Summary
Raster image correlation spectroscopy (RICS) is a new and novel technique for measuring molecular dynamics and concentrations from fluorescence confocal images. The RICS technique extracts information about molecular dynamics and concentrations from images of living cells taken on commercial confocal systems. Here we develop guidelines for performing the RICS analysis on an analogue commercial laser scanning confocal microscope. Guidelines for typical instrument settings, image acquisition settings and analogue detector characterization are presented. Using appropriate instrument/acquisition parameters, diffusion coefficients and concentrations can be determined, even for highly dynamic dye molecules in solution. Standard curves presented herein demonstrate the ability to detect protein concentrations as low as ~ 2 nM. Additionally, cellular measurements give accurate values for the diffusion of paxillin-enhanced-green fluorescent protein (EGFP), an adhesion adaptor molecule, in the cytosol of the cell and also show slower paxillin dynamics near adhesions where paxillin interacts with immobile adhesion components. Methods are presented to account for bright immobile structures within the cell that dominate spatial correlation functions; allowing the extraction of fast protein dynamics within and near these structures. Arunning average algorithm is also presented to address slow cellular movement or movement of cellular features such as adhesions. Finally, methods to determine protein concentration in the presence of immobile structures within the cell are presented. A table is presented giving guidelines for instrument and imaging setting when performing RICS on the Olympus FV300 confocal and these guidelines are a starting point for performing the analysis on other commercial confocal systems.
Keywords: correlation spectroscopy, FCS, ICS, RICS
Introduction
Recent advances in the speed and sensitivity of fluorescence confocal laser scanning microscopes (CLSMs) now allow the acquisition and analysis of fluorescence images from live cells. Properties such as protein dynamics and aggregation states can be extracted following a careful analysis of these images. New spatial-temporal fluorescence image correlation methods have been developed (Hebert et al., 2005;Digman et al.,2005a, 2005b; Brown et al., 2006) and go well beyond the initial conception of fluorescence correlation spectroscopy (FCS), a technique that was initially developed to measured time correlations of the fluorescence intensity fluctuations in a focal volume of a solution (Magde et al., 1974).
Intensity fluctuations within a focal volume in a biological system can arise from a number of phenomena including the entry and transit of molecules through the volume, protein conformational changes, and protein binding to immobile or large cellular structures. To capture and distinguish the subtle differences in the intensity fluctuations due to these cellular phenomena, the ideal correlation technique would have a spatial resolution limited only by light diffraction and would be sensitive to the very wide range of dynamics observed in living cells (Table 1).
Table 1.
Summary of characteristic times for various cellular processes and the techniques that can be used to measure these processes
| Process | Characteristic time |
Characteristic fluctuation amplitude |
Technique |
|---|---|---|---|
| Rotational diffusion | 1–10 µs | 1–3 counts | Polarized FCS |
| Blinking | 1–10 µs | 1–10 counts | FCS |
| Diffusion of a small molecule in solution | 0.2 ms | 1–10 counts | FCS, S-FCS, RICS |
| Diffusion of GFP in the cytoplasm | 1 ms | 1–10 counts | FCS, S-FCS, RICS |
| Lipid or lipid anchored protein diffusion | 10 ms | 1–10 counts | FCS, S-FCS, RICS, FRAP |
| Diffusion of a protein in a membrane | 100 ms | 1–10 counts | t-ICS, FRAP |
| Vesicle transport | 0.1–1 s | 100 to 1000 counts | t-ICS |
| Weak binding–unbinding | 0.1–10 s | 1–100 counts | FCS, S-FCS, t-ICS, TIRF t-ICS |
At present, an ideal method fulfilling these criteria has not yet been described in detail. Due to the nature of the scanning mechanism CLSM fluorescence images inherently capture a wide range of dynamic information potentially providing details about underlying biological processes within the cell. As the laser performs the raster scanning movement, it creates a space–time matrix of pixels within the image. Since the temporal and spatial sampling of the laser beam during the raster scanning is known, that is, the time the laser samples each pixel (pixel dwell time); time between scan lines and time between images. Therefore, the images contain information on the microsecond time scale for pixels along the horizontal scanning axis, millisecond time scale along the vertical scanning axis or between scan lines, and on the sub-second to second or longer time scale between successive images. Using the RICS correlation technique it is possible to generate spatial-temporal maps of dynamics occurring on the microseconds to milliseconds time scale across the cell. If the RICS analysis is performed in combination with temporal image correlation spectroscopy (t-ICS) (Wiseman et al., 2000) performed between images within an image time series on the seconds time scale (or longer) then the two methods provide dynamic information from microseconds to minutes or hours.
RICS can be performed on standard confocal images taken from commercial CLSMs opening up this type of analysis to a large number of researchers. In general, commercial CLSMs are sensitive, their operation is automatic and their performance has been optimized due to the large number of manufacturers, their large scale use and long term availability in the marketplace. However, in general the light detection of CLSMs is analogue in which the current of a photomultiplier detector is integrated and sampled at a specific frequency. In contrast, FCS instruments traditionally rely on photon counting detectors, where the pulse from individual detected photons is discriminated, and the number of photons detected in a certain time window is counted. More recently, detectors have been employed which allow the time interval between detected photons to be measured. In general, analogue detectors have largely been avoided in FCS analyses because the integration circuit in analogue detectors used before the digital sampling of the current can introduce unwanted correlations. In this paper we address the detection of spatial and temporal intensity fluctuation correlations using analogue detection on a commercial Olympus Fluoview 300 microscope. Guidelines for instrument settings, collecting images and performing the RICS analysis are provided. The data show that standard curves can be generated to determine protein concentrations both in solution and in the cell. In addition, methods are presented to show how large spatial structures within the cell or slow movement of these structures or the cell as a whole during image acquisition can be removed from spatial autocorrelation function (ACFs) so that protein concentrations and dynamics can be determined.
Materials and methods
Tissue culture
Cell culture, plasmids and sample preparation have been described in detail elsewhere (Laukaitis et al., 2001; Webb et al., 2004). Briefly, for EGFP expression cells were transfected with 1 µg of DNA and lipofectamine reagent (Invitrogen, Carlsbad, California, U.S.A.) 24–48 h prior to imaging, as previously described. EGFP expressing or stably transfected paxillin-EGFP cells were lifted with trypsin and plated in CCM1 medium (Hyclone, Utah, U.S.A.) buffered with 15 mM HEPES on homemade 35 mm glass bottomed dishes coated with 2 µg mL−1 fibronectin and maintained at 37°C during imaging with a Warner Instruments heated stage insert (Warner Instruments, Hamden, Connecticut) and a Bioptechs (Bioptechs, Butler, Pennsylvania, U.S.A.) objective heater.
RICS theory
Details of the theory for RICS have been published previously (Digman et al., 2005a). Briefly for raster scanning the spatial component of the correlation function decays due to movement of the laser beam scanning (S(ξ,ψ)) is expressed as:
| (1) |
Where ξ, ψ are the x and y spatial lags in pixels, δx and δy are the pixel size, τ p (typically 2–8 µs) is the pixel dwell time in x, τ l is the interline time in y (typically 1–4 ms), wo is the e−2 radius of the point spread function of the laser beam. The component of the ACF due to diffusion is the traditional correlation function:
| (2) |
Where γ is a shape factor due to uneven illumination across the focal volume and is 0.3535 for a 3D Gaussian under ideal conditions (Chen, 1999). Also for a 3D Gaussian profile the z-axis beam radius, wz, is set to 3wo.The overall ACF, Gs(ξ,ψ) is then given by:
| (3) |
Note: For two-photon excitation the factor in Equations 2 and 3 should be 8D rather than 4D.
Solution preparation and system calibration
The e−2 radius of the laser beam point spread function (PSF) was measured daily using a RICS analysis of images of a fresh 50 nM fluorescein solution in 100 mM Tris, pH 9. The stock concentration was calculated using A494 and ε = 75 000 cm−1M−1. ACFs from multiple images (typically 50– 100) taken at ~1 µm above the coverslip of a 35 mm glass bottomed dish were calculated using SimFCS (available at http://www.lfd.uiuc.edu/) and then averaged to give one ACF for the data set. The resulting ACF was fit to Eq. (3) while fixing D = 300 µm2 s−1 and determining wo and g(0,0). The beam radius typically varied from ~0.30–0.39 µm. The concentration of monomeric EGFP (mEGFP) (Zacharias et al., 2002), stock aliquots kept at −80°C) stock solution was calculated from A486 using ε = 61 000 cm−1 M−1. To avoid spontaneous adsorption of EGFP to the coverglass wells were pre-coated with 1% BSA for 1 h at 37°C. Stock solutions were then diluted in 100 mM Tris (pH 9) containing 1% bovine serum albumin (BSA) and were put into eight well coverglass bottomed Lab-Tek (Campbell, California, U.S.A.) chambers. BSA is required in the EGFP solutions or the EGFP rapidly exchanges with BSA adsorbed to the coverglass. Background counts were acquired for the EGFP and fluorescein measurements using 1% BSA or Tris buffer alone, respectively. ACFs were calculated from images of EGFP in solution using SimFCS and fitting to Eq. 3. Values for wo (determined from fluorescein samples) and the image parameters were input and the ACF was fit to determine g(0,0), D and g∞. The EGFP concentration (C) was calculated using Eq. 4:
| (4) |
where NA is Avogadro’s number, VPSF is the PSF volume (V = 3*π1.5ωo3), α is a correction factor for intensity contributions from the photomultiplier tube (PMT) background (α = 〈i〉2/〈i – ibg〉2), ibg was measured for a region of the image off the cell, or from an image of buffer alone for solution measurements.
