes of genes. Whereas genes associated with IRF4 (and PU.1) binding enriched for immune and inflammatory response categories, the IRF4 (not PU.1) associated genes enriched for cell biological categories including endoplasmic reticulum functions (Fig. S6D and Table S3). Many of these latter genes are important for the differentiation of specialized secretory cells suggesting that the IRF4 (not PU.1) targeting mode involving ISREs specifies the functional state of plasma cells.
Discussion
We have demonstrated an essential and cell intrinsic role for IRF4 in generating GC B cells. IRF4 does so in part by activating expression of the Bcl6 and Pou2af1 genes. Our combined genetic analyses involving conditional deletion of the Irf4 gene prior to B cell activation as well as its transient expression using the Irf4-inducible allele define the temporal requirement for IRF4 in promoting GC B cell fate to the first few days after antigen encounter. The results strongly suggest that IRF4 is required for initiation but not maintenance of the GC state, the latter is dependent on sustained expression of Bcl6. It follows that IRF4 is required for the activation but not maintenance of expression of the Bcl6 gene. In contrast expression of the Pou2af1 gene appears to continuously depend upon IRF4. Intriguingly, the latter gene along with IRF4 also functions in regulating plasma cell differentiation (Corcoran et al., 2005). Given that high and sustained expression of IRF4 antagonizes the GC state, these findings account for transient action of IRF4 in generating GC B cells and its down regulation in such cells. It will be interesting to determine whether in GC B cells negative feedback by Bcl6 serves to directly repress the Irf4 gene, as has been observed in cell lines (Alinikula et al., 2011). Our key conclusion that IRF4 functions in a cell intrinsic manner to regulate GC B cell differentiation differs from that reached by a recent report (Bollig et al., 2012). We note that our findings are based on the use of three distinct alleles of Irf4: germline, conditional and tet-inducible. Importantly, the Irf4-conditional allele (Irf4fl/fl) displays a GC B cell defect when CRE expression is driven by the CD19 locus but not by the Cγ1 locus. Finally, the tet-inducible Irf4 allele allowed us to unambiguously demonstrate that Bcl6 and AID expression can be induced in Irf4-/- B cells in an IRF4 dependent manner. Bollig et al. demonstrate a role for IRF4 in T follicular helper cell differentiation. We therefore propose that IRF4 may direct T follicular helper cell differentiation by directly activating Bcl6 expression, as is the case in B cells.
Using genome-wide analysis in a model system involving antigen-dependent B cell differentiation, three distinct modes of IRF4 binding to its target sequences have been delineated. The dominant mode (representing two thirds of the total) involves co-binding of IRF4 with PU.1 to EICE motifs. Two additional modes, both of which are PU.1 independent, involve IRF4 binding to either an ISRE or AICE motif. Co-occupancy of the EICE motif by PU.1 and IRF4 is associated with regulation of genes involved in B cell activation and function. Molecular redundancy between PU.1 and SpiB in the Ets family as well as between IRF4 and IRF8 in the IRF family can result in the targeting of the EICE motif by four distinct Ets-IRF ternary complexes (Eisenbeis et al., 1995). Accordingly, these complexes can play either redundant or unique roles in B cell development, activation and terminal differentiation. Importantly, although IRF4 and IRF8 function redundantly in the differentiation of pre-B cells (Lu et al., 2003), IRF4 is uniquely required for the GC response and plasma cell differentiation (Feng et al., 2011; Klein et al., 2006; Sciammas et al., 2006). Given that SpiB is uniquely important for the differentiation of GC B cells along with the observation that IRF8 is expressed at high levels in GC B cells, it will be interesting to determine whether occupancy of the EICE motif in this cellular context shifts to these factors.
