Abstract
The membrane-associated serine hydrolase, monoacylglycerol lipase (MGL), is a well-recognized therapeutic target that regulates endocannabinoid signaling. Crystallographic studies, while providing structural information about static MGL states, offer no direct experimental insight into the impact of MGL's membrane association upon its structure–function landscape. We report application of phospholipid bilayer nanodiscs as biomembrane models with which to evaluate the effect of a membrane system on the catalytic properties and conformational dynamics of human MGL (hMGL). Anionic and charge-neutral phospholipid bilayer nanodiscs enhanced hMGL's kinetic properties [apparent maximum velocity (Vmax) and substrate affinity (Km)]. Hydrogen exchange mass spectrometry (HX MS) was used as a conformational analysis method to profile experimentally the extent of hMGL–nanodisc interaction and its impact upon hMGL structure. We provide evidence that significant regions of hMGL lid-domain helix α4 and neighboring helix α6 interact with the nanodisc phospholipid bilayer, anchoring hMGL in a more open conformation to facilitate ligand access to the enzyme's substrate-binding channel. Covalent modification of membrane-associated hMGL by the irreversible carbamate inhibitor, AM6580, shielded the active site region, but did not increase solvent exposure of the lid domain, suggesting that the inactive, carbamylated enzyme remains intact and membrane associated. Molecular dynamics simulations generated conformational models congruent with the open, membrane-associated topology of active and inhibited, covalently-modified hMGL. Our data indicate that hMGL interaction with a phospholipid membrane bilayer induces regional changes in the enzyme's conformation that favor its recruiting lipophilic substrate/inhibitor from membrane stores to the active site via the lid, resulting in enhanced hMGL catalytic activity and substrate affinity.
Keywords: conformation, hydrogen exchange, mass spectrometry, phospholipid bilayer nanodisc, serine hydrolase
Introduction
A member of the α/β serine-hydrolase superfamily, monoacylglycerol lipase (MGL) is an esterase that features a characteristic serine (Ser122)–histidine (His269)–aspartic acid (Asp239) catalytic triad.1 MGL is primarily responsible for deactivating 2-arachidonoylglycerol (2-AG), the predominant endocannabinoid signaling lipid in the central nervous system, which is synthesized on-demand from membrane phospholipid precursors.2,3 Additionally, hydrolysis of 2-AG by MGL links endocannabinoid and eicosanoid signaling circuits by generating arachidonic acid precursor for biosynthesis of prostaglandins and other oxygenated lipid mediators.1,4 Since 2-AG acts as a full agonist capable of activating both principal 7-transmembrane cannabinoid receptors, designated cannabinoid receptor 1 (CB1R) and cannabinoid receptor 2 (CB2R), MGL represents a major control point for 2-AG-mediated cannabinergic transmission that influences a number of (patho)physiological processes, from psychobehavioral status to energy metabolism.5,6 Increased tissue 2-AG levels consequent to pharmacological or genetic MGL ablation have been associated with preclinical therapeutic benefit against pain,7,8 inflammation,9,10 neurodegenerative disorders,11 psychological stressors,12 nausea/emesis,13 and cancer pathogenesis.14,15 Although protracted MGL ablation invites functional CB1R desensitization in rodents,16 the salutary results of small-molecule, active site-directed MGL inhibitors in preclinical disease models have helped validate MGL as a drug target, focusing interest on temporally tuned human MGL (hMGL) inhibitors as medicines capable of elevating 2-AG tone (and, indirectly, CB1R transmission) to therapeutic levels with less risk of inciting the adverse events observed with systemic application of direct CB1R agonists.17,18 Recent demonstrations that MGL inhibition can limit arachidonic acid flux into proinflammatory and tumorigenic eicosanoid pathways suggest the use of MGL inhibitors for therapeutic purposes against inflammatory disorders and cancer.10,14,15
Lipases are acyl hydrolases that cleave long-chain triacylglycerols at the boundary between aqueous and lipid–substrate phases, and their biocatalysis is activated at the interface. Interfacial potentiation of lipolysis has been attributed to factors such as increased local substrate concentration in close proximity to the enzyme and optimized orientation of scissile triacylglycerol fatty-ester bonds.19 Many lipases feature a lid domain that regulates substrate access to the binding pocket/active site,20 and crystallographic data have supported inference that lipase structural changes upon association between the enzyme's lid region and the boundary of a lipid matrix also contribute to interfacial lipase activation. For example, atomic-level analysis of Rhizomucor miehei lipase has suggested that this enzyme's activation-associated conformational change reflects a hinge-type motion involving displacement of its lid domain so as to enhance the hydrophobic area for both enzyme interaction at the lipid interface and substrate binding.21 The mechanism of other lipases appears to involve more complex motions of multiple enzyme helices upon association with supramolecular assemblies containing triacylglycerol substrate.22 Several studies have demonstrated that MGL activity is found at varying proportions between membrane and soluble tissue subfractions, depending upon cell/tissue type. In mouse brain, MGL activity is primarily (∼90%) membrane-associated,3 whereas in rat macrophages and gastrointestinal tract it is enriched in the cytosol.23,24 The enzymatic properties of cytosolic and membrane-associated MGL differ as well; for example, in rat gastrointestinal tissue, the latter is less sensitive to pharmacological inhibition.24 These collective data for both lipases in general and MGL specifically have invited the hypothesis that, in situ, MGL interacts reversibly with cell membranes, allowing the enzyme to extract 2-AG substrate from membrane-associated pools and into its hydrophobic substrate-binding pocket containing the catalytic triad, thereby facilitating substrate engagement.1
Recent X-ray analyses of apo and covalently labeled hMGL species offer a putative mechanistic rationale for this hypothesis.25–27 Reminiscent of many other lipid hydrolases, the (h)MGL active site is gated by a flexible lid domain positioned to shield the entrance to the enzyme's substrate-binding pocket and thereby regulate substrate access to the catalytic center. A comparison of the crystal structures of apo-hMGL and hMGL in complex with a reversible inhibitor has supported the view that the hMGL lid domain also participates in anchoring the enzyme reversibly to the cell membrane during the catalytic cycle and in structural rearrangements upon inhibitor binding, eliciting a shift from an “open” apo-enzyme to a “closed,” ligand-bound form in which the active site is shielded.25,27 In contrast to this purported mechanism, a crystal structure of hMGL covalently bound to a serine-reactive carbamylating agent displayed an open—not closed—conformation.26 Thus, although static representations of unique states of apo- and liganded-hMGL variants are available at atomic resolution, ambiguities remain concerning the structural and functional ramifications of hMGL-membrane interaction.