Simulations
Computer simulations were used to model ideal expected results for different transport properties and conditions to allow direct comparison with the experimental results. An IDL program was written to simulate RICS data that would be obtained by laser scanning microscopy of point emitters in a 2D system under defined settings of instrument collection and particle mobility. The program allowed a wide range of system parameters to be defined including diffusion of the simulated particles on the 2D surface, densities of multiple populations of particles, laser beam size and shape characteristics, pixel dwell time and line-to-line dead time, image size, pixel size, the number of images collected for analysis and the time interval between images. Periodic boundary conditions were used at the image edges, and discrete displacements in x and y were computed at every time step for each particle using normally-distributed, floating-point, pseudo-random numbers having a mean of zero and standard deviation The simulations were run on a desktop PC.
Confocal microscopy
All images were collected on an Olympus Fluoview 300 (FV300) confocal microscope (Olympus, Japan) built around an IX70 inverted microscope fitted with a 60X PlanApo (1.40 NA) oil immersion objective. Details of the instrument and image acquisition have been previously described (Wiseman et al., 2004). For these specific experiments, the PMT was operated at 800 V with 1X gain, and 0% offset. Images were typically collected at 1024 × 1024 resolution, zoom 5X (0.046 µm pixels) or 10X (0.023 µm pixels), with a clip box of 256 × 256 pixels. Pixel dwell times for the FV300 are 2 µs/pixel (fast scan), 4 µs/pixel (med scan) or 8 µs/pixel (slow scan), and the time between lines was 1.608 ms (fast scan), 2.12ms (medium scan), and 3.15ms (slow scan). The pinhole was always set to 4 (200 µm) on the FV300 scale allowing more light to be detected when performing acquisitions on living cells without sacrificing the axial resolution too much. A pinhole setting of 5 (300 µm) does not result in a very large increase in intensity at the expense of a loss in axial resolution. The PSF diameter was estimated using fluorescein in solution and setting the diffusion constant to 300 µm2s−1. Under these image acquisition settings the PSF was measured to be 0.29– 0.35 µm in diameter (wo) and the z-dimension was estimated at 3 wo. Similar values were obtained for the PSF dimensions using sub-resolution (0.1 µm diameter) fluorescent beads.
Immobile removal algorithm
The immobile removal algorithm has been described previously (Digman et al., 2005a). Basically, spatial correlations due to immobile structures within the images are removed by taking the image of the average intensity for the entire time series and subtracting that average image from each image frame. The average intensity of the individual image frame is then added back to avoid having negative pixel intensities. This method works well for determining protein dynamics from the shape of the ACF, however it does not accurately provide the ACF amplitude which is normalized to the average intensity. This is because the average intensity of the image frame that is added back takes into account both the bright immobile component and the mobile component while the RICS ACF only measures the fluctuating dynamic component. Therefore, the average intensity of the mobile component is overestimated by simply using the average intensity of the image. To accurately determine the concentration of mobile proteins, the average intensity of a reference area of the cell that does not contain immobile proteins is added back to the images so that the intensity of the mobile component is not overestimated. It is important to choose a reference area close to the region of interest and in the same time frame since protein concentrations in the cell are heterogeneous in both space and time.
When structures within the cell are moving or the cell itself is dynamic then a moving average immobile removal algorithm is applied. Basically, for removal of a 20 frame time window the average of frames 1–20 would be subtracted from frame 11, and the average of frames 2–21 would be subtracted from frame 12 and so on with a moving average through the entire image stack. The first 10 frames and the last ten frames of the data set would be ignored because there is not enough data to perform the running average calculation properly. However, if a running average of ten frames was used only the first and last five frames would be ignored and so on. This eliminates artefacts in the ACF due to cellular or sub-cellular (e.g. adhesions, filaments, vesicles) movements when applying the immobile removal algorithm using the entire image time series.
Results and discussion
Simulations of RICS
Correlation measurement or fluctuation analysis requires that there is adequate data sampling to have enough statistics to recover the correlation function from the noise. The correlation function (in the simplest form) is characterized by two parameters, the amplitude of the function and the characteristic decay or correlation time. These two parameters are measured with different statistical errors which are dependent upon the number of molecules in the excitation volume. For an accurate statistical determination of the correlation time from the ACF it is necessary to measure ~10 000 fluctuation events (e.g. 10 000 events where molecules move in and out of the laser beam). To determine concentration information from the amplitude of the ACF, fewer events are required and depends largely on the number of particles within the focal volume of the focused laser beam; that is, the concentration. For example, for a temporal analysis like point FCS of a process with a characteristic correlation time of 100 µs, and a sampling rate of 10 µs (about ten times faster than the characteristic time), a series of at least 105 data points corresponding to about 1 s of data acquisition or longer is required. If this measurement is repeated at each point in an image in order to map out dynamics across the cell, it will require prohibitively long times, inappropriate for live cell imaging. In turn, when an image is collected, if each pixel is independent, there can be about 105 or more readings in a single image. If only the average dynamics across the image is required, then only a few images (or sometimes one) are sufficient to obtain the average amplitude and characteristic time of the fluctuations with good statistics. For dynamic measurements at a higher spatial resolution, or to distinguish mobile and immobile populations more images must be analysed. In the majority of the cases 50 to 100 images are sufficient to reach a good level of signal-to-noise (S/N).
Simulations of point sources moving in solution were used to optimize conditions for RICS analyses (details in Materials and methods). The simulations used an average point source density of 25 points per simulated laser beam spot, and diffusion coefficients (D) representative of hindered diffusion of transmembrane proteins, such as integrins (Wiseman et al., 2004) or membrane receptors (Hegener et al., 2004), ≤0.1 µm2 s−1, 0.1 µm2 s−1 for simulation, membrane lipid diffusion, 1 µm2 s−1 (Webb et al., 1981), cytosolic protein diffusion, 10 µm2 s−1 (Ruan et al., 2002) and free protein diffusion (i.e. EGFP) in solution, 100 µm2 s−1 (Ruan et al., 2002). In addition, a pixel size of 0.03 µm, a pixel dwell time of 8 µs, an interline time of 7 ms, an image to image time of 0.86 s and a laser beam radius of 0.30 µm were used. These conditions approximate the settings on the Fluoview 300 when using the slow scan speed, zoom ten and a1024×1024 pixel image for the Fluoview 300.
