Abstract
Epsins are eukaryotic, endocytic adaptor proteins primarily involved in the early steps of clathrin mediated endocytosis. Two epsins exist in Saccharomyces cerevisiae, Ent1 and Ent2, with single epsin knockouts being viable, while the double knockout is not. These proteins contain a highly conserved Epsin N-terminal homology (ENTH) domain that is essential for cell viability. In addition, overexpression of the ENTH domain of Ent2 (ENTH2) was shown to play a role in cell division by interacting with the septin organizing, Cdc42 GTPase activating protein, Bem3, leading to increased cytokinesis failure. In contrast, overexpression of the ENTH domain of Ent1 (ENTH1) does not affect cytokinesis, despite being 75% identical to ENTH2. An ENTH2N112D, S114E, E118Q mutant that switches residues in loop 7 to those found correspondingly in ENTH1 was incapable of inducing the cytokinesis phenotype. In order to better understand the role of loop 7 in the ENTH2-induced phenotype at a molecular level, X-ray crystallography was used to elucidate the structures of yeast ENTH2WT and ENTH2DEQ. Our results indicate that mutations did not affect the conformation of loop 7, but rather introduce an increased negative charge on a potential interaction interface. Morphological analysis of cells overexpressing ENTH2 loop 7 mutants showed that the cytokinesis failure phenotype was abolished by the single mutants N112D, E118Q, and to a lesser extent by S114E. Taken together, our results indicate that the interaction surface that contains loop 7 and the specific nature of these residues are crucial for ENTH2 involvement in cytokinesis. This research provides insight into a molecular mechanism by which ENTH2, but not ENTH1, overexpression in yeast leads to cell division defects. Structural data of WT and mutant ENTH2 domains along with in vivo phenotypic analysis of ENTH2 overexpressing cells indicate that the biochemical nature of three loop 7 residues is crucial for its role in cytokinesis.
Keywords: ENTH domain structure, epsin, cytokinesis failure, septin mislocalization
Introduction
Epsins are endocytic adaptor proteins that consist of a conserved epsin N-terminal homology (ENTH) domain followed by a C-terminal region that contains an array of binding motifs for ubiquitin, clathrin and other endocytic proteins.1–3 Initially named for their interaction with Eps15,4 epsins (Eps15 interacting) are most notably known for their involvement in the early steps of clathrin-mediated endocytosis in which the ENTH domain binds to phosphatidylinositol-4,5-bisphosphate (PIP2).5 PIP2 binding of ENTH at the membrane leads to the formation of a N-terminal amphipathic α-helix, termed helix 0, which inserts itself into the membrane causing membrane curvature.6–9 Five epsins are present in Saccharomyces cerevisiae—Ent1 and Ent2, which are involved in endocytosis and Ent3, Ent4, and Ent5, which are involved in intracellular trafficking. The ENTH domains of Ent1 and Ent2 share 75% sequence identity, while Ent3-5 domains share only 24–34% identity and appear to represent a different Ent subclass.10
Cells lacking either Ent1 or Ent2 were shown to be viable, however the double knockout was not; viability could be restored by introducing the ENTH domain from Ent1 or Ent2, but not the ENTH domain from Ent3-5.10 Other studies show that Ent2 also plays a critical role in a cell division signaling pathway and that its ENTH domain interacts with the septin organizing Cdc42 GTPase activating protein, Bem3.11 Upon overexpression of the isolated ENTH2 domain, cells exhibited septin mislocalization and cytokinesis failure. However, ENTH1 does not affect cytokinesis, despite 75% identity to ENTH2 and displays a lower apparent affinity for Bem3.11,12 Differential lipid binding is unlikely to underlie this cytokinesis phenotype, as the PIP2 binding determinants are the same in Ent1 and Ent2. In addition, overexpression of an ENTH2 mutant that is unable to bind membranes (ENTH2R62L, H72L) still induces the overexpression phenotype.11 An ENTH2N112D, S114E, E118Q (ENTH2DEQ) mutant, in which three residues from loop 7 were changed to the residues found at the same position in ENTH1, produced a protein that mimics ENTH1 and no longer leads to cytokinesis failure upon overexpression. ENTH2DEQ also had decreased binding efficiency towards Bem3 compared to WT ENTH2.11 Nevertheless, it is not clear whether these results reflect an alteration in the conformation of loop 7 or simply a change in the nature of specific amino acids at a functional domain interface.