In the current study, we have chosen to examine this issue experimentally by using purified recombinant hMGL and phospholipid bilayer nanodiscs. A nanodisc is a discoidal form of high density lipoprotein composed of a nanometer-sized phospholipid bilayer surrounded by two α-helical, amphipathic membrane scaffold proteins (MSPs).28 Nanodiscs represent a unique, water-soluble biomimetic system for studying membrane-associated proteins, since monodisperse nanodiscs do not suffer from the propensity for aggregation, geometric distortion, and heterogeneity characteristic of other structured lipid platforms such as micelles, bicelles, and liposomes.29,30 In conjunction with a nanodisc-hMGL reconstitution system, we have employed hydrogen exchange mass spectrometry (HX MS) as an experimental conformational analysis method. The results were elaborated using molecular dynamics (MD) simulation to gain insight into membrane influence on hMGL structure, ligand engagement, and catalytic activity. Peptide-level HX MS involves experimental quantification of protein amide-hydrogen exchange with heavier deuterium isotope from D2O. After the exchange at neutral pH is quenched by acidification, the extent and kinetics of deuteration in peptide fragments is analyzed and reflects the regional solvent accessibility and hydrogen bonding of the protein. Both of these parameters serve as a proxy for protein conformational state and dynamics.31 MD simulation is a computational approach toward modeling protein structure, dynamics, and interactions.32
We provide data indicating that hMGL interaction with the nanodisc phospholipid bilayer involves a transition from a solution to a membrane-associated conformation, as evidenced primarily by structural changes in the major helical component (helix α4) of the hMGL lid domain and neighboring regions (especially helix α6). These conformational changes may help stabilize an open hMGL conformation at the membrane–water interface, facilitate hMGL-ligand (substrate, inhibitor) interaction, and enhance catalysis. Our results further suggest that engagement of lipophilic ligand by the hMGL catalytic site involves the ligand's lateral diffusion within the membrane bilayer to access the hMGL substrate-binding channel through the enzyme's membrane-associated lid domain.
Results and Discussion
Phospholipid bilayer nanodisc characterization
Nanodiscs are formed through a controlled self-assembly process from a defined molar ratio of phospholipid micelles in detergent and stabilizing MSPs to yield monodisperse, nanometer-size discoidal phospholipid bilayers, each encircled by two MSPs whose length defines the diameter of the disc.28,33 For this study, purified membrane scaffold protein 1D1 (MSP1D1)28 was mixed with micelles composed of sodium cholate and either 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) only or a 3:2 molar ratio of POPC and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG). From an initial MSP1D1:phospholipid:sodium cholate molar ratio of 1:78:200, gradual detergent removal was used to effect component assembly. The resulting nanodiscs, as isolated by fast protein liquid chromatography (FPLC) on a calibrated size-exclusion column, evidenced the expected28,34 Stokes hydrodynamic diameter of ∼10 nm [Supporting Information Fig. S1(a), red] and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) contained MSP1D1 as the sole protein component [Supporting Information Fig. S1(b)], substantiating the purity and integrity of our nanodisc preparation.
Phospholipid bilayer nanodiscs enhance hMGL catalytic activity and substrate affinity
Diacylphosphoglycerides such as POPC and POPG are not MGL substrates,1,4 and the charged head groups of bilayer phospholipids typically endow the surface of biological membranes with a net negative charge.35 These considerations, along with MGL's membrane association in various tissues,3,23,24 led us to examine initially whether anionic phospholipid bilayer nanodiscs containing a 3:2 POPC:POPG molar ratio might influence the kinetic properties of hMGL, the recombinant enzyme expressed and purified as previously detailed by this laboratory.36
As compared to hMGL in aqueous Tris buffer, the presence of POPC/POPG nanodiscs enhanced by ∼2.5-fold the enzyme's apparent affinity (Km) for the fluorogenic reporter substrate, arachidonoyl 7-hydroxy-6-methoxy-4-methylcoumarin ester (AHM MCE),36,37 and increased by approximately threefold the enzyme's apparent rate of hydrolysis (Vmax) of this substrate (Table I). The POPC/POPG nanodiscs elicited a similar enhancement of hMGL kinetic properties for hydrolysis of the enzyme's natural endocananbinoid substrate, 2-AG (Table I). Nanodisc enhancement of hMGL substrate turnover and affinity was independent of phospholipid bilayer charge, since charge-neutral POPC bilayer nanodiscs decreased hMGL Km and increased the enzyme's Vmax to extents comparable to the effect of anionic POPC/POPG nanodiscs (Table I). Although the nonionic surfactant Triton X-100 can serve as a hydrophobic phase that helps solubilize both lipases and their triacylglycerol substrates to elicit apparent lipase activation,38 we found that Triton X-100 at a final concentration of 0.5 mM (i.e., approximately twofold its critical micelle concentration38) did not affect hMGL substrate affinity and increased hMGL substrate turnover by ∼1.5-fold relative to the enzyme in buffer alone (Table I). Thus, Triton X-100 micelles affected hMGL's Vmax only modestly as compared to the enhanced effect of either negatively charged or charge-neutral phospholipid bilayer nanodiscs on both hMGL Km and Vmax.
Table I.
Kinetic Parameters for Hydrolysis of AHMMCE and 2-AG by hMGL in the Presence and Absence of Detergent or Phospholipid Bilayer Nanodiscs
| Substrate | ||||
|---|---|---|---|---|
| AHMMCE | 2-AG | |||
| Incubation conditions | Km (μM) | Vmax (μM/min/mg) | Km (μM) | Vmax (μM/min/μg) |
| Buffer | 18.3 ± 2.8 | 191.4 ± 10.5 | 41.9 ± 5.9 | 28.3 ± 1.1 |
| Buffer + POPC/POPG nanodiscs | 7.5 ±1.8a | 565.4 ± 52c | 11.9 ± 1.8c | 57.3 ± 3.2c |
| Buffer + POPC nanodiscs | 7.9 ± 2c | 590 ± 45a | n.d. | n.d. |
| Buffer + Triton X-100 | 18.3 ± 4 | 295 ± 37.2a | n.d. | n.d. |
For AHMMCE substrate, buffer was 50 mM Tris–HCl, pH 7.4. For 2-AG substrate, TME buffer (25 mM Tris base, 5 mM MgCl2, and 1 mM EDTA, pH 7.4) was used. Apparent Km and Vmax values are the mean ± SD for triplicate determinations across three independent enzyme preparations. Statistical significance of group-mean differences was evaluated by a two-sample independent t-test, the significance level set at P ≤ 0.05. n.d., not determined.