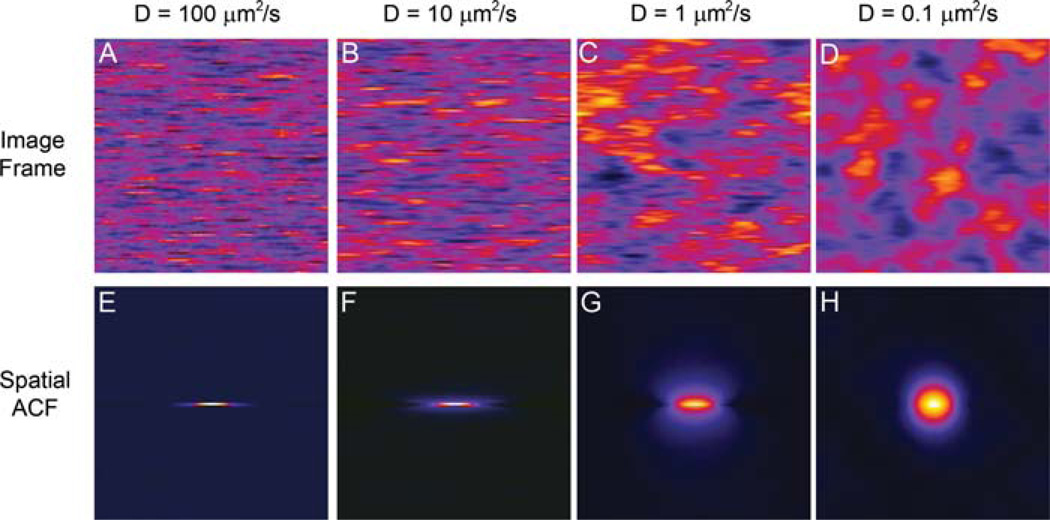
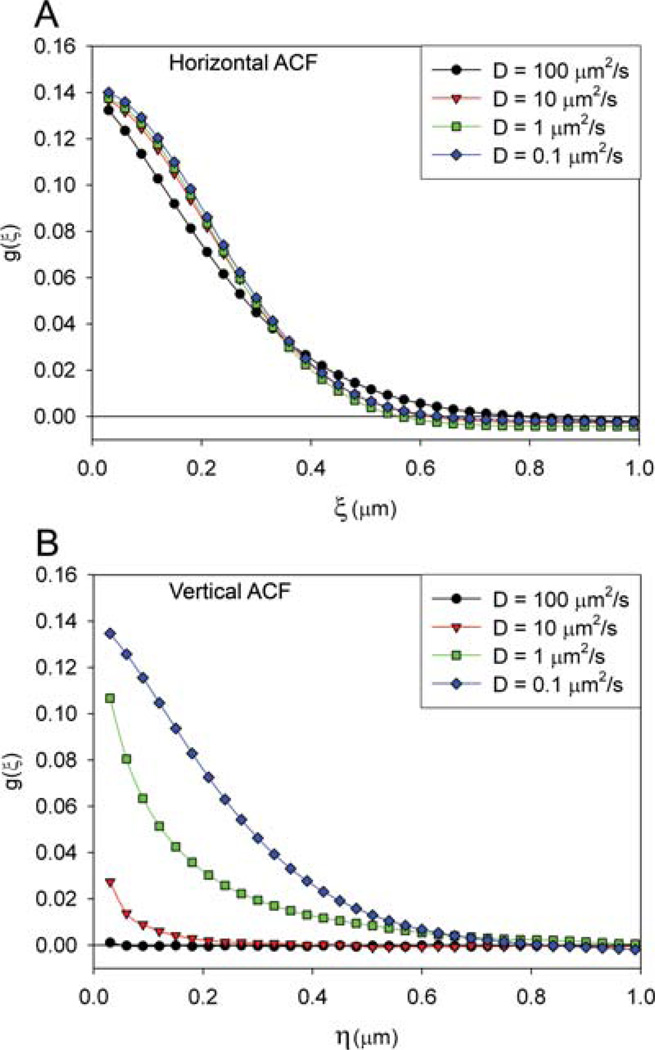
Figure 1 shows a representative image of the simulated point sources for the four diffusion coefficients (Figs. 1A – D), and the average ACF function calculated for 300 128 ×128 pixel images. It is clear from images of the point sources moving at different speeds that the shape of the spatial ACF is a reflection of the particle motion (Figs. 1E – H). When the point sources are moving rapidly compared to the line to line scan time of 7 ms (D = 100 µm2 s−1), the point source appears as horizontal streaks within the image (Fig. 1A). This is because as the laser beam scans the sample the point source will be in a given pixel location, but by the time the laser beam gets back to the adjacent pixel location one scan line down, or 7 ms later, the point source is no longer in that location. As a result, the spatial ACF appears elongated along the horizontal axis with little height along the vertical axis (Fig.1E). In order to visualize subtle changes in the 2D spatial ACF, the 1D profile of the ACF along the horizontal scan x-axis (horizontal ACF) or the vertical interline y-axis (vertical ACF) is plotted (Figs 2A and B, respectively). For fast diffusion, the horizontal component of the ACF is well defined while there is no vertical component (compare Figs 2A and B, black lines). As diffusion rates decrease, the spatial ACF broadens along the vertical axis because point sources have a higher probability of still being in the same or similar pixel location following the 7 ms interline time (Fig. 1B and Fig. 2B, red line). For point sources that are moving very slowly (0.1 µm2 s−1), the ACF approaches the shape of the laser beam PSF seen in spatial image correlation spectroscopy (ICS) (Fig. 1H, Figs 2A and B, blue curves, (Wiseman et al., 2000). It is important to note that if the molecules are moving slower than the laser beam is scanning, the spatial ACFs will be insensitive to differences in diffusion rates and the ACF will look the same (i.e. will have the shape of the PSF) and will not see differences in dynamics (Fig. 2A, e.g. horizontal plots for D = 1 (green curve) and D = 0.1 (blue curve) µm2/s have a very similar shape). In this case, t-ICS analysis on an image time series collected on a time scale matching the rate of the molecular dynamics of interest will reveal the different diffusion rates (Wiseman et al., 2004; Wiseman et al., 2000). Consequently, from a single image time series using a combination of the two approaches, RICS and t-ICS,a very wide range of dynamic events occurring on a wide range of time scales (microseconds to seconds or longer) can be measured.
Fig. 1.
Images and spatial ACFs from simulations of point sources with a density of 25 per beam, moving in solution in 2D with variable diffusion coefficients. The imaging conditions were 0.03 µm pixels, 8 µs pixel dwell time, 7 ms between lines, 0.86 s between images and a laser beam radius of 0.30 µm. Spatial ACF are an average of 300 spatial ACFs from 300 image frames. Conditions are similar to the conditions when using the Fluoview 300 with slow scan speed, zoom 5 and a 256 × 256 clip box on the 1024 × 1024 image setting.
Fig. 2.
Horizontal (η = 0) (A) and vertical (ξ = 0) (B) components of the ACF for simulations of point source movement with variable diffusion coefficients. ACFs were calculated for each of 300, 128 × 128 pixel image frames and averaged. Pixel dwell time is 8 µs, interline time was 7 ms, 0.03 µm pixels.
Characterization of the fluoview 300 analogue PMT: pixel to pixel correlations
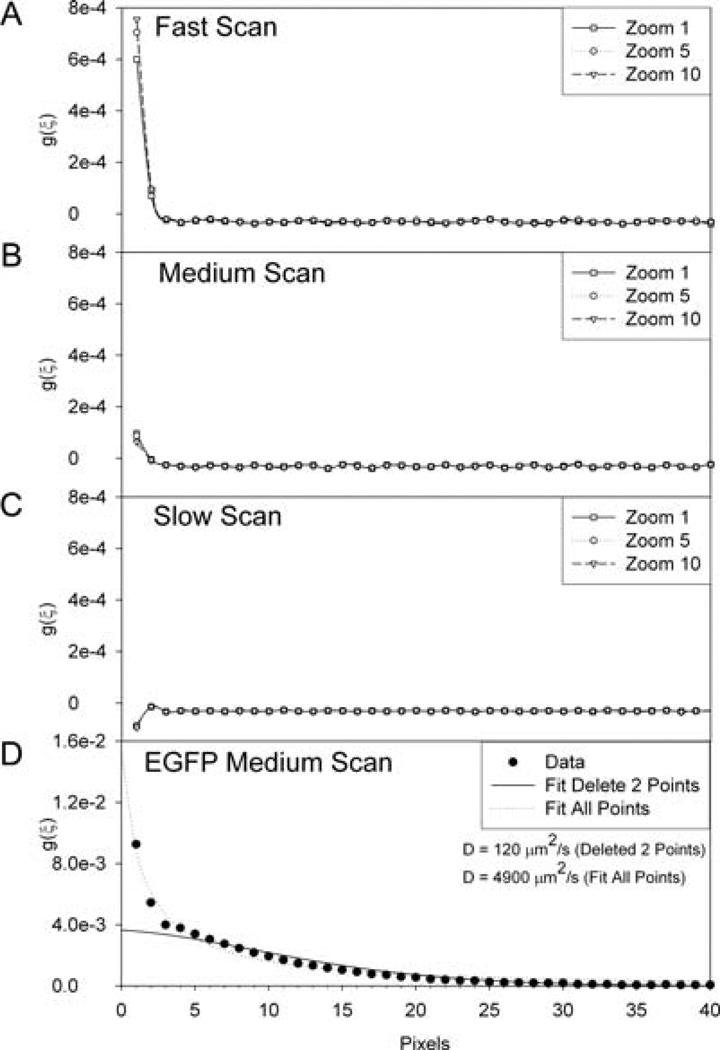
To characterize time correlations in herent in the PMT detector, images were collected with no sample in place and the microscope set to the eyepiece so that no light can enter the sideport of the microscope or the PMTs. These images contain the dark current (shot noise) of the detector and depend only on the digitization rate of the detector (i.e. scan speed), and not other instrument settings. Images were collected at the fast, medium and slow scan rates, corresponding to pixel dwell times of 2, 4, and 8 µs, respectively. From these images the 2D ACF was calculated for the shot noise component of the analogue PMT. ACFs along the x-axis (y pixel shift, η = 0), that is, the fast laser beam scanning axis, show spatial correlations in the first two pixels (Fig. 3). A similar correlation was not observed between pixels along the y-axis (x pixel shift, ξ = 0), or interline axis, where the time between adjacent pixels is on the milliseconds time scale (not shown). When using the slow scan speed when the PMT reads high countrates there appears to be an overshoot of the intensity measurement followed by a negative intensity reading likely due to the amplifier, resulting in a negative correlation (Fig. 3C). The reason for this phenomenon is not clearly understood at this time, however, by deleting the first two pixels in the ACF there is minimal effect On the analysis since this overshoot occurs rapidly. As expected for correlations that depend only on the integration time of the detector, the magnitude of the correlation was independent of the image resolution or optical zoom (Fig. 3, compare Zoom 1, 5 and 10). However, if the digitization rate was changed by increasing the pixel sampling time (i.e. using slower scan speeds), the magnitude of the detector correlation decreased (Fig. 3, compare fast, medium and slow scan speeds). Thus, on a 2-µs time scale there is not enough time for the analogue detection to reset itself before collecting the next data point. This leaves a residual signal from the previous data point that will be correlated with the following data point. To eliminate this ‘bleed through noise’ from the RICS analysis, the first two points along the x-axis of the ACF as well as the zero lag point in the correlation function (due to the large shot noise peak at zero spatial lag)were deleted before curve fitting. The bleed through noise strongly affects the spatial and temporal resolution of the technique. For example, if the initial image resolution is 0.1µm after deleting the first two data points, then the actual initial data point along the scan axis in the ACF is at 0.3 µm, or 24 µs for the slow scan speed. It is crucial to remove these bleed through noise data points, before fitting the ACF because they can result in a lack of convergence of the fitting functions and the resulting error in g(0,0) can be on the order of 100% or more as seen for EGFP in solution (Fig. 3D). This is especially important when trying to measure fast dynamics (i.e. dye or protein in solution) when the ACF decays rapidly along the horizontal scan axis. The data shown here is specific for the Olympus Fluoview 300 (FV300), and therefore, detectors for other commercial systems will need to be characterized before RICS analysis can be performed. Preliminary data shows that the analysis can be performed on the FV1000 from Olympus, and the pixel bleed through is not observed. However, when possible it is best to use the photon counting setting of the FV1000 PMTs which avoids a number of analogue detector issues. For the Olympus system, it is best to use the 8 µs pixel dwell time to increase the S/N and minimize the bleed through noise while only slightly reducing the temporal resolution. On the FV1000 even longer pixel dwell times are available and should be used.