To understand the role of ENTH2 loop 7 in cytokinesis, the structures of ENTH2 and ENTH2DEQ were solved by x-ray crystallography and analyzed. The structures show that the N112D, S114E and E118Q mutations clearly alter the distribution of charge on the interaction interface without changing the loop conformation of the ENTH2 domain. Analysis of the morphological characteristics of yeast cells overexpressing ENTH2 loop 7 mutants indicated that all three ENTH2-specific (N112, S114, and E118) residues are required for this domain to induce its characteristic cytokinesis phenotype.
Results and Discussion
ENTH2 structure
S. cerevisiae wild type ENTH2 and ENTH2DEQ mutant were cloned with N-terminal 6His tags, expressed in E. coli, and purified. Crystals were obtained by sitting drop vapor diffusion at 20°C and optimized via microseeding. Diffraction data were collected using both synchrotron radiation and an in-house Rigaku R4200 rotating anode X-ray generator. Crystals of both proteins belonged to space group P212121 with unit cell parameters of a = 41.2 Å, b = 53.2 Å, c = 70.3 Å, α = β = γ = 90° for ENTH2 WT and a = 41.7 Å, b = 54.2 Å, c = 70.7 Å, α = β = γ = 90° for ENTH2DEQ. Phases for ENTH2 WT were obtained by molecular replacement with the full structure of rat ENTH1 (PDB: 1EYH), while phases for ENTH2DEQ were obtained via molecular replacement with the full structure of wild type yeast ENTH2. As seen in Table 1, ENTH2 WT and ENTH2DEQ were refined to 1.30 and 1.75 Å, respectively, with Rwork/Rfree values of 0.210/0.230 and 0.205/0.238. The coordinates of ENTH2 WT and ENTH2DEQ have been deposited in the Protein Data Bank (accession codes 4GZC and 4GZD, respectively). The overall structures can be seen in Figure 1 superimposed via a least squares method including all backbone and C-alpha atoms with a resulting Rmsd = 0.24 Å.
Table 1.
Structural Statistics of the ENTH2 WT and ENTH2DEQ Crystal Structures
| ENTH2 WT | ENTH2DEQ | |
|---|---|---|
| Wavelength (Å) | 1.00 | 1.54 |
| Space group | P212121 | P212121 |
| Resolution (Å) | 35.6–1.30 | 29.9–1.75 |
| (1.33–1.30)a | (1.79–1.75) | |
| Completeness (%) | 95.7 (74.0) | 99.1 (87.7) |
| Reflections | 191,724 (1,958) | 102,382 (904) |
| Redundancy | 5.1 (4.2) | 6.1 (4.1) |
| I/sigI | 22.5 (2.1) | 31.9 (3.4) |
| Rsym | 0.057 (0.75) | 0.052 (0.39) |
| Refinement | ||
| Nreflections | 35,623 | 15,701 |
| Nfree | 1,873 | 976 |
| Rwork | 0.210 | 0.205 |
| Rfree | 0.230 | 0.238 |
| Rmsd bond length (Å) | 0.008 | 0.011 |
| Rmsd bond angle (°) | 1.21 | 1.53 |
| Average B-factor (Å2) | 18.8 | 23.0 |
| Total number of atoms | ||
| Protein | 1,136 | 1,224 |
| Water | 110 | 81 |
| Cl− | 1 | 1 |
| 2−propanol | − | 1 |
| Ramachandran | ||
| Favored (%) | 100 | 98.5 |
| Allowed (%) | 0 | 1.5 |
| Disallowed (%) | 0 | 0 |
Values in parentheses refer to the corresponding values of the highest resolution shell.