P ≤ 0.01 versus buffer.
bP ≤ 0.02 versus buffer + Triton X-100.
P ≤ 0.001 versus buffer.
dP ≤ 0.002 versus buffer + Triton X-100.
The coincident enhancement of hMGL activity and substrate affinity induced by phospholipid bilayer nanodiscs suggests that these biomembrane mimetics facilitate hMGL interaction with AHMMCE or 2-AG in a manner distinct from merely serving as a solubilization depot for hydrophobic substrate, since quantitatively parallel effects in hMGL kinetic properties were not induced by detergent micelles. From the observation that both anionic and charge-neutral nanodiscs enhanced hMGL activity and substrate affinity (Table I), it is tempting to hypothesize that hydrophobic interactions between the structured nanodisc phospholipid bilayer and hMGL may form and establish an interfacial microenvironment that enhances hMGL kinetic properties and facilitates AHMMCE/2-AG diffusion from the membrane into the enzyme's open, hydrophobic substrate-binding pocket.
hMGL interacts with phospholipid bilayer nanodiscs
Our hypothesis that a hydrophobic association between hMGL and phospholipid bilayer nanodiscs enhances the enzyme's kinetic properties accords with the concept that physical interaction between the hydrophobic lid region of lipases with an α/β hydrolase fold and their lipid-phase substrates helps govern lipase catalysis by influencing substrate supply/accessibility, orientation, and partitioning from the lipid phase to the enzyme's substrate-binding pocket.19,39 The paradigm for interfacial effects on lipase activity encompasses conformational rearrangement of the enzyme's lid domain as a mechanism for gating substrate access to the active site.20 These concepts led us to probe experimentally whether hMGL associates with the nanodiscs. For this purpose, we analyzed the POPC/POPG nanodisc, hMGL preparations and an hMGL–nanodisc mixture by size-exclusion FPLC. The results demonstrate that two distinct nanodisc-containing populations with similar, but differentiable, Stokes hydrodynamic diameters are resolvable from free hMGL [Supporting Information Fig. S1(a), green], whereas the hMGL–nanodisc mixture [Supporting Information Fig. S1(a), black] evidences a mean Stokes diameter greater than the nanodiscs alone [Supporting Information Fig. S1(a), red]. Only the hMGL–nanodisc mixture contained both MSP1D1 and hMGL protein [Supporting Information Fig. S1(b)]. These data offer provisional evidence that hMGL associates with nanodiscs to form hMGL–nanodisc complexes having a mean Stokes diameter greater than either the enzyme or the unassociated nanodiscs.
hMGL interaction with nanodiscs modifies enzyme regional conformation
We next investigated experimentally whether phospholipid bilayer nanodiscs influence hMGL conformation in the two regions of α/β lipid hydrolases most critical for substrate interaction and turnover: the lid and the substrate-binding pocket/active site domains.19,20,39 For this purpose, we used HX MS to compare the kinetics with which these hMGL regions exchange amide hydrogens for heavier deuterium isotope (i.e., the degree of solvent accessibility) when the intact, functional enzyme is incubated in D2O medium in the presence or absence of POPC/POPG nanodiscs. Deuterium uptake into hMGL was performed over incubation periods from 10 s to 4 h, at which times the labeling reaction was quenched. Pepsin hydrolysates were then generated and analyzed by MS to determine the extent of hMGL deuteration in peptides within the enzyme's lid domain and substrate-binding pocket. At a given pH and temperature, deuteration rate is modulated by protein conformational properties: rapid deuterium exchange is characteristic of more disordered, solvent-exposed protein regions. Conversely, slower, limited exchange indicates a more compact, hydrogen-bonded and solvent-shielded protein state.31 The difference between deuterium incorporation into hMGL peptides in the absence or presence of phospholipid bilayer nanodiscs serves as proxy for nanodisc structural impact on the enzyme. Since peptide-level HX MS readout is based on the masses of peptides assignable to the protein under study (here, hMGL),31 the presence of MSP1D1 in some hMGL samples did not interfere with HX MS analysis of the enzyme itself.
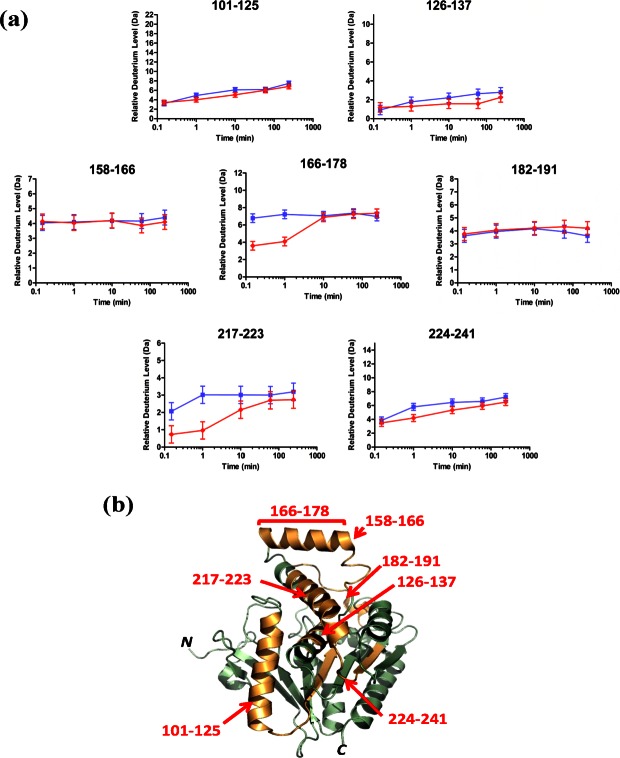
Supporting Information Table SI lists the common peptic peptides identified through electrospray ionization-MS/MS from three independent hMGL preparations for enzyme in the absence or presence of nanodiscs, along with the maximum possible deuterium incorporation for each respective peptide. This subfamily includes peptic peptides representing the hMGL lid and substrate binding-pocket/active site domains that are the focus of the current study. Specifically, residues 101–137 (designated for 6-His-hMGL) have been localized to the bottom of the substrate-binding pocket and encompass the catalytic-triad serine.25,26 Residues 158–191 span what has been considered apo-hMGL's lid region, with residues 166–178 encompassing helix α4, which is flanked by residues 158–165 (leading from sheet β6 into helix α4) and residues 182–191 (loop conjoining helices α4 and α5).25,26 Residues 217–223 encompass helix α6 which, along with residues 224–241, are in the vicinity of the active site.25,26