A second mode of IRF4 binding is observed on composite AP-1-IRF motifs (AICE). This unusual motif has been observed by us in the context of T cells, where it functions as the dominant mode of IRF4 genomic targeting, given that these cells do not express PU.1 or SpiB (Glasmacher et al., 2012). We have demonstrated that this composite motif directs cooperative binding of IRF4 with BATF heterodimers belonging to the AP-1 family. As Batf-/- B cells partially phenocopy Irf4-/- B cells (Betz et al., 2010; Ise et al., 2011), it will be informative to analyze the molecular consequences of IRF4-BATF family complexes that assemble on AICE motif containing genes. In accord with a signal-dependent mode of gene regulation by AP-1 family members, the AICE motif is observed at high incidence in DNA bound regions at the Day 1 time point, soon after BCR signaling initiated by antigen encounter. At this time point B cells are forming blasts and initiating the gene regulatory programs necessary for subsequent differentiation. The integration of IRF4 with the AP-1 system at this stage, both of which are controlled by BCR signaling, suggests that they could be important for effecting the downstream transcriptional responses that are triggered by differential BCR signaling; i.e., low or high affinity antigen or levels of co-receptor engagement.
The third mode of IRF4 binding in the B cell genome involves classical ISREs. We demonstrate that IRF4 binds the ISRE as a homodimer and this interaction is of lower affinity than the PU.1-IRF4 interaction with the EICE motif. Accordingly, binding to the ISRE is preferentially observed at the Day 3 time point of B cell differentiation when IRF4 protein levels peak. The Day 3 time point represents a stage where a majority of the B cells in the culture system have undergone differentiation into plasma cells. Intriguingly, the increased usage of the ISRE in plasma cells suggests an association of this regulatory element with structural genes important for their differentiation. Indeed, such target genes are enriched for those that encode secretory functions. Although the concept of differing concentrations of a transcription factor regulating distinct cell fates has been suggested to be widely operative in mammalian cell differentiation, see (DeKoter and Singh, 2000), its molecular basis has proven difficult to elucidate. Our results provide an appealing molecular explanation for the requirement of higher concentrations of IRF4 in regulating plasma cell differentiation by enabling occupancy of low affinity ISRE motifs that are associated with plasma cell genes. Based on experimental and mathematical analyses, we have proposed a mechanism of kinetic control in which the initial rate of IRF4 accumulation, driven by the strength of BCR signaling, controls a gene regulatory network that orchestrates cell fate decisions between cells undergoing CSR, a “GC-like” state, versus cells differentiating into plasma cells (Sciammas et al., 2011). In this model, low affinity or avidity antigen interactions with the BCR favor a longer duration of a “GC-like” state before plasma cell differentiation. In contrast, high affinity or avidity antigen interactions with the BCR limit the period of time in which AID is expressed and therefore promote plasma cell generation accompanied with relatively low levels of CSR and SHM. Herein, we corroborate and extend this regulatory model. Specifically, as evidenced by the “pulsed” experiment, we propose that transient expression of IRF4 in GC B cells (IRF4loBcl6+) serves to “reset” the mechanism of kinetic control initiated by the first exposure to antigen. Hence, kinetic control would be reinstated during positive selection of somatically hypermutated GC B cells upon their interaction with antigen displayed by follicular dendritic cells. Specifically, those clones accumulating mutations that confer higher affinity to their BCR would induce increased levels of IRF4 expression, both as a function of BCR signaling and CD40 signaling by T helper cells, and thus differentiate into plasma cells. In support of this notion, it has been found that post-GC plasma cells are preferentially enriched for high affinity SHM-dependent mutations (Phan et al., 2006; Smith et al., 1997). In contrast, those clonotypes exhibiting lower affinity conferring mutations would induce lower levels of IRF4 expression and differentiate into memory B cells or undergo new rounds of SHM and selection (Smith et al., 1997; Victora et al., 2010). Given that GC B cells express high amounts of Bcl6, it will be interesting to determine whether selection into the high affinity cell pool involves a steeper magnitude of IRF4 induction to overcome repression by Bcl6.