Fig. 3.
Characterization of the Olympus Fluoview 300 photomultiplier tube (PMT). Images were taken of the dark current reaching the PMT with the microscope set to the eyepiece so that no light can enter the sideport of the microscope and using variable scan speeds. Plots show the x-axis positive spatial co-ordinate for the ACF. The correlation in the first two points is due to bleed through noise from pixel to pixel. The effect is minimized by slowing down the scan speed which increases the detector integration time. Pixel dwell times are (A) fast = 2 µs; (B) medium = 4 µs; (C) and slow =8 µs. (D) Horizontal ACF for EGFP insolution with medium scan speed, 0.023 µm2 pixels, 2.12 ms between lines. ACF is the average of 500 ACFs calculated for 256 × 256 images.
Spatial resolution of RICS
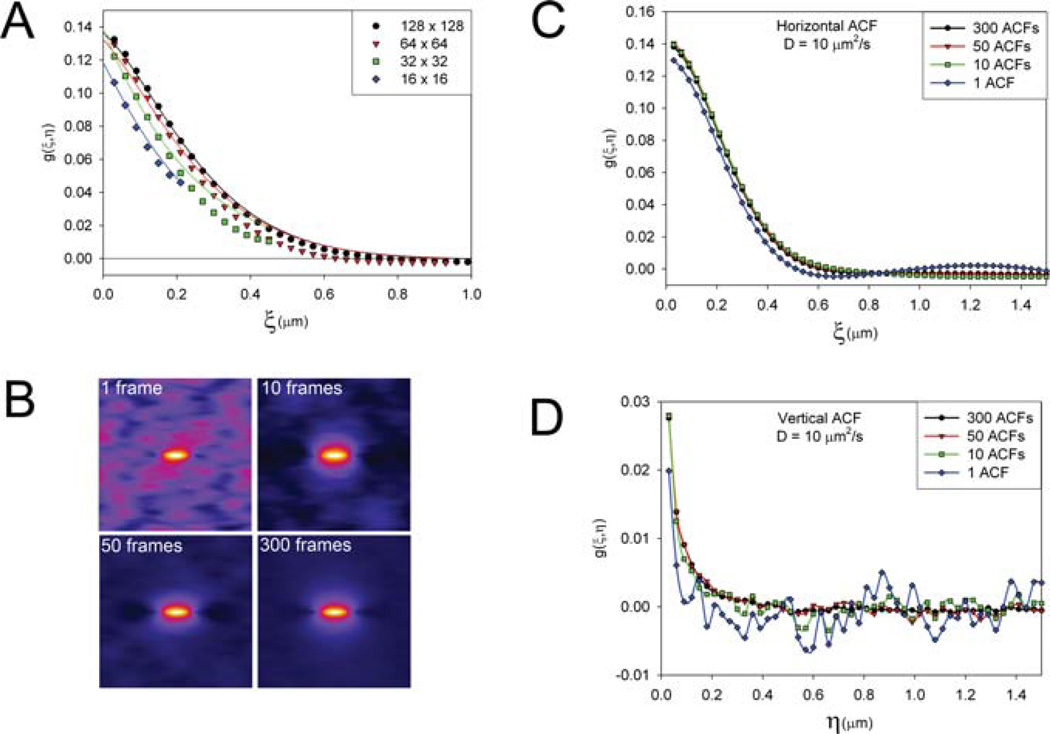
To determine the optimum and minimum image area for RICS analysis, the simulated data with D = 100 µm2s−1 were analysed using variable box sizes from 128×128 pixels (3.84 × 3.84 µm) to 16 × 16 pixels (0.48 × 0.48 µm). The correlation function along the x-axis is well defined for the 128×128 region, however with smaller regions; the correlation function no longer decays to zero due to under-sampling (Fig. 4A). This incomplete decay of the ACF, which is characteristic of under-sampling in spatial correlation analyses, results in an overestimation of D (Table 2). However, the y-intercept, or g(0,0), is only modestly affected by the under-sampling. Even for a very small, 16 × 16 sampling region, the N value calculated underestimated g(0,0) by only ~7% (Table 2). Thus, fairly accurate protein concentrations can be determined even on small areas (~0.25 µm2) of the cell as long as the extrapolated g(0,0) intercept value is used and the offset of the function (g∞), which is large due to under-sampling, is ignored.
Fig. 4.
Spatial limits and how many images need to be analysed for the RICS analysis. (A) Horizontal profile of the autocorrelation function (ACF) for D = 100 µm2 s−1 using variable sizes for the region of analysis. Solid lines are the RICS fit to the data. (B–D) Variable numbers of ACFs are averaged showing with fewer ACFs noise peaks appear at long correlation distances. Shown as intensity peaks around the periphery of ACF images (B),or shown as intensity peaks at long correlation distances along the horizontal (C) and vertical (D) ACFs. The simulated images were created with the following conditions: 0.03 µm pixels, 8 ms pixel dwell time, 7 ms between lines, 0.86 s between images and a laser beam radius of 0.3 µm.
Table 2.
RICS fitting values for variable box sizes for simulated data. Expected D is 100 µm2s−1 and expected g(ξ, ψ) is 0.140. Four separate ROIs were analysed and the data was averaged to get the values shown in the table
| ROI Size | g(ξ, ψ) | D µm2s−1 | % Error g(ξ,ψ) | % Error D |
|---|---|---|---|---|
| 128 | 0.1375 ± 0.0001 | 101 ± 1 | 1.8 | 0.6 |
| 64 | 0.134 ± 0.001 | 120 ± 10 | 4 | 21 |
| 32 | 0.142 ± 0.002 | 240 ± 10 | 1 | 137 |
| 16 | 0.131 ± 0.003 | 250 ± 20 | 7 | 152 |
Number of image frames for analysis
The optimal number of images needed to provide the statistics required to accurately determine and fit the ACF was investigated. As the number of frames decreases (from 300 to 1), the correlation function shape does not change markedly; however the error in each data point increases and the precision of the fit decreases (Figs 4C and D). The origin of the error is due to contributions from random correlation peaks within the ACF at large ξ,ψ values. When only one frame is used to calculate the ACF there are many random correlation peaks seen throughout the ACF in regions away from the central correlation peak where the function should decay to zero (Fig. 4B, particularly one frame analysis). These peaks are due to random overlap of signals when the images are shifted in x and y. The location of these peaks is truly random so averaging a few ACFs causes them to cancel out and the function decays to zero as expected. Averaging as few as ten ACFs drastically decreases the magnitude of the random correlation peaks (Fig. 4B). The random correlation peaks are more prevalent in the vertical ACF (Fig. 4D) where the amplitude of the ACF is smaller than the horizontal ACF (Fig. 4C). Using fewer frames for the analysis is advantageous when working with live cells because it reduces exposure to laser light, and increases the temporal resolution of the RICS measurement making it useful for following changes in protein concentrations and dynamics over time. Of course, the number of frames is tightly tied to the analysis box size; for a larger region fewer frames can be used increasing temporal resolution at the expense of spatial resolution, or vice versa. The balance will depend on the specific application and questions that need to be addressed.