The peptide-level HX MS results for the hMGL lid domain and substrate-binding pocket/active site region are plotted in Figure 1(a), and the peptides considered are highlighted in the wild-type hMGL structure representation [protein data bank (PDB) ID: 3JW8]26 (http://www.rscb.org/pdb/explore.do?structureId=3JW8) in Figure 1(b). The limited, gradual deuteration of peptides 101–125 and 126–137 indicates that the distal region of the substrate binding pocket containing catalytic Ser129 maintains only modest solvent exposure whether nanodiscs are present or not and is likely a stable structural element of the enzyme without significant breathing/unfolding motions. Peptide 224–241 was likewise shielded from solvent, but appeared somewhat dynamic, since it acquired significant deuterium over time to become labeled to 46% of maximum by 10 min, regardless of the presence of nanodiscs. Deuterium uptake into hMGL lid-domain peptides 158–166, 166–178, and 182–191 was extremely rapid, indicative of high solvent exposure. Within the lid domain, however, deuteration of helix α4 (i.e., peptide 166–178) was markedly suppressed before 10 min in the presence of nanodiscs and increased thereafter to parallel the peptide's deuterium uptake in the absence of nanodiscs. Yet the deuterium exchange in hMGL peptides 158–166 and 182–191 flanking helix α4 was unaffected by the nanodiscs. A nanodisc-induced suppression of deuterium uptake into peptide 217–223 (i.e., helix α6) was also evident with an even more protracted approach to maximal deuteration than was observed for the influence of nanodiscs on lid-domain helix α4. Especially as compared to hMGL helices α4 and α6, the relatively modest deuterium uptake by other hMGL peptic peptides identified was unaffected by phospholipid bilayer nanodiscs (data not shown). This latter observation is consistent with the ordered, shielded nature of the core of eukaryotic α/β-hydrolases across species,40 a characteristic shared by apo-hMGL, as evident in a refined homology model of the enzyme41 and the finding that wild-type apo-hMGL and unliganded variants crystalize as compact globular proteins.25,26
Figure 1.
Localization of comformational changes in hMGL induced by phospholipid bilayer nanodiscs. (a) Deuterium incorporation curves derived from HX mass spectra for the peptides generated from hMGL pepsin digestion and designated by their amino acid residue numbers in 6-His-hMGL. Relative deuterium incorporation (Da) is plotted versus time of hMGL incubation in D2O in the absence (blue lines,  ) or presence (red lines,
) or presence (red lines,  ) of POPC/POPG bilayer nanodiscs. The maximum amount of deuterium incorporation possible for each respective peptide is designated on the y-axis of each kinetic plot. Data were obtained through peptide level HX MS analysis of hMGL. The error of peptide HX MS mesurements with this experimental setup was ±0.50 Da, as determined by replicate analysis of peptide standards in prior HX MS work with this instrumentation.51,52 (b) The hMGL peptides for which deuterium uptake curves are presented in panel (a), above, are highlighted (orange) in the wild-type hMGL structure representation derived from PDB ID: 3JW8.26 Residue designations in red type correspond to 6-His-hMGL.
) of POPC/POPG bilayer nanodiscs. The maximum amount of deuterium incorporation possible for each respective peptide is designated on the y-axis of each kinetic plot. Data were obtained through peptide level HX MS analysis of hMGL. The error of peptide HX MS mesurements with this experimental setup was ±0.50 Da, as determined by replicate analysis of peptide standards in prior HX MS work with this instrumentation.51,52 (b) The hMGL peptides for which deuterium uptake curves are presented in panel (a), above, are highlighted (orange) in the wild-type hMGL structure representation derived from PDB ID: 3JW8.26 Residue designations in red type correspond to 6-His-hMGL.
The nanodisc-induced suppression of deuterium uptake by hMGL helix-α4 peptide 166–178 along with the nanodisc's enhancement of hMGL reaction velocity and substrate affinity suggest that this region of the enzyme's lid domain associates with the nanodisc phospholipid bilayer, the interaction serving both to shield this lid region from solvent and facilitate access of hydrophobic lipid substrate to the enzyme's lid-gated ligand-binding pocket. By analogy with other lipases,20 it has been speculated that, in situ, hMGL substrate diffuses into the enzyme's binding site from a biomembrane pool by virtue of hMGL-membrane association.1,3,4,25–27 To the authors' best knowledge, the present study provides the first experimental evidence supporting such a mechanism for hMGL. As observed for hMGL helix-α4 peptide 166–178, nanodiscs also acutely suppressed deuteration of helix-α6 peptide 217–223 localized nearer to the active site,25,26 suggesting that hMGL interaction with the nanodisc phospholipid bilayer hinders the solvent accessibility of this enzyme region as well. Interaction of the phospholipid bilayer with hMGL may also be appreciated from the less dynamic and more protected nature of peptide 224–241 in the active site region (as indicated by its attenuated deuterium uptake) in the presence of nanodiscs. Collectively, these data indicate that hMGL association with phospholipid bilayer nanodiscs influences regional hMGL conformation within the enzyme's lid domain and in the vicinity of its active site.
Covalent hMGL inhibitor modulates enzyme conformation in the presence of nanodiscs
Potent MGL inhibitors share intrinsic lipophilicity with both hMGL endocannabinoid substrate, 2-AG, and reporter substrate, AHMMCE.1,4,5,8,9,14,17,18,42 In light of our demonstration that phospholipid bilayer nanodiscs enhance hMGL kinetic properties and induce enzyme lid-domain and active site conformational changes consonant with hMGL–nanodisc association, we used HX MS to determine the potential influence of a small-molecule inhibitor on these hMGL regions in the presence of POPC/POPG nanodiscs. For this purpose, we selected AM6580 [Fig. 2(a)] as being representative of the principal class of covalent hMGL inhibitors, agents with the potential to carbamylate the enzyme's catalytic serine.1,18 We previously demonstrated that AM6580 inhibits hMGL with nanomolar potency37 and now provide evidence from matrix-assisted laser desorption ionization-time of flight (MALDI-TOF/TOF) MS to substantiate that AM6580's fluorenyl piperazine moiety covalently modifies hMGL catalytic Ser129, resulting in the expected hMGL mass increase of 277 Da (Supporting Information Fig. S2).