Experimental Procedures
Mice
The generation of the Irf4-/- and Irf4-inducible mice has been previously described (Mittrucker et al., 1997; Sciammas et al., 2011). The B1-8i gene targeted mice were a generous gift of K. Rajewsky. SWHEL mice have been previously described (Phan et al., 2005) and were used to cross to Irf4-/- mice and Irf4-inducible mice. Conditional Irf4fl/fl mice (Klein et al., 2006) were crossed to CD19+/CRE mice. Mice were housed in specific pathogen-free conditions and were used and maintained in accordance of the Institutional Animal Care and Use Committee guidelines.
ChIP and ChIPseq
ChIP was performed using anti-IRF4, -PU.1, -H3K4me1, -H3K27Ac, control IgG antibodies (Santa Cruz Biotech. and Abcam) (Sciammas et al., 2006). For massively parallel sequencing, 200ug of chromatin fragments were immunoprecipitated using anti-IRF-4 and anti-PU.1 antibodies and DNA libraries were prepared. DNA was sequenced using the Illumina GA2, data was processed with the Solexa pipeline package, and sequences were aligned to the mouse genome (mm9) using ELAND software. Further details are available in the Supplement.
Generation of mixed bone marrow chimeric mice and SRBC immunization
Bone marrow was collected from the long leg bones of wild type CD45.1/2 heterozygous mice and from Irf4+/+ or Irf4-/- (both CD45.2) mice and mixed together in equal numbers. Cells were injected into irradiated (2 x 550 rads) wild type CD45.1 mice. Eight weeks following adoptive transfer, mice were immunized with sheep RBC (Lampire Biologicals) i.p. and the GC response was quantified by flow cytometry seven days later. Conditional Irf4fl/fl mice were bred to CD19C+/CRE, immunized with SRBC, and spleens were analyzed 14 days later. Further details are available in the Supplement.
Adoptive transfer of SWHEL B cells
SWHEL mice were crossed to Irf4-inducible or Irf4-/- mice and used for adoptive transfer experiments using established methods (Phan et al., 2005). Doxcycline (DOX, Sigma) was administered in drinking water that contained 1% (w/v) sucrose, at a concentration of 0.5mg/mL; the control group was fed sucrose water only and the “pulse” group was changed to sucrose water only after the initial DOX treatment. Further details regarding CFSE labeling, numbers of transferred cells, SRBC-conjugated antigens, and sorting are available in the Supplement.
Flow cytometry and ELISpot assays
RBC-depleted spleen cell suspensions were prepared, stained, analyzed on the LSR II flow cytometer and the resulting data was processed using FlowJo software (Tree Star, Inc.). HEL+ cells were identified as described (Phan et al., 2005). Detection of Bcl6 was performed by fixing and permeabilizing cells with Fix/Perm staining kit (eBioscience) and staining with anti-Bcl6 (BD). Detection of IRF4 was performed as previously described (Sciammas et al., 2011). For ELISpot analysis, total splenocytes were cultured for 6hrs on plates coated with HEL (Sigma) and processed with anti-IgM and anti-IgG antibodies as previously described (Sciammas et al., 2006). Further details are available in the Supplement.
RNA preparation, microarray and RT-PCR analysis
For transcriptome analysis, total RNA was prepared from triplicate cell samples using Trizol and processed for hybridization to Affymetrix mouse 430A chips using standard procedures. For RT-qPCR analysis of sorted SWHEL B cells, total RNA was prepared by sorting 5,000 cells directly into RLT buffer from the RNeasy Micro Kit (Qiagen). Further details are available in the Supplement.
Supplementary Material
Highlights.