Figure 1.
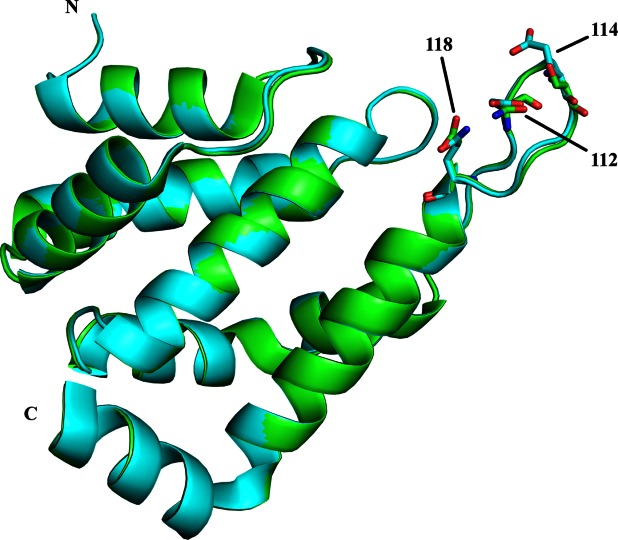
Superposition of ENTH2 WT and ENTH2DEQ (residues 15–146). ENTH2 WT and ENTH2DEQ are colored in green and cyan, respectively. Mutated residues 112, 114, and 118 in loop 7 are shown as stick representations.
Consisting of 8 α-helices connected by loops of varying lengths, wild type ENTH2 and ENTH2DEQ maintain the typical fold found in other ENTH domains. Helices 1-2, 3-4, and 6-7 form hairpin structures that stack on top of each other, creating a tight globular structure. Both structures have the first 14 residues disordered, which is consistent with the idea that these residues only form an ordered helix 0 upon phosphoinositide binding.6 Additionally, the last 4 residues (147–150) in helix 8 of both structures were disordered.
Both wild type ENTH2 and ENTH2DEQ are well-defined structures with average B-factors of 18.8 and 23.0. However the side chains of Loop 7 seem to be highly flexible, requiring refinement with reduced occupancy in order to see 2Fo-Fc density necessary to fit the side chains. If the occupancy of these residues is not reduced, the resulting B-factors jump to the high 60s, which is significant considering the average B-factor for both proteins, is so low. Specifically, both structures are well ordered through residue N/D112, at which point the backbone remains clear but the density for some of the residue side chains is weak through residue F116 (Fig. 2). The 2Fo-Fc electron density map at 1σ clearly shows density for the backbone atoms throughout the loop. The structures reveal that Loop 7 is partially stabilized via a single hydrogen bond between residues 112 and 118 for both wild type ENTH2 and ENTH2DEQ as seen in Figure 3(a). The hydrogen bond is conserved in rat ENTH1, suggesting that it might be important for maintaining the conformation of this loop and therefore possibly important for the overexpression phenotype.13 However, none of the 20 models in the ensemble of the NMR structure of human ENTH1 by Koshiba et al. contain this hydrogen bond.14 In these data, the would-be hydrogen bonding residues (D113 and Q119) either face opposite directions or are too far apart to form a bond, and often flank the opening of a charged pocket between loops 3 and 7, as seen in Figure 3(b).
Figure 2.
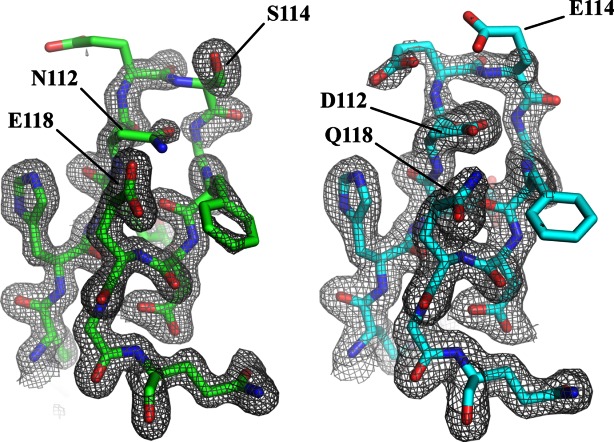
Observed electron densities of loop 7 for ENTH2 WT and ENTH2DEQ. 2Fo-Fc electron densities are shown at 1σ for residues 109–120. (Left) ENTH2 WT shown in green. Corresponding sequence is RHENESGFDEGQ. (Right) ENTH2DEQ shown in cyan. Corresponding sequence is RHEDEEGFDQGQ.
