Following our recent global survey of all structures deposited in the PDB,1 we focused our attention on the models with sequences different from that reported by the standard genetic analysis. One of the examples belonging to this class is N10-formyltetrahydrofolate synthetase from Moorella thermoacetica. In a recent Protein Science article, the authors concluded their studies with a proposal for this enzyme's catalytic mechanism.2 The structures presented in the article reiterated the initially proposed two amino-acid deletion that was proposed nearly 12 years ago to explain the experimental densities derived at that time. Here, we investigated the divergence between the reported sequence in M. thermoacetica N10-formyltetrahydrofolate synthetase and the sequence derived from the crystal structure of this enzyme. The refinement of the recently published structures2 using high-resolution data showed four fragments of the model that were mistraced in the vicinity of the active site [Fig. 1(A, B)]. The mistraced portion constitutes nearly 10% of the structure. These errors resulted in a cascade of inaccuracies that raised concerns about main findings of this recent article.2
Figure 1.
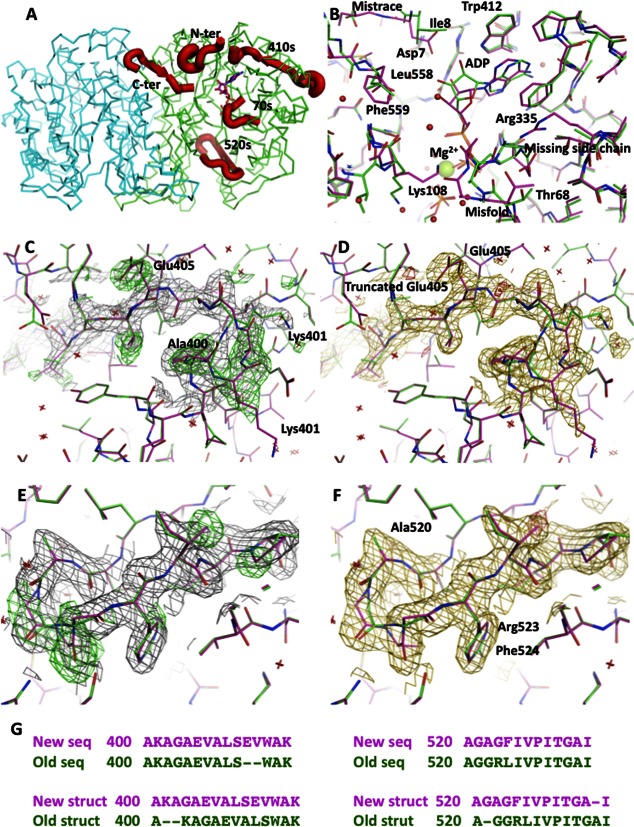
(A) Model of the dimer of N10-formyltetrahydrofolate synthetase in carbon alpha representation with four misthreaded fragments marked in thick red ribbon. The stick representation of ADP marks the active site. (B) Superposition of the original and corrected structure with ADP at 2.5 Å resolution. The corrections lead to a significant repositioning of the ADP and a proper characterization of the metal ion as Mg2+. (C) The original model in green in the 400–410 region superimposed on the new model in purple, covered with the electron density calculated from the original model. The positive difference electron density contoured at 2.8 sigma level is in green. (D) The superimposed models at the same region as in (C) covered with the electron density calculated from the new model. The difference electron density contoured at 2.8 sigma level is in red. (E) The original model in green in the 520–530 region superimposed on the new model in purple, covered with the electron density calculated from the original model. The positive difference electron density contoured at 2.8 sigma level is in green. (F) The superimposed models at the same region as in (C) covered with the electron density calculated from the new model. The difference electron density contoured at 2.8 sigma level is in red. (G) The sequence corrections for 410 and 520 regions indicate the sequence variant for a clone ATCC 39073. Original sequence is in green and the new sequence is in purple, which corresponds to colors used in panels B–F. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
The two amino-acid deletion was proposed to explain insufficient density to fit the full reported sequence. However, the site of deletion contained highly conserved Ser residue and was located at the beta strand. In the original report, it was claimed that “there was no density for residues 410 and 411.”3,4 Motivated by the need to reconcile the sequence derived from the crystal structure with the gene sequences of these protein, and to clarify this issue, we retrieved the model coordinates and the diffraction data for the recently reported, the highest resolution structure of this enzyme determined at 2.2 Å resolution (3PZX), and initiated a refinement.