Selection of temporal and spatial settings
In general, for spatial fluorescence correlation measurements oversampling at higher than the Nyquist Criterion is required. More specifically, the pixel size needs to be small enough to have a minimum of ~10 data points within the spatial-temporal decay of the ACF to allow accurate fitting of the data. For highly dynamic cytosolic proteins, the ideal pixel size is ≤0.05 µm for the FV300. The reason the pixel size has to be so small is partly because the first two data points along the scan axis have to be deleted due to detector bleed through noise discussed previously. Under these conditions the first data point in x is actually at 0.15 µm. On the FV300, for example, this pixel size can be obtained at various resolutions from 512 × 512 to 2048 × 2048 image sizes using the zoom feature and clip boxes to select regions of interest. If there is no detector bleed through (such as on the FV1000) the pixel size can be as high as 0.15 µm. The resolution can also be in this range for slower moving molecules where there is not a lot of movement during the pixel dwell time and the correlation function is truly 2D being less dependent on the horizontal fast scan axis data. In our experience the FV300 2048 × 2048 setting scans too slowly to be practical and results in extensive photobleaching and phototoxicity to the cells. Most of the experiments presented here were done using the 1024 × 1024 resolution setting at zoom 5 with a 256 × 256 clip box. In all cases, when optimizing image acquisition setting and ACF fitting parameters the spatial correlation function should be calculated and visualized to ensure there are enough data points, and that there is a good fit to the ACF.
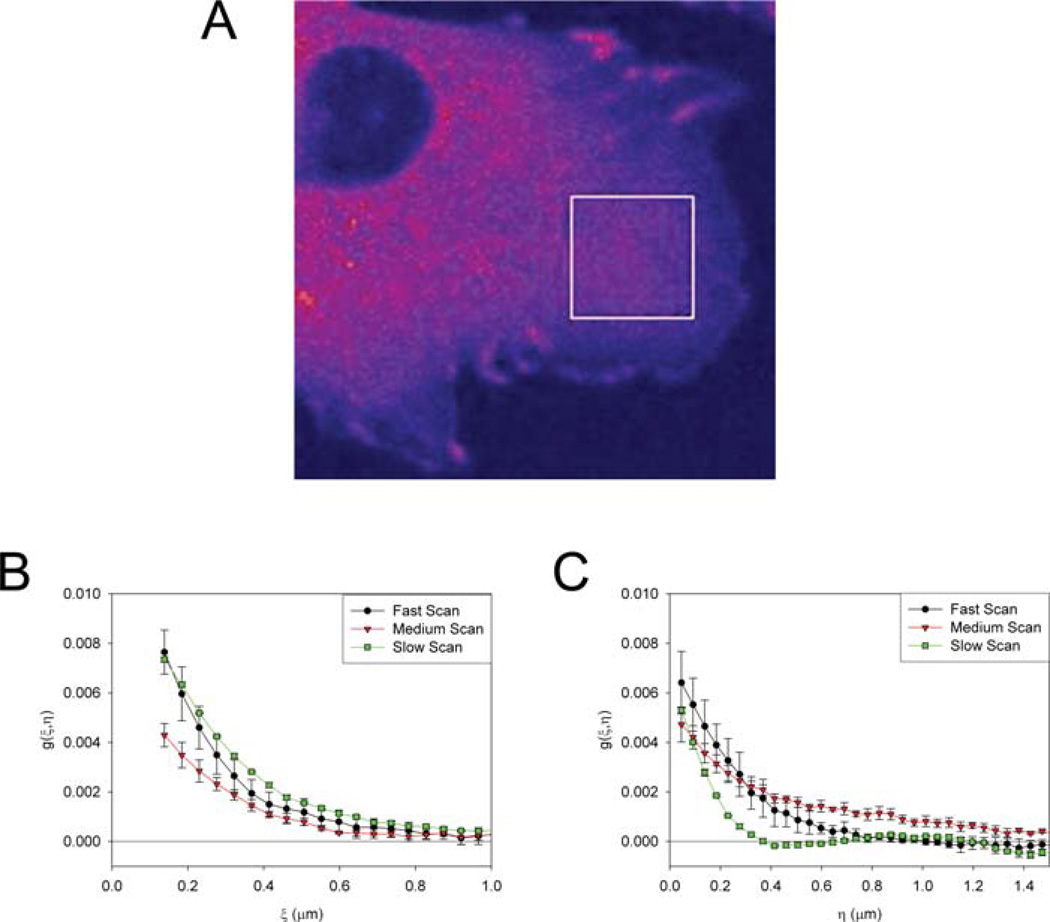
For the FV300 the three available scan speeds are fast, medium and slow, corresponding to 2, 4 and 8 µs pixel dwell times, respectively. At slower scan speeds the S/N increases due to the increased integration time, however there is a corresponding increase in the interline time. At faster scan speeds there are more data points before the ACF decays to zero, but due to the decreased S/N there is more error in each data point. To demonstrate these issues images of paxillin in the cytosol (Fig. 5A) were collected at the three scan speeds and analysed. The ACF data points (Figs 5B and C) for the fast scan (filled circles) have much larger error when compared to the medium (open triangle) or slow(filled square) scan speeds. However, all three scan speeds give enough data to accurately describe the ACF and determine the dynamics of paxillin in the cytosol. Note that these images were collected from three different cells so it is not surprising that the amplitudes and shapes of the ACFs, and hence the concentration and dynamics of paxillin differ somewhat. From the vertical ACFs is it clear that the ACF for the slow scan data decays quite rapidly due to the long interline time (Fig. 5C, filled square). In some cases, such as measurement of diffusion in solution, it may be advantageous to use a faster scan speed with more error in each data point in order to more accurately determine the 2D shape of the ACF along the scan axis. In this case, the interline time is of little significance because there is no correlation between lines and the ACF is essentially only along the horizontal axis (Fig.1E,Figs 2A and B black line). However, for slower moving molecules in the cytosol or the membrane of the cell where the dynamics are much slower than in solution it is better to go to slower scan speeds (8 µs pixels dwell time or longer) and improve the S/N. The S/N can also be increased by increasing the laser illumination power, however when working with cells it is best to keep the laser power low and the detector sensitivity (PMT gain) high to avoid adverse affects on the cell. The images may appear noisy; however, this thermal noise from the high detector gain does not contribute to the ACF because it is truly random. For living cells it is desirable to work with ~0.1–0.2% of the incident power of a 40 mW Ar ion 488 nm laser line. On other instruments and the newer FV1000 instrument scan speeds slower than 8 µs/pixel are available and should be used for cellular measurements. For measuring cytosolic protein movements a pixel dwell time of 20 µs is ideal while for slower motions such, as binding events, diffusion of large protein aggregates or membrane-associated proteins a pixeld well time of 100 µs is recommended.
Fig. 5.
Various scan speeds and the ACF. Calculations were performed on 50, 128 × 128 pixel images from four regions of interest and averaged. Error bars are SD. Images were of paxillin-EGFP in the cytosol of CHOK1 cells and were imaged at 1024 × 1024 resolution, zoom 5 with a 256 × 256 clip box. Pixel dwell time is 2 µs (fast), 4 µs (medium), 8 µs (slow), interline time was 1.608 ms (fast), 2.12 ms (medium), 3.152 ms (slow), 0.046 µ pixels. A representative Chinese hamster ovary (CHO) cell expressing paxillin-EGFP showing a typical 256 × 256 region of interest (A), horizontal ACF (B) and vertical ACF (C) for various scan speeds.