Figure 3.

Loop 7 morphology in yeast ENTH2 WT and ENTH2DEQ and human ENTH1. A: Loop 7 of yeast ENTH2 WT and ENTH2DEQ are shown in green and cyan, respectively. Residues 112 and 118 in both structures form a hydrogen bond. B: Loop 7 of yeast ENTH2 WT and ENTH2DEQ and the first reported solution structure from the family of PDB's corresponding to human ENTH1 are shown in green, cyan, and magenta, respectively. Structures are superimposed using residues 110–120. Loop 7 of human ENTH1 is also shown as a surface representation, depicting an open pocket flanked by charged residues.
In-vivo ENTH2 overexpression
Previous reports have demonstrated that while overexpression of wild type ENTH2 leads to dramatic cell division defects, overexpression of ENTH1 or ENTH2DEQ does not, suggesting that N112, S114, and E118 are required for the ENTH2 phenotype.11 To determine the relative role of the individual residues in loop 7 for the cell division phenotype, we generated a set of ENTH2 mutants: ENTH2N112D, ENTH2S114E, ENTH2E118Q, ENTH2N112D, E118Q, ENTH2N112D, S114E. The ability of each mutant, ENTH2DEQ and ENTH2WT to induce a cytokinesis defect is shown in Figure 4, expressed as the frequency of observation in a four-category phenotype classification (see Materials and Methods,11). None of the mutants induced severe cytokinesis defects (classified as cells exhibiting the category 4 phenotype) upon overexpression. The frequency of multinucleated cells with concatenated buds (i.e., Category 3) was significantly higher (P ≤ 0.008) in the ENTH2S114E mutant compared to ENTH2DEQ. For all the mutants, frequency of Category 2 cells with buds emerging from a common neck underwent a decrease, but cells with minor cell wall defects, representing the Category 1 phenotype, were present in comparable abundance for all of the mutants. Only wild type ENTH2 showed a large fraction of cells with cytokinesis defects in all of the categories.
Figure 4.
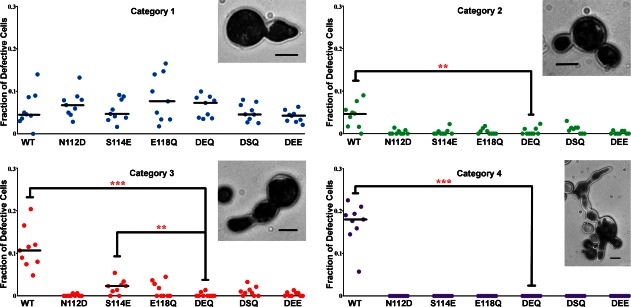
Analysis of cytokinesis defects in yeast caused by overexpression of wild type and mutant ENTH2 domains. Cells displaying defects representative of each category are shown for reference (bars = 5 μm). Nine independent experiments were performed for each construct with more than 200 cells counted each time. Horizontal lines represent the median, but are omitted for clarity if equivalent to zero. Statistical significance of the difference with respect to cells transformed with ENTH2DEQ was estimated using the Wilcoxon test with a correction for significant threshold according to the number of tests performed (α= 0.05/6 ∼ 0.008). **P < 0.008; ***P < 0.001.
Overall our results indicate that each of the individual residues is important for inducing the cell division phenotype, particularly residues N112 and E118. Figure 4 shows that the ENTH2S114E mutant still yields a statistically significant number of cells displaying a Category 3 phenotype, while the N112D and E118Q ENTH2 single mutants do not.