After 10 cycles of LSQ refinement in Refmac,5 an inspection of the electron density showed clearly that the existence of additional residues at the end of the beta strand containing the deletion. Moreover, a closer inspection of the proposed deletion site showed that some side chains (in particular Glu405), which were directed to the interior of the protein, were truncated in the deposited model to avoid a discrepancy with the electron density. During the refinement, the pattern of positive and negative difference electron density patches disappeared after introduction of the two missing residues (SE) that suggested a correct sequence modeling [Fig. 1(C, D)]. The resulting model also provided a better fit to the electrostatic signature of the protein surface with Glu405 radiating to the exterior of the molecule and complementing other surface charges. The introduction of the additional two residues shifted the sequence of 10 residues and created an additional turn at the preceding helix.
This successful correction, and achieved reconciliation, with existing NCBI sequences encouraged us to carry out a detailed inspection of the entire model. A careful inspection of the entire model showed that the N- and C-terminal portions, as well the region 520–533, were surrounded by significantly elevated difference densities. Subsequent refinement showed that all the mentioned regions contained misthreaded and missing residues. The sequence of the N-terminal region was shifted by a single residue up to Lys15. Additionally, after the corrections of the initially displaced residues, the density appeared to allow for introduction of two more residues at the beginning of the sequence. The final model has all the residues from Lys3 to Lys15 in full atom representation [Fig. 1(A)].
A similar story happened with the C-terminal portion of the model. The residues starting at Asp552 were misaligned with the electron density. A correction allowed for a full tracing of the entire C-terminal portion including Phe 559. A slightly different course of action was needed to correct the fourth fragment (520–533). The electron densities suggested that the sequence of the entire fragment was shifted by a single residue [Fig. 1(E, F)]. However, the required shift was placing the Arg residue at the site, where Gly was initially refined. A closer inspection of the electron densities suggested a significant change of the sequence. A fragment GGRL was becoming GAGF and the single amino acid shift was placing a Phe residue at the site, where previously Arg side chain was refined [Fig. 1(G)]. The new placement with a new sequence provided a far superior fit to the electron densities, therefore, it was deemed correct. A closer look at the original model showed that the Arg side chain was refining as a replica of an aromatic residue that had the guanidine group in a semicircular conformation before [Fig. 1(E, F)].
An inspection of the available sequences at the NCBI website provided clues to explain all these changes. The new sequence represented a different gene variant of the M. thermoacetica N10-formyltetrahydrofolate synthetase. This sequence was not available in 2000, when the original crystal structure was published.6,7 The new variant that was published in 2008 fit the electron densities and explained all the residual electron density features. These corrections are of certain importance as the N- and C-terminal portions protrude into the active site cavity, and the introduced C-terminal fragment plays a role in substrate placement by immobilizing the strand responsible for the formation of the active site. All the above corrections to the mistraced sequences precipitated further changes that upon correcting, allowed for identification of conformational changes associated with binding of substrates/inhibitors.
Discovery and resolution of problems in the model of the enzyme at the highest resolution, prompted the reevaluation of other accompanying models (3QB6, 3SIN, 3RBO) containing the substrates and/or inhibitors. We present in Table I the refinement parameters for the original and rerefined models. Corrections improved the refinement statistics for all the models, but a particular improvement was made for the 2.5 Å resolution model of the enzyme with the substrate ADP and phosphate (3RBO). The placement of the ADP was significantly influenced by the corrections, and there is strong evidence supporting the presence of formylphosphate, but in a post-catalytic (rotated conformation). The originally refined conformation was in agreement with the in-line attack mode of the activated formate. In our model, a water molecule occupies this axial position. The electron density and the resulting temperature factors are more consistent with a rotated position of the formylphosphate that is needed for the next step of the proposed reaction. Additionally, the metal ion refined as sodium was refined as Mg2+ not Na+ ion. This change was suggested by the coordination scheme, the distances in a strict octahedral coordination, and the resulting temperature factors, that fully support this choice of the metal ion.