Measuring diffusion and concentration in solution from images
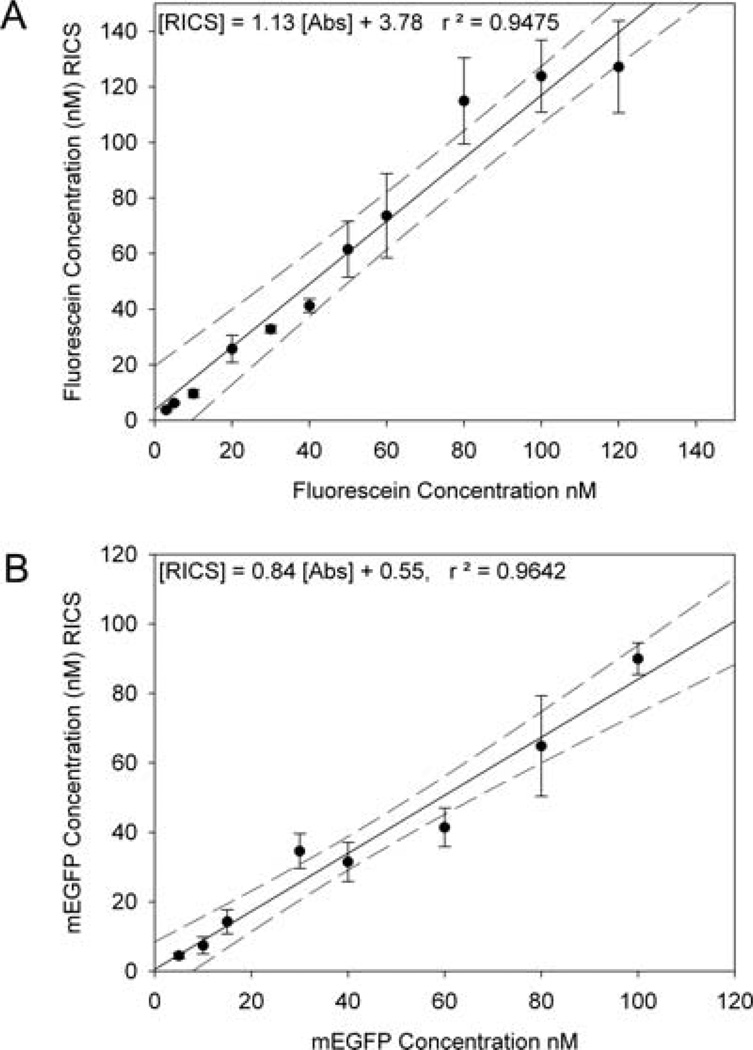
The accuracy of the RICS approach to determine concentrations and diffusion coefficients was assessed using the Fluoview 300 system with solutions of fluorescein and EGFP. Fluorescein concentrations (3 nM and 200 nM) were determined by absorbance and compared to concentrations determined by the RICS analysis (Fig. 6A; slope 1.13; r2 = 0.9475). Concentrations at and above 80 nM and below3 nM have higher errors associated with them, and the data points often lie outside of the 95% confidence intervals (Fig. 6A). At concentrations higher than 80 nM there are simply too many molecules within the excitation volume (>80) to accurately determine the magnitude of the fluctuations when molecules move into or out of the focal volume. For concentrations lower than 3 nM more frames need to be acquired for sufficient statistics to accurately determine and fit the ACF. If only the data between 3 nM and 80 nM are plotted there is a higher r2 value for the fit but the slope doesn’t change significantly (not shown). A 50 nM sample of fluorescein was used daily to measure the effective laser beam radius wo for calculating EGFP concentrations.
Fig. 6.
Standard curves from the RICS analysis for fluorescein (results from four separate experiments) (A) and mEGFP (B) in solution. Fluorescein data were collected with 0.023 µm pixels, 2 µs pixel dwell time, 1.608 ms between lines, at the fast scan speed, while mEGFP data were collected 0.023 µm pixels, 4 µs pixel dwell time, 2.12 ms between lines, at the medium scan speed. Error bars are SD for multiple experiments and 95% confidence intervals are shown by the dotted lines.
EGFP solutions ranging from 1–100 nM were prepared, imaged (Fig. 7A) and ACFs were calculated using SimFCS (Fig. 7B). A standard curve with the concentration of EGFP measured by the RICS analysis relative to a concentration measurement by absorbance showed a slope of 0.84 and an r2 of 0.9642 (Fig. 6B). Thus, RICS effectively measures protein concentration in the 1–100 nM range. However, the EGFP concentration is consistently under-estimated by 10– 20% from RICS. The focal volume used to determine the EGFP concentration is measured based on the laser beam radius, wo, determined from the fitting of fluorescein solution data and the assumption that the focal volume is a 3D Gaussian. It is possible that there is a systematic error in the focal volume measurement resulting in an underestimate of the protein concentration. The correlation techniques tend to give larger than theoretical PSFs or focal volumes which would result in an underestimate of the protein concentration.In addition, the underestimate of the concentration determined by RICS could also be due to the presence of some denatured (aggregated) or non-fluorescent EGFP, the tendency of EGFP to bind to the glass and the sides of the chamber, or localized photobleaching of the protein.
Fig. 7.
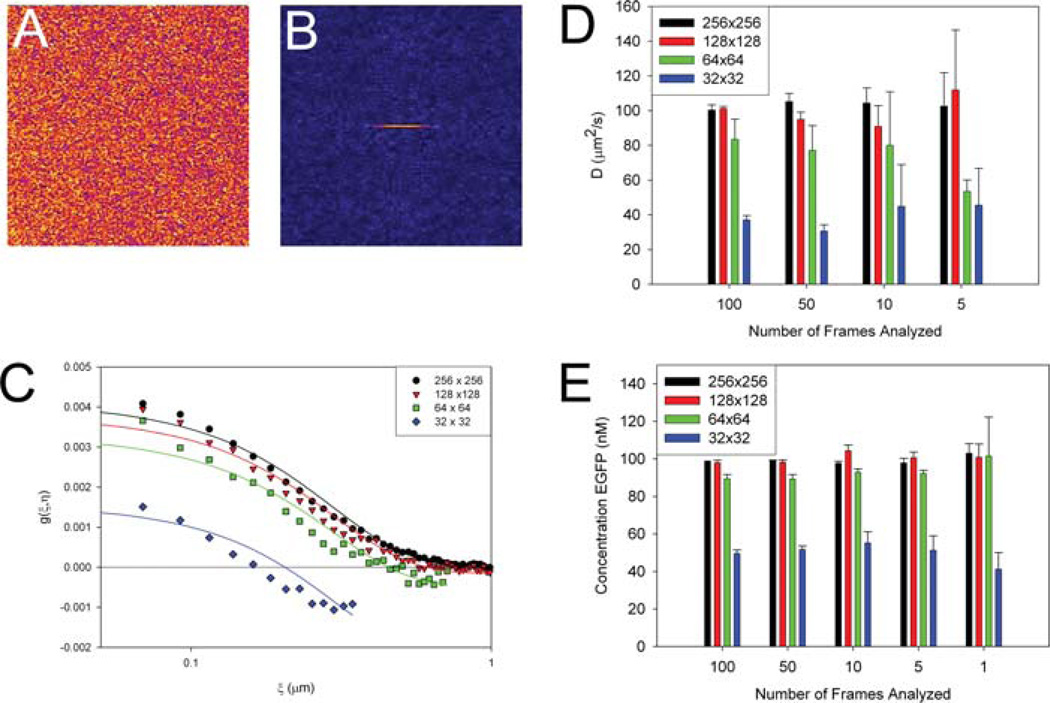
Image of EGFP in solution taken at fast scan speed (A), average spatial ACF for 100 images of EGFP in solution (B), fits to the horizontal spatial ACF for variable box sizes for the region of analysis (C). When under-sampled (i.e. box is too small) the ACF goes negative at longer correlation distances. Effect of ROI size and number of image frames analysed on the measured diffusion coefficient (D) and concentration (E) of EGFP in solution. Pixel size was 0.023 µm, 4 µs pixel dwell time, 2.12 ms interline time and 0.542 s between image frames. A laser beam radius of 0.295 µm was used for diffusion constant and EGFP concentration calculations.
The analysis shows that using smaller box sizes results in an ACF that underestimates the D value due to under-sampling. For example, with a 32 × 32 box (1.5 × 1.5 µm) the ACF becomes negative at longer correlation distances and is not fit well with the diffusion equations (Fig. 7C). For the fast motion of EGFP in solution the D value is more precise when a large region of interest (ROI) and number of frames are analysed (Fig. 7C) and compares well with published data (~100 um2 s−1, (Digman et al., 2005a)). When smaller regions of interest are used, the correlation functions often become negative and do not completely level off at long distances (Fig. 7C). However, a reasonable estimate of the D value (with an error of 10–20%) can also be obtained with as few as 5–10 image frames when a large ROI (at least 128 × 128 or 2.94 × 2.94 µm) is used (Fig. 7D).
In contrast to the diffusion measurements, the concentration of EGFP determined from the amplitude of the correlation function, G(0,0), and Eq. (4) is much less sensitive to the ROI size and the number of frames analysed (Fig. 7E), as seen in the simulations. The concentration measurement for a 100 nM EGFP solution is accurate to a single frame for the large ROIs (256 × 256 and 128×128). Furthermore, as few as five frames with regions as small as 64 × 64 (2.94 × 2.94 µm) still give useful concentration estimates (Fig. 7E). Thus, it possible to follow changes in concentration over time in small regions of the cell with a time resolution of only ~5 image frames. Using small ROIs can give concentrations on a sub-second time scale for this kind of measurement (up to eight frames per second for 64 × 64 box with fast scanning for the FV300).