Figure 2.
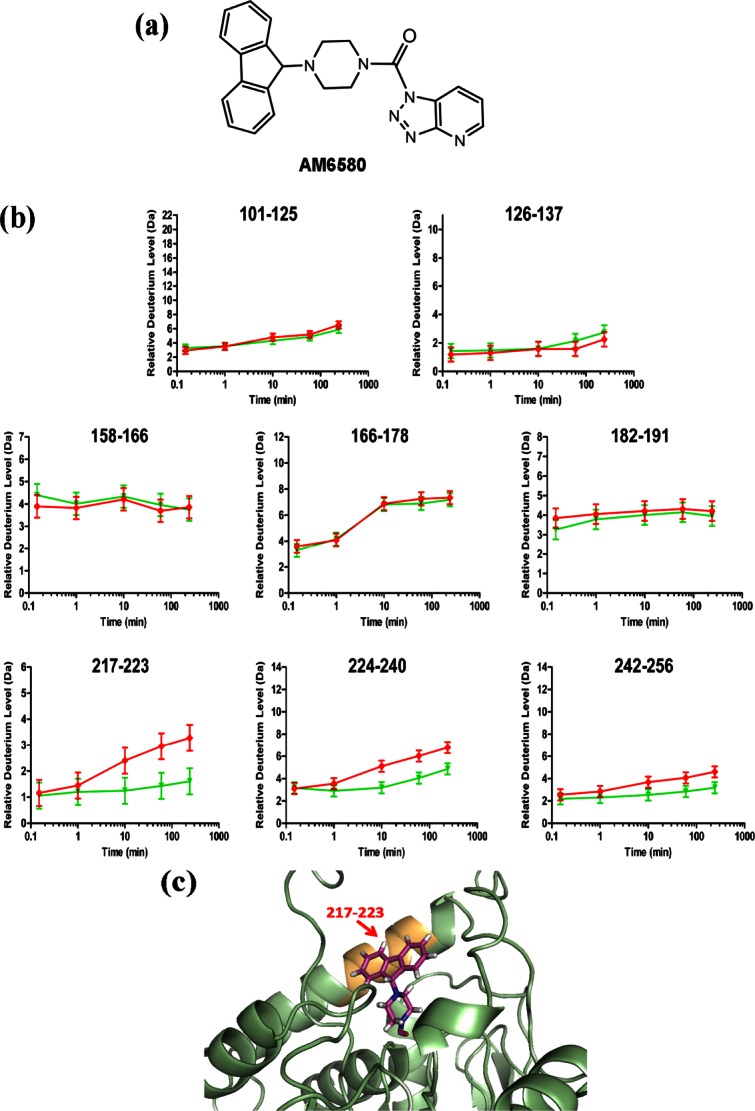
Localization of conformational changes in hMGL induced through active-site Ser carbamylation by the covalent inhibitor, AM6580. (a) Structure of AM6580. (b) Deuterium incorporation curves derived from HX mass spectra for peptides generated from hMGL pepsin digestion and designated by their amino acid residue numbers in 6-His-hMGL. Relative deuterium incorporation (Da) is plotted versus time of hMGL incubation in D2O in the presence of POPC/POPG phospholipid bilayer nanodiscs and without (red lines,  ) or with (green lines,
) or with (green lines,  ) AM6580. The maximum amount of deuterium incorporation possible for each respective peptide is designated on the y-axis of each kinetic plot. Data were obtained through peptide-based HX MS analysis. The error of peptide HX MS mesurements with this experimental setup was ±0.50 Da, as determined by replicate analysis of peptide standards in prior HX MS work with this instrumentation.51,52 (c) Schematic depicting the docking of the AM6580-derived fluorenyl piprazine group covalently attached to hMGL active-site Ser122 (i.e., Ser129 for 6-His-hMGL) in a portion of the hMGL structure representation derived from PDB ID: 3JW8.26 Helix-α6 peptide 217–223 (orange) is shielded by the carbamylating-group modification at Ser122.
) AM6580. The maximum amount of deuterium incorporation possible for each respective peptide is designated on the y-axis of each kinetic plot. Data were obtained through peptide-based HX MS analysis. The error of peptide HX MS mesurements with this experimental setup was ±0.50 Da, as determined by replicate analysis of peptide standards in prior HX MS work with this instrumentation.51,52 (c) Schematic depicting the docking of the AM6580-derived fluorenyl piprazine group covalently attached to hMGL active-site Ser122 (i.e., Ser129 for 6-His-hMGL) in a portion of the hMGL structure representation derived from PDB ID: 3JW8.26 Helix-α6 peptide 217–223 (orange) is shielded by the carbamylating-group modification at Ser122.
In the presence of phospholipid bilayer nanodiscs, carbamylation of hMGL Ser129 by AM6580 significantly reduced deuterium uptake in the vicinity of the enzyme's active site [i.e., peptides 217–223 (helix-α6), 224–240, and 242–256 (from sheet β7 to helix α7 and including catalytic-triad aspartic acid)]25,26 [Fig. 2(b)]. Interaction between the AM6580-derived fluorenyl moiety and hydrophobic residues in carbamylated hMGL may be responsible for shielding helix-α6 peptide 217–223, as suggested by docking the fluorenyl piprazine moiety from AM6580 into the active site of the hMGL structure representation PDB ID: 3JWE26 [Fig. 2(c)]. The fluorenyl piprazine group covalently attached to hMGL Ser129 may also render the enzyme's active site region more rigid and less dynamic. In contrast, AM6580 did not influence deuterium uptake into hMGL peptides either at the bottom of the enzyme's substrate-binding pocket and encompassing the catalytic-triad serine (i.e., peptides 101–125 and 126–137) or within the hMGL lid domain [i.e., peptides 158–166, 166–178, and 182–191; Fig. 2(b)]. The lack of effect of nanodiscs on deuterium uptake by the helix-α4 lid peptide (residues 166–178) whether hMGL was inhibited or not suggests that AM6580 enters the enzyme's substrate-binding pocket from the nanodisc phospholipid bilayer via the membrane-associated lid, the inhibited enzyme itself remaining associated with the nanodisc in an open conformation.