IRF4 regulates GC B cell differentiation by directly activating Bcl6 and Obf1 genes
Transient expression of IRF4 can induce the GC B cell fate
Sustained expression and high concentration of IRF4 induces plasma cells
At high concentrations IRF4 binds to ISRE motifs enriched in plasma cell genes
Acknowledgements
We thank Qiang Wang and Jing Harakh for animal husbandry and injections. We thank the UC Flow Cytometry Core Facility and the Bendelac laboratory for assistance with cell sorting and analysis, respectively. We are grateful to K. Michelini and J. Zekos for operating the Illumina Genome Analyzer II at the University of Chicago (supported by the Howard Hughes Medical Institute). Research was supported by the US Department of Energy (DOE) Computational Science Graduate Fellowship Program (M.M.-C.), CLL Global Research Foundation (U.K.), the National Institutes of Health (P50, GM081892 to A.R.D. and H.S.), the National Kidney Foundation, IL Division and the Illinois Department of Public Health, Juvenile Diabetes and Islet Transplantation Research Grant (R.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alinikula J, Nera KP, Junttila S, Lassila O. Alternate pathways for Bcl6-mediated regulation of B cell to plasma cell differentiation. Eur J Immunol. 2011;41:2404–2413. doi: 10.1002/eji.201141553. [DOI] [PubMed] [Google Scholar]
- Allman D, Lindsley RC, DeMuth W, Rudd K, Shinton SA, Hardy RR. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, Kim CH, Taparowsky EJ. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollig N, Brustle A, Kellner K, Ackermann W, Abass E, Raifer H, Camara B, Brendel C, Giel G, Bothur E, et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc Natl Acad Sci U S A. 2012;109:8664–8669. doi: 10.1073/pnas.1205834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. Embo J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran LM, Hasbold J, Dietrich W, Hawkins E, Kallies A, Nutt SL, Tarlinton DM, Matthias P, Hodgkin PD. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J Exp Med. 2005;201:1385–1396. doi: 10.1084/jem.20042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- Feng J, Wang H, Shin DM, Masiuk M, Qi CF, Morse HC., 3rd IFN regulatory factor 8 restricts the size of the marginal zone and follicular B cell pools. J Immunol. 2011;186:1458–1466. doi: 10.4049/jimmunol.1001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Dahl R, Rao S, Barton KP, Simon MC. PU.1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood. 2001;97:2908–2912. doi: 10.1182/blood.v97.9.2908. [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner C, Rutz S, et al. A Genomic Regulatory Element That Directs Assembly and Function of Immune-Specific AP-1-IRF Complexes. Science. 2012 doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maienschein-Cline M, Zhou J, White KP, Sciammas R, Dinner AR. Discovering Transcription Factor Regulatory Targets Using Gene Expression and Binding Data. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittrucker HW, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- Muto A, Ochiai K, Kimura Y, Itoh-Nakadai A, Calame KL, Ikebe D, Tashiro S, Igarashi K. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. Embo J. 2010;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin Immunol. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Paus D, Phan TG, Chan TD, Gardam S, Basten A, Brink R. Antigen recognition strength regulates the choice between extrafollicular plasma cell and germinal center B cell differentiation. J Exp Med. 2006;203:1081–1091. doi: 10.1084/jem.20060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Gardam S, Basten A, Brink R. Altered migration, recruitment, and somatic hypermutation in the early response of marginal zone B cells to T cell-dependent antigen. J Immunol. 2005;174:4567–4578. doi: 10.4049/jimmunol.174.8.4567. [DOI] [PubMed] [Google Scholar]
- Phan TG, Paus D, Chan TD, Turner ML, Nutt SL, Basten A, Brink R. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203:2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, Pernis A, Pasqualucci L, Dalla-Favera R. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Sciammas R, Li Y, Warmflash A, Song Y, Dinner AR, Singh H. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7:495. doi: 10.1038/msb.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Smith KG, Light A, Nossal GJ, Tarlinton DM. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. Embo J. 1997;16:2996–3006. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su GH, Chen HM, Muthusamy N, Garrett-Sinha LA, Baunoch D, Tenen DG, Simon MC. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. Embo J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.