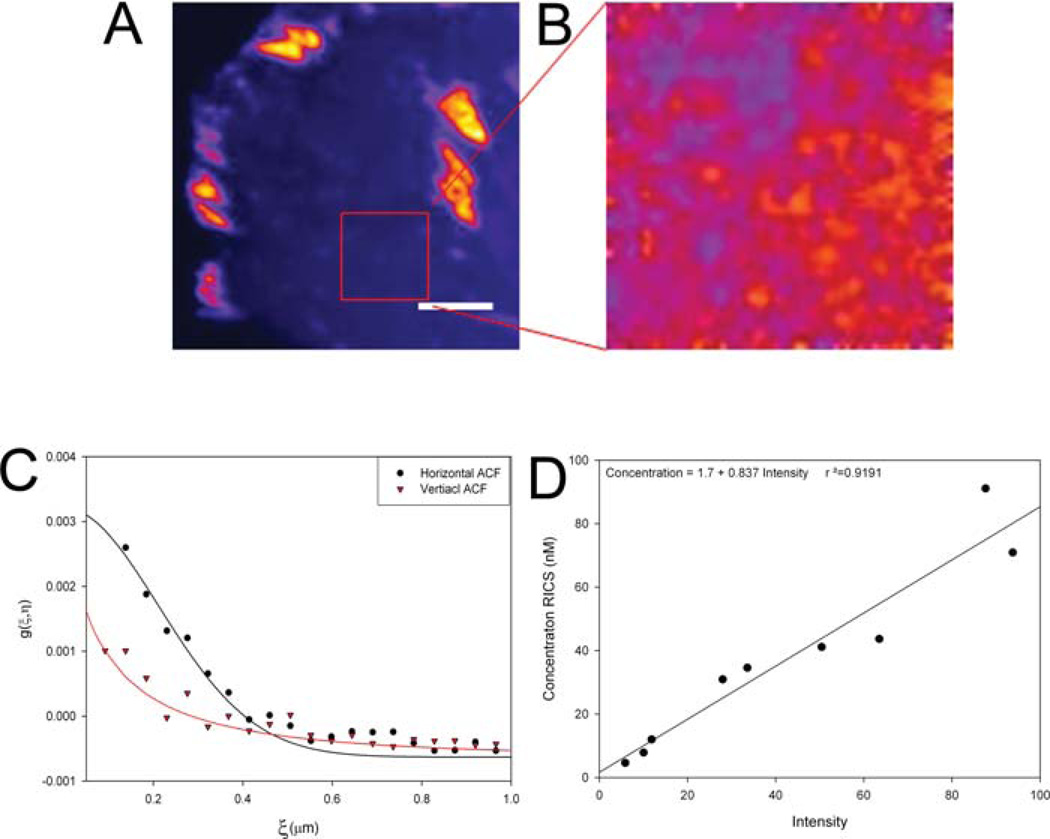
Measuring diffusion in a cell
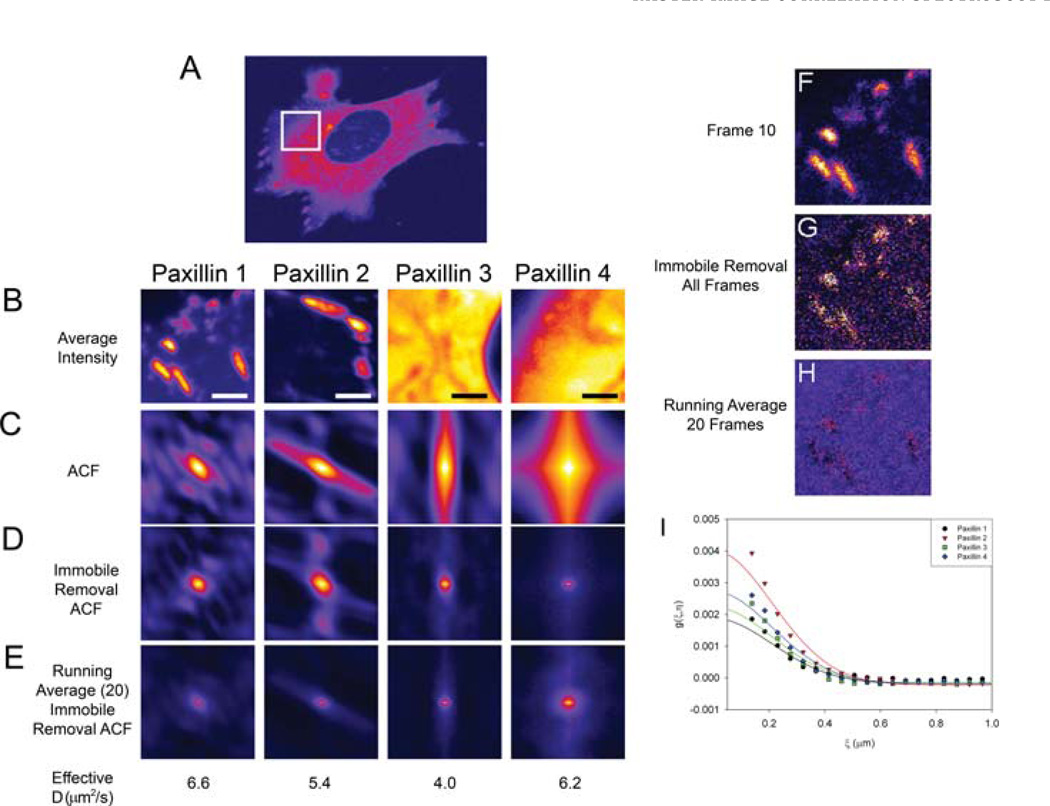
Measurements in cells are more challenging than those in homogenous solutions. Cells display many large-scale spatially correlated features (Fig. 8A), which include adhesions (Fig. 8B, Paxillin 1,2), stress fibres, microtubules, edges, organelles (Fig. 8B, Paxillin 4) and the nucleus (Fig. 8B, Paxillin 3). In addition, volume variations, due to the thickening of the cell as one moves from the edges toward the nucleus, can also present problems (Fig. 8B, Paxillin 4). These long-range spatial structures will appear as long-range spatial correlations and will dominate the spatial ACF (Fig. 8C). These immobile structures can be filtered from the images in several ways. One is to compute the average intensity of all the image frames and subtract the resulting average image from each of the other image frames. The average image intensity is then added back to each pixel to avoid negative pixel intensities (Digman et al., 2005a,b). For example, if the average intensity were 1000 units within an adhesion then as molecules move into and out of the region the intensity would fluctuates over a narrow range, for example, from 990 to1010. These fluctuations correspond to only 1% of the average signal. However, if the immobile removal algorithm is applied the average intensity of the mobile fraction is 20 units and the intensity fluctuations is from 10– 30 or up to 50% variation. This makes the ACF sensitive to changes in intensity due to molecular motions, similar to FCS. Using the average image of the stack to correct for the immobile fraction is functionally equivalent to subtracting the zero-order component using Fourier transform methods (Hebert et al., 2005).
Fig. 8.
RICS calculations were performed for paxillin-EGFP in CHO cells (A). Images were collected with a pixel size of 0.046 µm, 4 or 8 ms pixel dwell time, 2.12 or 3.15 ms interline time. The average intensity (B), spatial ACF with no immobile component removal (C) and with the immobile component removed for the time series average (D) or using a twenty frame moving average (E). Correlation functions were fit using one diffusion component fits and fitting 64 data points within the correlation curve. All images are presented as averages of 200 images (B) or 200 ACFs (C–E). Frame 10 from the time series of images of paxillin-EGFP in a CHO-K1 cell (F); frame 10 and after the immobile removal algorithm using all 200 image frames (G); after the immobile removal algorithm using 20 image frames (H). Horizontal ACF curves for the 2D fits are shown by the symbols with the fits as solid curves (I). Scale bar is 2 µm.
However, if the cell (see Paxillin 1, Supplemental movie 1) or its organelles (see Paxillin 4, Supplemental Movie 2) move during image acquisition, then subtracting an average of all frames within the movie will result in a broadening of the ACF (Fig. 8D). The broadening is particularly apparent when there are bright structures like adhesions present in the images (compare Fig. 8D, Paxillin 1,2 and Paxillin 3,4). This broadening is due to the fact that the movements of adhesions within the cell prevent complete removal of the immobile components. This can be visualized as adhesion patterns within a single plane that has been corrected by subtraction of the average intensity image (Figs 8F and G). However, if a moving average immobile removal algorithm is used so that the average of a few frames from the movie is subtracted (20 in this case) rather than the average for the whole image time series then the adhesions are no longer visible in the corrected image (Fig. 8H) and the broadening artifact is not observed in the ACF (Fig. 8E) because only small fluctuations due to the mobile protein are analysed. The ACFs calculated using the running average immobile correction (Fig. 8E) were fit to a single diffusing component and gave diffusion constants for paxillin (4–7±1 µm2 s−1) that compare well with FCS measurements (Digman et al., in press). However, the ACFs reflect the complex, non-homogenous dynamics of paxillin and thus are not adequately described by the single component model used to fit the ACF from solution measurements (Fig. 8I).