MD simulations predict hMGL interaction topology with nanodiscs
As a computational approach for characterizing further the conformational impact of hMGL membrane interaction, we next performed MD simulations with POPC/POPG nanodiscs and hMGL modeled from the X-ray structure of wild-type hMGL (PDB ID: 3JW8)26 in an open conformation. Following 10 ns of MD simulation, significant components of the hMGL lid region were found to interact with the nanodisc phospholipid bilayer. Lid-domain helix α4 penetrated into the phospholipid bilayer, whereas helix α6 in the vicinity of the lid and closer to the active site evidenced a less intimate membrane association, whether the enzyme was in its unliganded apo form [Fig. 3(a)] or carbamylated by inhibitor AM6580 [Fig. 3(b)].
Figure 3.
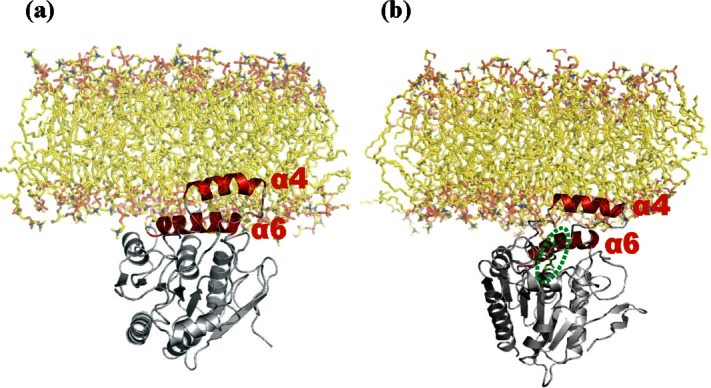
MD simulation of hMGL interaction with membrane phospholipid bilayer. Snapshots of hMGL (structure derived from PDB ID: 3JW8)26 with a phospholipid bilayer membrane of the same composition as in the experimental nanodiscs (POPC: POPG, 3:2 molar ratio) after 10 ns of MD simulation. The enzyme is depicted: (a) unliganded as apo-hMGL and (b) occupied with carbamylating inhibitor, AM6580, in the active site. Helix α4 in the lid domain and nearby helix alpha 6 are depicted in red, and AM6580 (b) is highlighted within the green oval and depicted in green.
Based on Cα positions, superposition of the starting hMGL model (PDB ID: 3JW8)26 with hMGL after 10 ns of MD simulations revealed that the majority of Cα atoms were indeed superimposable, whereas helices α4 and α6 displayed large variations after 10 ns simulations [Fig. 4(a)]. Helix α4 rotated 31° counterclockwise in the same plane away from the hMGL active site opening, resulting in the helix α4 ends being separated by a distance of 5.5 Å [Fig. 4(b)]. Helix α4 rotation was accompanied by simultaneous motion of the loop region connecting helices α4 and α5 (residues 177–192).25,26 In particular, residues Leu179 and Ile182 shifted and were present as different rotamers, substantially altering the hMGL active site conformation. Helix α6 rotated 22° counterclockwise in the same plane away from the active site. The net results of these collective conformational changes are increased opening of the hMGL substrate-binding pocket and enhanced active site accessibility. Although the 10-ms timescale used in our MD simulations is relatively brief and may not sample fully the dynamics of, especially, integral-membrane proteins such as transporters, ion channels, and seven transmembrane receptors,43 the computational data support our experimental observations of functionally relevant hMGL membrane interactions and local motions involving helices α4 and α6 and the loop conjoining helices α4 and α5.
Figure 4.
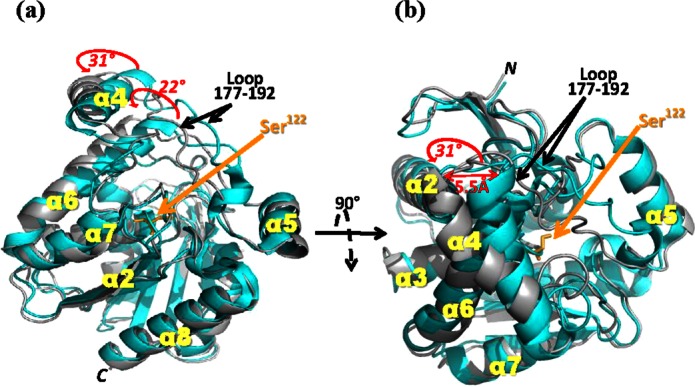
Schematic superposition of the X-ray structure of hMGL (cyan, PDB ID: 3JW8)26 and the model of hMGL obtained after 10 ns of MD simulation at a water/phospholipid interface (gray). (a) Side view of hMGL; (b) top view of hMGL rotated 90° as indicated from the orientation in (a). The structures are virtually identical, except for helices α4 and α6 and the loop region from amino acid residues 177–192 connecting helices α4 and α5. The movements of helices α4 and α6 induced by a phospholipid bilayer (POPC:POPG, 3:2 molar ratio) are indicated by red arrows and red numerals specifying the extent of movement. Active site Ser122 (i.e., Ser129 for 6-His-hMGL) is highlighted.
Conclusions
Crystallographic analyses of various apo and liganded hMGL variants25–27 have furnished atomic-level structural detail on a few static conformational states of an enzyme that has garnered intense interest as a therapeutic target.1,5,17,18 Despite documentation that MGL exists in situ as a membrane-associated enzyme3,23,24 and observations that lipase association with supramolecular lipid assemblies activates these enzymes,19,20 direct experimental information regarding the potential influence of membrane association on hMGL molecular properties is lacking. Our biochemical, HX MS, and computational analyses of the impact of a well-recognized biomembrane mimetic, the phosphoplipid bilayer nanodisc, on hMGL kinetic properties and conformation constitute the first detailed study to address this subject. The collective data provide evidence that subdomains within and proximal to the hMGL lid region associate intimately with the membrane phospholipid bilayer through hydrophobic interaction, creating an interfacial microenvironment that enhances the enzyme's kinetic properties. Our HX MS and MD simulation data indicate that the process of hMGL membrane association involves hMGL lid-domain helix α4 and, somewhat more distally, helix α6 and is accompanied by dynamic regional alterations in the conformation of these helices and the enzyme's active site region that would help stabilize an open hMGL conformation at the lipid/water boundary receptive to substrate/inhibitor partitioning from the membrane bilayer and into the enzyme's substrate-binding pocket. The shielding of select hMGL regions in proximity to the active site upon Ser129 carbamylation by AM6580 demonstrates directly that considerable conformational plasticity is associated with the enzyme's active site region in response to covalent inhibitors. Our data suggest a mechanism of hMGL–substrate/inhibitor interaction according to which the ligand diffuses laterally within the membrane bilayer and accesses the hMGL binding pocket by entering through the enzyme's lid, which is in close continuity with the nanodisc membrane. The results are also in-line with an earlier publication from our laboratory in which we proposed that hydrophobic ligands gain access to the cannabinoid-receptor binding domain by a lateral diffusion process within a bilayer membrane system.44 The emergence of covalent enzyme inhibitors as potential drug candidates for various diseases45 and the identification of serine-reactive carbamylating agents as the lead chemical class of hMGL inhibitors1,18 make our observations particularly relevant to the design and targeting of hMGL inhibitors as potential medications. More generally, the present study demonstrates the suitability of peptide-level HX MS combined with nanodisc technology for investigating the structure–function correlates of enzyme–membrane interaction.