Table I.
Crystallographic Data and Refinement Statistics for the Original and Newly Refined Models of M. thermoacetica N10-Formyltetrahydrofolate Synthetase
| Ligands native | FTHFS | ADP/XPO | ZD9/XPO | Folate |
|---|---|---|---|---|
| Original PDB code | 3PZX | 3RBO | 3SIN | 3QB6 |
| Space group | R32 | P21212 | R32 | R32 |
| Unit cell dimensions | ||||
| a (Å) | 161.20 | 91.17 | 162.37 | 160.99 |
| b (Å) | 161.20 | 212.97 | 162.37 | 160.99 |
| c (Å) | 256.92 | 53.44 | 258.07 | 256.61 |
| Resolution (Å) | 2.2 | 2.50 | 2.67 | 3.0 |
| Average redundancy | 4.8 | 3.8 | 4.5 | 9.7 |
| Average I/r | 10.3 | 8.0 | 9.4 | 7.8 |
| Completeness (%) | 99.8 | 91.5 | 91.1 | 98.8 |
| (Outer shell) (%) | (99.9) | (64.8) | (87.4) | (100) |
| Total R-merge (%) | 7.6 | 8.5 | 4.9 | 16.5 |
| R-value | ||||
| Old R-value (%) | 20.7 | 23.3 | 20.5 | 23.0 |
| New R-value (%) | 18.0 | 17.3 | 19.4 | 22.0 |
| Old Rfree-value (%) | 26.0 | 30.0 | 29.3 | 28.3 |
| New Rfree-value (%) | 23.0 | 25.7 | 27.7 | 29.0 |
| RMSD | ||||
| Old bond lengths (Å) | 0.02 | 0.02 | 0.02 | 0.01 |
| New bond lengths (Å) | 0.018 | 0.011 | 0.011 | 0.010 |
| Old bond angles (deg.) | 2.2 | 2.0 | 1.7 | 1.6 |
| New bond angles (deg.) | 1.93 | 1.65 | 1.56 | 1.49 |
| Subunit A average B-factors | ||||
| Old B-values (Å2) | 29.6 | 41.1 | 45.2 | 24.8 |
| New B-values (Å2) | 29.6 | 37.9 | 52.8 | 23.6 |
| Compounds (occupancy) | SO4 | ADP, XPO | ZD, XPO | Folate (0.75) |
| Old B-values (Å2) | 52.3 | 51.0, 67.6 | 73.8, 48.2 | 71.9 |
| Compounds (occupancy) | SO4 | ADP, XPO | ADP(0.6), ZD9(0.4), XPO(0.4) | |
| New B-values (Å2) | 36.4 | 32.2, 44.5 | 67.7, 70.5, 76.8 | 50.2 |
| Subunit B average B-factors | ||||
| Old B-values (Å2) | 49.9 | 36.8 | 67.8 | 50.6 |
| New B-values (Å2) | 50.5 | 34.7 | 74.8 | 55.6 |
| Compounds (occupancy) | SO4 | ADP, XPO | ||
| Old B-values (Å2) | 63.4 | 51.2, 59.5 | ||
| Compounds (occupancy) | SO4 | ADP, XPO | ADP(0.6) | |
| New B-values (Å2) | 67.5 | 31.8, 43.3 | 70.8 | |
| New PDB code | 4IOJ | 4IOK | 4IOL | 4IOM |
An even more radical change was required in the model of the complex with the inhibitor 2,7-dimethyl-6-[(prop-1-yn-1-ylamino)methyl]quinazolin-4(3H)-one (ZD9). There is substantial evidence that the density represents a mixture of the ADP and the inhibitor. Our refinement converged with ADP in 0.6 occupancy, and superposed with the ZD9 in 0.4 occupancy in an active site of subunit A. The situation is more complex in subunit B that has less well-defined electron density, but still showed a clear density for ADP and phosphate. It is possible that the inhibitor molecule is bound in a mixed occupancy state, but the generally weaker density makes it impossible to model it with any level of certainty. In general, the models in the R32 space group show the electron density being much weaker and the temperature factors much higher in the B subunit of the enzyme than in subunit A (3PZX, 3SIN, 3RBO). The close packing contacts of subunit B, high temperature factors, and fuzzy densities suggest that the R32 symmetry is most likely violated and that true symmetry of the lattice is R3. This conclusion is partially supported by the excessively high Rmerge in one of the presented crystal structures (16% in 3QB6) that contains a folate. This crystal structure was determined at 3 Å resolution.