Measuring concentration in the cell
Measuring protein concentrations in the cell can be difficult due to the large spatial features discussed above. In addition, the scaling of the amplitude of the ACF changes when applying the immobile removal algorithm, and the G(0,0) value can no longer be used as a direct measure of the protein concentration. Simulations show that having immobile structures within the images result in the underestimation of G(0,0) values, and an overestimation of the concentration. This arises because G(0,0) is normalized by the average intensity squared (Wiseman et al., 2000). Due to the intensity dependence of G(0,0) the error is systematic and depends on the total brightness of the immobile component, not the physical size of the structures (not shown). As expected, there is little effect on the diffusion coefficient determined with an immobile component because dynamics are not dependant on the image intensity (not shown). This allows the motion of molecules of interest to be characterized in a very complex environment.
To correctly determine the concentration of soluble paxillin within cells in the presence of immobile structures the immobile removal algorithm needs to be modified. Normally, the average intensity of the image before immobile removal is added back to each pixel to avoid having negative pixel intensities.However,a large part of the average image intensity is due to intensity coming from large bright adhesions, and therefore in regions of the cell where there is only cytosolic mobile protein, the intensity is overestimated. However, if the average intensity in a region of the cell with no immobile structures is added back to the image overestimation of the intensity due to bright immobile structures will be avoided and the ACF will be properly normalized and the concentration of protein can be determined. This technique can be used to determine concentrations of mobile EGFP-labelled proteins within the cell. While this method can be used to determine protein concentrations across the cell it will only apply to mobile protein populations. In paxillin-EGFP expressing cells using multiple reference regions leads to ~7% variation (standard deviation) in the mobile protein estimate. This is due to heterogeneity of paxillin concentrations across the cell that we have also observed in FCS experiments (Digman et al., in press). There is also variability in protein concentration overtime for a given location within the cytosol (~14%). So it is best to choose a reference region with no immobile protein near the region of interest and also during the time frame of interest.
To estimate protein concentrations within large immobile structures like adhesions, the system can then be calibrated by generating a standard curve similar to Fig. 6B using EGFP in solution or in cells with variable expression levels of soluble EGFP. Then concentrations can simply be estimated using fluorescence intensity as long as all instrument settings are kept constant between calibration and data collection and that the laser power of the system is stable.
To demonstrate that concentrations of mobile paxillin-EGFP can be measured, (Fig. 9A) a cytoplasmic region of the cell with little or no immobile fraction was chosen (Fig. 9B, corresponding to the box in Fig. 9A) and RICS was performed without using the immobile algorithm giving a paxillin concentration of 44 nM (Fig. 9C). However, paxillin monomers or clusters within the region of interest as long as it is mobile. The total concentration of paxillin monomers (equivalents) was determined using the intensity data for this region of the cell and the solution EGFP standard curve (Fig. 9D), giving a concentration of 120 nM. These two measurements are complimentary; taken together they reveal that on average paxillin resides in aggregates of 2–4 proteins in this region of the cell. Indeed, photon counting histogram (PCH) of paxillin in the cytosol of these same cells shows that it is often found in mixed populations of monomers and clusters (Digman et al., in press).
Fig. 9.
RICS calculations were performed for paxillin-EGFP in CHO cells (A). Images were collected with a pixel size of 0.046 µm, 2 ms dwell time, and 1.608 ms interline time. RICS was done on 200 frames from a 64 × 64 pixel square region (red box) shown in Fig. (B). The ACF was fit using a one diffusion component fit. Horizontal ACF is shown with black symbols and a solid black line for the fit (C); the vertical ACF is shown with red symbols and a red solid line shows the fit (C). The EGFP intensity and concentration curve was used to estimate the absolute concentration of paxillin EGFP from the average intensity of the region of interest (D). Scale bar is 2 µm.
Conclusion
With attention to proper instrument settings and fitting algorithms, RICS provides a very powerful tool for measuring the dynamics of proteins, lipids and other molecules in cells using commercial confocal microscopes. This paper presents general guidelines for performing RICS measurements and illustrates their use on an Olympus FV300 laser scanning confocal microscope. A summary of the recommend instrument settings and analysis parameters is presented (Table 3). In general, RICS can be performed on any confocal system, but care must be taken to characterize the nature of the analogue detectors to ensure the instrument is not introducing correlation by filtering the data. This can be done simply by looking at the ACF for images taken when no signal is present. Basically, no patterns should be detected in the ACF and all of the data points should be close to zero aside from the central g(0,0) noise peak. When possible it is best to work with photon counting detectors to avoid issues with analogue detection. While these analyses have focused on diffusion and concentration, other dynamics accessible by correlation methods, such as binding interactions, will be accessible with modifications of the fitting models presented here. Correlation tools now provide a means to generate highly quantitative maps of molecular properties during cellular phenomena and the fact these measurements can be done on commercial CLSMs make them very accessible to researchers in the life sciences.
Table 3.
Recommended instrument setting and analysis parameters for the RICS analysis
| Parameter | Recommended |
|---|---|
| Magnification | 60× or 100× (1.4 NA) |
| Pixel dwell time | ≥8 µs |
| Pixel size | ≤0.05 µm (0.1 µm for FV1000) |
| Laser power | ≤2% |
| (30–40mW488nmor or argon ion laser) |
|
| PMT voltage | ~800 V (use photon counting mode on FV1000) |
| Offset | ~0 (or set to ensure no pixels read 0 intensity) |
| Number of images | ≥5 (high S/N) |
| ≥10 (low S/N) | |
| Analysis region of interest size | ≥ 2 µm × 2 µm |
Acknowledgements
These studies were supported by the Cell Migration Consortium U54GM64346 (CB,RD,MD,AH,EG). GM23244 (CB,RH). BH was supported by a PGS fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada. The Laboratory for Fluorescence Dynamics (LFD) at the University of California, Irvine (UCI) is supported jointly by the National Center for Research Resources of the National Institutes of Health (PHS 5 P41-RR003155) and UCI.
References
- Brown CM, Hebert B, Kolin DL, Zareno J, Whitmore L, Horwitz AR, Wiseman PW. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J. Cell. Sci. 2006;119(Pt 24):5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- Chen Y. PhD Thesis. University of Illinois; 1999. Analysis and applications of fluorecence fluctuation spectroscopy. [Google Scholar]
- Digman MA, Brown CM, Horwitz AF, Mantulin WW, Gratton E. Paxillin Dynamics Measured During Adhesion Assembly and Disassembly by Correlation Spectroscopy. Biophys. J. doi: 10.1529/biophysj.107.104984. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys. J. 2005a;89(2):1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Sengupta P, Wiseman PW, Brown CM, Horwitz AR, Gratton E. Fluctuation correlation spectroscopy with a laser-scanning microscope: exploiting the hidden time structure. Biophys. J. 2005b;88(5):L33–L36. doi: 10.1529/biophysj.105.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert B, Costantino S, Wiseman PW. Spatiotemporal image correlation Spectroscopy (STICS) theory, verification, and application to protein velocity mapping in living CHO cells. Biophys. J. 2005;88(5):3601–3614. doi: 10.1529/biophysj.104.054874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegener O, Prenner L, Runkel F, Baader SL, Kappler J, Haberlein H. Dynamics of beta2-adrenergic receptor-ligand complexes on living cells. Biochemistry. 2004;43(20):6190–6199. doi: 10.1021/bi035928t. [DOI] [PubMed] [Google Scholar]
- Laukaitis CM, Webb DJ, Donais K, Horwitz AF. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell. Biol. 2001;153(7):1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D, Elson EL, Webb WW. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13(1):29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Chen Y, Gratton E, Glaser M, Mantulin WW. Cellular characterization of adenylate kinase and its isoform: two-photon excitation fluorescence imaging and fluorescence correlation spectroscopy. Biophys. J. 2002;83(6):3177–3187. doi: 10.1016/S0006-3495(02)75320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell. Biol. 2004;6(2):154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- Webb WW, Barak LS, Tank DW, Wu ES. Molecular mobility on the cell surface. Biochem. Soc. Symp. 1981;46:191–205. [PubMed] [Google Scholar]
- Wiseman PW, Brown CM, Webb DJ, Hebert B, Johnson NL, Squier JA, Ellisman MH, Horwitz AF. Spatial mapping of integrin interactions and dynamics during cell migration by image correlation microscopy. J. Cell. Sci. 2004;117(Pt 23):5521–5534. doi: 10.1242/jcs.01416. [DOI] [PubMed] [Google Scholar]
- Wiseman PW, Squier JA, Ellisman MH, Wilson KR. Two-photon image correlation spectroscopy and image cross-correlation spectroscopy. J Microsc. 2000;200(Pt 1):14–25. doi: 10.1046/j.1365-2818.2000.00736.x. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296(5569):913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]