Materials and Methods
Materials
SDS-PAGE supplies, SM-2 biobeads, and Bio Spin columns were purchased from Bio-Rad (Hercules, CA). MS-grade trypsin (Trypsin Gold) was from Promega (Madison, WI). AM6580 and AHMMCE were synthesized at the Center for Drug Discovery, Northeastern University (Boston, MA) by standard routes. High-performance liquid chromatography (HPLC) grade acetonitrile, ethylenediaminetetraacetic acid (EDTA) (99%), and 85% phosphoric acid were purchased from Fisher Scientific (Pittsburgh, PA). Arachidonic acid was from Nu-Check Prep (Elysian, MN), and 2-AG was a generous gift from the National Institute on Drug Abuse (Bethesda, MD). Fatty acid-free bovine serum albumin, magnesium chloride tetrahydrate, and Trizma base were purchased from Sigma-Aldrich (St. Louis, MO). The plasmid expressing MSP1D1 was from Add Gene (Cambridge, MA). POPC and POPG were purchased from Avanti Polar Lipids (Alabaster, AL) as stock solutions in chloroform. 1,2-Deuterium oxide (>99%) was purchased from Cambridge Isotope Laboratories (Andover, MA).
Purification of recombinant wild-type hMGL
Recombinant 6-His-tagged, wild-type human hMGL was expressed in Escherichia coli and purified by cobalt affinity chromatography, as previously detailed.36 Chromatographic fractions were pooled and dialyzed against 50 mM Tris–HCl, pH 8.0, containing 100 mM NaCl, and protein concentration was estimated by absorbance at 280 nm. Purity was monitored by SDS-PAGE.
MSP1D1 purification
MSP1D1 was expressed and purified as described.46,47 The protein was isolated by cobalt affinity chromatography, and purity was monitored by SDS-PAGE. Fractions containing MSP1D1 were pooled and dialyzed against 20 mM Tris–HCl, pH 7.4, containing 100 mM NaCl, 0.5 mM EDTA, and 0.01% NaN3. Protein concentration was estimated by absorbance at 280 nm.
Nanodisc self-assembly and isolation
Purified MSP1D1 in 20 mM Tris–HCl, pH 7.4, containing 100 mM NaCl, 0.5 mM EDTA, and 0.01% NaN3 was added to a solubilized mixture of either POPC/sodium cholate or POPC-POPG (3:2 molar ratio)/sodium cholate, and the final molar ratio was adjusted to 1:78:200 (MSP1D1:phospholipid:sodium cholate). The mixture was incubated for 1 h on ice, and cholate detergent was removed during a gentle 10-h rotation with SM-2 biobeads at 4°C. Nanodiscs were purified by size-exclusion chromatography with an Amersham-Pharmacia ÄKTA FPLC Protein Purifier System (GE Healthcare Life Sciences, Pittsburgh, PA) on a Superdex 200 10/300 column eluted with 20 mM Tris–HCl, pH 7.4, containing 100 mM NaCl and 0.5 mM EDTA at 0.5 mL/min. Column eluate absorbance was monitored at 280 nm, and SDS-PAGE was used to evaluate the purity of the nanodisc-containing fractions. FPLC fractions containing purified nanodiscs were collected and concentrated for experimental use. Nanodisc concentration was calculated form the absorbance at 280 nm and the MS1D1 molar extinction coefficient, accounting for two MSP1D1 molecules per disc.
FPLC analysis of nanodiscs incubated with hMGL
Purified, detergent-free hMGL was incubated with purified nanodiscs for 30 min at room temperature at an hMGL:nanodisc molar ratio of 1:2. Samples of hMGL alone and the hMGL–nanodisc coincubation were analyzed by FPLC as detailed above for nanodiscs.
Determination of hMGL kinetic properties
Kinetic constants for hMGL were determined by measuring the hydrolysis of either natural substrate (2-AG) or fluorogenic reporter substrate (AHMMCE) with methods adapted from our prior work.36 In brief, 2-AG at varying concentrations (10–400 μM) was incubated at 37°C with either 2.9 nM hMGL alone or 2.9 nM hMGL with nanodiscs at an MGL:nanodisc molar ratio of 1:2 in TME buffer (25 mM Tris base, 5 mM MgCl2, and 1 mM EDTA, pH 7.4) in a total reaction volume of 300 μL. Reaction samples (50 μL) were taken immediately at the start of the incubation and after 4 min, diluted 1:2 by volume with acetonitrile, and centrifuged at 20,000g for 5 min at 4°C. A 20-μL aliquot of each supernatant was subjected to reverse-phase HPLC on an Agilent Zorbax XDB-C18 column (4.6 mm × 150 mm, 3.5 µm; Agilent Technologies, Santa Clara, CA). Mobile phase A was 100% acetonitrile, and mobile phase B consisted of 8.5% aqueous phosphoric acid/acetonitrile (60:40, v/v) with the following gradient at a flow rate of 1 mL/min: 100% mobile phase B for 2 min, 5% mobile phase B for 5 min, 100% mobile phase B for 1 min followed by a 5-min injection delay. In an 8-min run, 2-AG was eluted at 4.2 min, and arachidonic acid at 5.0 min, allowing the reaction to be followed by either substrate utilization or product formation.
A fluorogenic hMGL assay based upon the conversion of AHMMCE reporter substrate to coumarin fluorophore was conducted as described.36 Reaction samples at room temperature included various concentrations of AHMMCE incubated with either 2.9 nM hMGL alone, 2.9 nM hMGL with 0.5 mM Triton X-100, or 2.9 nM hMGL with 5.8 nM nanodiscs in 50 mM Tris–HCl buffer, pH 7.4, at a reaction volume of 200 μL. Fluorescence readings at 360 nm/460 nm (λexcitation/λemission) were taken every 15 min for up to 2 h, and relative fluorescence units were converted to the amount of coumarin flurophore formed based upon a coumarin standard curve.