One explanation for the large impact of the N112D and E118Q single mutants is that there would no longer be a hydrogen bond between the two residues as seen in the x-ray structures. This might destabilize the loop region, affecting its interaction with other molecules (e.g., Bem3). It was thought that creating the ENTH2N112D, E118Q double mutant, thus preserving the hydrogen bond between them as seen in these two structures, would yield cells with an overexpression phenotype similar to wild type ENTH2. However, this was not the case. Both single and double mutants of N112 and E118 abolish the ENTH2 overexpression phenotype and appear to be required.
The high similarity between the ENTH2 WT and ENTH2DEQ structures suggests that the ENTH2 overexpression phenotype is dependent on the specific nature of residues at positions 112 and 118, and to a lesser extent at 114. The addition of a negative charge in loop 7 in ENTH2DEQ (EDDEGIQG vs ENESGFEG) appears to result in a modified interface for interaction with proteins involved in cytokinesis. Given that ENTH2DEQ does not bind to Bem3 as well as ENTH2 WT11, these residues may be crucial in contacting Bem3 and affecting its activity and downstream signaling.
Materials and Methods
Plasmids and DNA manipulations
DNA constructs were made using standard techniques. A high copy (2 μ) URA3 plasmid containing ENTH2 WT under the control of the MET25 promoter was used to generate the mutants.15 Site directed mutagenesis was performed using the QuikChange Lightning kit (Agilent Technologies). The mutants constructed were: ENTH2N112D, ENTH2S114E, ENTH2E118Q and ENTH2N112D, E118Q and ENTH2N112D, S114E, E118Q.
Yeast culture conditions and transformation procedures
ENTH2 WT or mutants were transformed into the S. cerevisiae strain W303. The transformations were performed using the lithium acetate protocol according to the Clontech yeast handbook. Yeast strains were grown at 30°C in synthetic medium with dextrose, lacking uracil for plasmid maintenance, for 3–4 days. For liquid culture assays, ∼105 cells were inoculated in 10 mL medium lacking uracil and methionine, and incubated overnight at 30°C.
Expression and purification
Yeast ENTH2 (1-150) WT and ENTH2DEQ were cloned into the pET-28 expression vector and transformed into Rosetta (DE3) E. coli. Cells were grown to an OD600 of 1.5 in Terrific Broth at 37°C in the presence of 30 mg/mL kanamycin and 34 mg/mL chloramphenicol. Overexpression of ENTH2 was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.4 µM. Cells were grown for an additional 16 h at 16°C before being pelleted by centrifugation and stored at −80°C. The frozen cell pellet was resuspended in 50 mM Tris pH7.4, 500 mM NaCl, and 10 mM imidazole (Buffer A). Cells were lysed via French Press and mixed with 100 μL Protease Inhibitor Cocktail set III from CalBiochem. Cell lysate was pelleted by centrifugation at 50,000g and 4°C for 75 min. The supernatant was loaded onto a 5 mL Ni-NTA affinity column which was pre-equilibrated with Buffer A. 5 mL fractions were collected throughout the purification. The column was washed with 10% Buffer B (50 mM Tris pH7.4, 500 mM NaCl, 500 mM imidazole, 5% glycerol) with an overall imidazole concentration of 60 mM. ENTH2 was eluted off the column by increasing the imidazole concentration to 260 mM (50% Buffer B) in a single step. Aliquots from fractions obtained during the purification were loaded onto an SDS gel and electrophoresed. Fractions containing ENTH2 were pooled and concentrated to 2 mL using a 10 kDa cutoff spin concentrator. The sample was then loaded onto a HiPrep 26/60 Sephacryl S-200 size exclusion column that was pre-equilibrated with a buffer containing 20 mM Tris pH8 and 200 mM NaCl. Aliquots from the 1 mL fractions which were collected throughout the purification were analyzed via SDS-PAGE. Fractions containing ENTH2 were pooled and concentrated to 8–13 mg/mL.