During our investigation, we have inspected all the corresponding models in PDB-Redo.8 To our disappointment, all of these models contain the errors described above. Despite the laudable effort that goes into validation and model checking procedures we, as a community, do not have an effective mechanism for spotting and correcting errors like these described in this presentation. Significantly different, but only modestly better refinement parameters, do not allow for immediate discrimination of correctly determined structures, leading to many notable recent corrections.9
In conclusion, the derived model represents a different sequence variant from the original structure. The refinement with the correct sequence leads to numerous model improvements including significant displacements or changes in identities of the bound at the active site ligands. The most significant implication of this study is that these errors were not detected by the newest tools and services including PDB-Redo and that the authors, despite improvements in technology over the last 10 years since the first structure was published, failed to detect and correct the structural errors. These results underscore the growing need for better methodology for structure validation and its application before publishing. As this work clearly demonstrates, we have an urgent need for devising such methods, as many high profile results that are presented at lower resolution can only gain in its early validation and save many hours wasted for unsuccessful reproductions.9
REFERENCES
- 1.Prasad BVLS, Zhang Y, Godzik A, Stec B. Global distribution of conformational states in redundant models in the PDB points to non-uniqueness of protein structure. Proc Natl Acad Sci USA. 2009;106:10505–10510. doi: 10.1073/pnas.0812152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celeste LR, Chai G, Bielak M, Minor W, Lovelace LL, Lebioda L. Mechanism of N10-formyltetrahydrofolate synthetase derived from complexes with intermediates and inhibitors. Protein Sci. 2012;21:219–228. doi: 10.1002/pro.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radfar R, Shin R, Sheldrick GM, Minor W, Lovell CR, Odom JD, Dunlap RB, Lebioda L. The crystal structure of N(10)-formyltetrahydrofolate synthetase from Moorella thermoacetica. Biochemistry. 2000;39:3920–3926. doi: 10.1021/bi992790z. [DOI] [PubMed] [Google Scholar]
- 4.Radfar R, Leaphart A, Brewer JM, Minor W, Odom JD, Dunlap RB, Lovell CR, Lebioda L. Cation binding and thermostability of FTHFS monovalent cation binding sites and thermostability of N10-formyltetrahydrofolate synthetase from Moorella thermoacetica. Biochemistry. 2000;39:14481–14486. doi: 10.1021/bi001577w. [DOI] [PubMed] [Google Scholar]
- 5.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 6.Lovell CR, Przybyla A, Ljungdahl LG. Primary structure of the thermostable formyltetrahydrofolate synthetase from Clostridium thermoaceticum. Biochemistry. 1990;29:5687–5694. doi: 10.1021/bi00476a007. [DOI] [PubMed] [Google Scholar]
- 7.Pierce E, Xie G, Barabote RD, Saunders E, Han CS, Detter JC, Richardson P, Brettin TS, Das A, Ljungdahl LG, Ragsdale SW. The complete genome sequence of Moorella thermoacetica. Environ Microbiol. 2008;10:2550–2573. doi: 10.1111/j.1462-2920.2008.01679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joosten RP, Joosten K, Murshudov GN, Perrakis A PDB_REDO: constructive validation, more than just looking for errors. Acta Cryst D. 2012;68:484–496. doi: 10.1107/S0907444911054515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang G, Roth CB, Reyes CL, Pornillos O, Chen YJ, Chen AP. Retraction Science. 2006;314:1875. doi: 10.1126/science.314.5807.1875b. [DOI] [PubMed] [Google Scholar]