Crystallization, data collection, and refinement
Crystallization trials were conducted using commercially available crystallization screens from Hampton, JCSG, and Emerald Biosystems and were set up in 96-well sitting drop vapor diffusion trays with a Genomic Solutions crystallization robot. Wells contained reservoir volumes of 50 μL and droplets with a protein:reservoir ratio of 1 μL:1 μL.
Initial crystals were obtained for wildtype ENTH2 after 30 days under conditions containing buffers in a pH range of 7–9.5, various percentages of either PEG 3000, PEG 8000, or PEG 10,000, and varying amounts of sodium chloride. After two sequential rounds of micro-seeding, large single crystals grew to (1 × 0.3 × 0.3 mm) in 48 h under the following conditions: 100 mM imidazole pH 8.0, 20% PEG 8,000, and 400 mM NaCl. Crystals were cryoprotected by transfer into mother liquor containing 15% PEG-400 for 30 s. X-ray diffraction data were collected at the Advanced Photon Source at Argonne National Labs on beam line ID 23-D. Data were integrated and scaled using HKL200016 and the CCP417 suite of programs. The Matthews coefficient indicated one molecule per asymmetric unit with a solvent content of 41%.18 Phases were obtained via molecular replacement using BALBES19 using the published crystal structure of rat epsin ENTH (PDB: 1EYH) as a search model. The model was built using Coot20 and refined with Refmac5.21 Side chain occupancies of some residues in loop 7 were reduced due to high B-factors.
Crystals of ENTH2DEQ grew after 10 days from the JCSG Core Suite 2 well B11 that contains 85 mM HEPES pH7.5, 17% PEG 4000, 8.5% 2-propanol, and 15% glycerol. One round of micro-seeding using identical conditions produced single crystals (0.5 × 0.1 × 0.1 mm) in 48 h. X-ray diffraction data were collected using a Rigaku R4200 rotating anode X-ray generator with an RAXIS IV++ image plate. Data were integrated and scaled using HKL200016 and the CCP417 suite of programs. The Matthews coefficient indicated one molecule per asymmetric unit with a solvent content of 41%.18 Phases were obtained via molecular replacement using the previously solved structure of ENTH2 wildtype as a search model. The model was built using Coot20 and refined with Refmac5.21 To help reduce phase bias from the wildtype structure, the occupancies of residues in Loop 7 (107–120) were reduced to zero during molecular replacement and the first two rounds of refinement. For later rounds of refinement, side chain occupancies of some residues in loop 7 were reduced due to high B-factors.
Microscopy
Cell images were acquired using a Zeiss Axiovert 200M in white-field mode. Image processing was performed with ImageJ software. For quantification of the cytokinesis defect, cells were fixed with 3.7% formaldehyde, washed with sterile water and resuspended in 100μl media containing 30% (v/v) Methylene blue solution. Methylene blue stains the cytoplasm of fixed cells, therefore, cell wall abnormalities can be easily detected.
Quantification of the cell-division phenotype
After slide preparation, each coverslip was divided into nine imaginary quadrants, followed by acquisition of three images per quadrant.22 This was followed by quantification and characterization of the cells into four different phenotypic categories depending on the severity of cytokinesis defect. Category 1 includes morphologically normal cells that are mono-/ bi-nucleated but display abnormal septa and/ or ectopic cell wall deposition. Cells grouped under category 2–4 cells are multinucleated and are morphologically abnormal. Cells under category 2 form branched buds emerging from a common neck; those under category 3 form linear chains of buds, while cells classified under category 4 cells contain multiple branched buds.11
The frequency of abnormal cell division events was determined using the calculations described in Mukherjee et al.11. Briefly, the number of failed division attempts was described as Σ[(number of nuclei counted for a multinucleated cell) –1]. The percentage of failed division attempts = 100 × number of failed division attempts/total number (successful + failed) of division attempts. At least 200 cells were quantified for each experiment. Statistical analysis was performed using Wilcoxon's nonparametric test.
Acknowledgments
The authors thank Dr. C. Nicklaus Steussy for helpful advice and discussions on structure refinement.
Glossary
- ENTH1
epsin 1 N-terminal homology domain
- ENTH2
epsin 2 N-terminal homology domain
- ENTH2DEQ
ENTH2 bearing N112D, S114E, E118Q mutations
References
- 1.Kay BK, Yamabhai M, Wendland B, Emr SD. Identification of a novel domain shared by putative components of the endocytic and cytoskeletal machinery. Protein Sci. 1999;8:435–438. doi: 10.1110/ps.8.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Camilli P, Chen H, Hyman J, Panepucci E, Bateman A, Brunger AT. The ENTH domain. FEBS Lett. 2002;513:11–18. doi: 10.1016/s0014-5793(01)03306-3. [DOI] [PubMed] [Google Scholar]
- 3.Sen A, Madhivanan K, Mukherjee D, Aguilar RC. The epsin protein family: coordinators of endocytosis and signaling. Biomol Concepts. 2012;3:117–126. doi: 10.1515/bmc-2011-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, DiFiore PP, De Camilli P. Epsin is an EH-domain–binding protein implicated in clathrin mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 5.Itoh T, Koshiba S, Kigawa T, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 6.Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 7.Stahelin RV, Long F, Peter BJ, Murray D, De Camilli P, McMahon HT, Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and Epsin N-terminal homology (ENTH) domains. J Biol Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- 8.Kweon DH, Shin YK, Shin JY, Lee JH, Lee JB, Seo JH, Kim YS. Membrane topology of helix 0 of the epsin N-terminal homology domain. Mol Cell. 2006;21:428–435. [PubMed] [Google Scholar]
- 9.Capraro BR, Yoon Y, Cho W, Baumgart T. Curvature sensing by the epsin N-terminal homology domain measured on cylindrical lipid membrane tethers. J Am Chem Soc. 2010;132:1200–1201. doi: 10.1021/ja907936c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee D, Coon BG, Edwards DF, Hanna CB, Longhi SA, McCaffery JM, Wendland B, Retegui LA, Bi E, Aguilar CR. The yeast endocytic protein Epsin 2 functions in a cell-division signaling pathway. Cell Sci. 2009;122:2453–2463. doi: 10.1242/jcs.041137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilar RC, Longhi SA, Shaw JD, Yeh LY, Kim S, Schön A, Freire E, Hsu A, McCormick WK, Watson HA. Epsin N-terminal homology domains perform an essential function regulating Cdc42 through binding Cdc42 GTPase-activating proteins. Proc Natl Acad Sci USA. 2006;103:4116–4121. doi: 10.1073/pnas.0510513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman J, Chen H, DiFiore PP, DeCamilli P, Brunger AT. Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF) J Cell Biol. 2000;149:537–546. doi: 10.1083/jcb.149.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koshiba S, Kigawa T, Kikuchi A, Yokoyama S. Solution structure of the epsin N-terminal homology (ENTH) domain of human epsin. J Struct Funct Genom. 2002;2:1–8. doi: 10.1023/a:1011397007366. [DOI] [PubMed] [Google Scholar]
- 15.Aguilar RC, Wendland B. Endocytosis of membrane receptors: two pathways are better than one. Proc Natl Acad Sci USA. 2005;102:2679–2680. doi: 10.1073/pnas.0500213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Methods in Enzymology, Volume 276: Macromolecular Crystallography, part A. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 17.Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Cryst D50. pp. 760–776. [DOI] [PubMed]
- 18.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 19.Long F, Vagin A, Young P, Murshudov GN. BALBES: a molecular replacement pipeline. Acta Cryst D. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee D, Sen A, Aguilar RC. Analysis of the development of a morphological phenotype as a function of protein concentration in budding yeast. J Vis Exp. 2010;37 doi: 10.3791/1863. Available at: http://www.jove.com/details.php?id=1863, doi: 10.3791/1863. [DOI] [PMC free article] [PubMed] [Google Scholar]