Michaelis–Menten kinetic parameters were derived with Prism software (GraphPad, San Diego, CA). Apparent Km and Vmax values are the mean ± standard deviation (SD) for triplicate determinations across three independent enzyme preparations. Statistical significance of group-mean differences was evaluated by a two-sample independent t-test, the significance level set at P ≤ 0.05.
Peptide-level HX MS analysis
Continuous labeling hydrogen-deuterium exchange experiments48 were initiated by diluting 10-fold 12 μL of an hMGL–nanodisc mixture (5 μM hMGL and 10 μM nanodiscs) into 99% D2O buffer (50 mM Tris–HCl containing 100 mM NaCl, pD 7.6) at room temperature. For inhibited enzyme samples, AM6580 was added to the hMGL–nanodisc mixture at an inhibitor:hMGL molar ratio of 5:1 and was incubated for 1 h at room temperature prior to dilution into D2O buffer. At selected times ranging from 10 s to 4 h after the introduction of D2O, samples of the exchange reaction were taken and immediately quenched by acidifying to pH 2.5 with formic acid and placed on ice to limit back exchange.49 After quenching, nanodiscs were rapidly disassembled with the addition of ice-cold sodium cholate in a 25:1 molar ratio of sodium cholate:nanodisc phospholipid.50 Porcine pepsin (1.2 μL of a 10 mg/mL stock solution) was added to digest the sample during a 5-min incubation on ice. In the last minute of digestion, ZrO2 beads were added to the digestion mixture to facilitate phospholipid removal. The sample was filtered through a prechilled, 0.45-μm cellulose acetate membrane by centrifugation at 18,000g and 4°C for 1 min to trap both the pepsin and ZrO2 beads. The flow through was injected without delay into a precolumn trap (Waters VanGuard C18, 2.1 mm × 5 mm, 1.7 μm) and desalted with 0.05% formic acid in water for 5 min. The trap was placed in-line with a second identical precolumn directly connected to the analytical column (Waters BEH C18, 1.0 mm × 100 mm, 1.7 μm). HX data were acquired on a Waters nanoAcquity UPLC with HDX Technology. Peptides originating from hMGL pepsinolysis were identified from triplicate analyses of undeuterated control enzyme samples using Waters ProteinLynx Global Server 2.4. All reported peptide-based HX MS data were derived from triplicate hMGL preparations, each analyzed in triplicate. The error of peptide HX MS measurements was ±0.50, as determined by replicate analyses of peptide standard and prior HX MS data from this experimental setup.51,52
MALDI-TOF/TOF MS analysis of hMGL covalent modification by AM6580
Purified hMGL was incubated with AM6580 (molar ratio 1:5, enzyme:inhibitor) at room temperature for 1 h, at which time the enzyme was verified by direct biochemical assay to be inhibited (above). The incubation was terminated by desalting using a Bio-Spin 6 column and 25 mM ammonium bicarbonate buffer, pH 8.0, containing 0.05% CYMAL. The desalted enzyme sample was digested overnight with 200 ng Trypsin Gold. Equal volumes of the trypic digest and α-cyano-4-hydroxycinnamic acid matrix (5 mg/mL in aqueous 50% acetonitrile–0.1% trifluoroacetic acid; 0.5 μL each) were cocrystallized. MS characterization of hMGL covalent modification by AM6580 were acquired on a 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA) fitted with a 200-Hz solid-state ultraviolet laser (wavelength 355 nm) from samples spotted on Opti-TOF 384-well plate inserts.
Computational methods
MD simulation setup
A 128-lipid, racemic POPG bilayer neutralized with 128 Na+ counterions and hydrated with 3527 water molecules was used as the basis of a POPC/POPG bilayer.53 All water molecules and ions were removed. Since the experimental nanodisc contained POPC and L-POPG in a 3:2 molar ratio, all D-POPG and 11 L-POPG lipid molecules were replaced with POPC, and three L-POPG lipid molecules were removed. This resulted in a phospholipid bilayer containing 75 POPC and 50 L-POPG lipid molecules, which was solvated with 4270 water molecules and neutralized with 50 Na+ counterions. The system was fully minimized with steepest descent, and 40 ns of unrestrained MD were then performed to produce a fully solvated, fully equilibrated POPC/POPG bilayer (simulation procedure below). The potential energy reached a stable plateau after 3 ns. After 10 ns, the area of the xy plane per lipid fluctuated around 69.7 Å2, a value within the experimental range for POPC (63.0–68.3 Å2) and probable range for POPG (64–70 Å2).53,54 Two simulation systems were established: one with unliganded, wild-type hMGL (PDB ID: 3JW8),26 the other with hMGL covalently carbamylated by AM6580. The carbamylating moiety of AM6580 was docked to wild-type hMGL (PDB ID: 3JWE)26 using Glide at the XP level,55 and a covalent bond was made between this moiety and catalytic Ser122 (i.e., Ser129 for the 6-His-hMGL). To enable MD simulation of a covalently bound ligand, a custom residue was defined as Ser122 carbamylated by AM6580 and was parameterized.56
MD simulation procedure
Unmodified and AM6580-carbamylated hMGL were placed with the enzyme's lid domain partially embedded in the POPC/POPG bilayer, as given by the “Orientations of Proteins in Membranes” database.57,58 Any lipid or water molecules clashing with the enzyme were removed. The system was made electrically neutral and minimized with steepest descents to relax unfavorable intermolecular contacts. To stabilize the lipid environment during production dynamics, equilibration MD was performed for 2 ns, with all heavy atoms in the protein positionally restrained with a force constant of 1000 kJ/mol/nm. Unconstrained production MD was performed for 10 ns on each system. All MD simulations were performed in the NPT (isothermal–isobaric) ensemble with periodic boundary conditions. A temperature of 300 K was maintained with time constant 0.1 ps by an extension of the Berendsen thermostat to which a properly constructed random force was added.59 Semi-isotropic pressure coupling was used, with the reference pressure set at 1.0 bar and time constant at 5 ps. Coulomb and short-range neighbor list cutoffs were both set to 0.9 nm, and Lennard-Jones cutoffs were set to 1.2 nm. The electrostatic interactions were computed using the Particle-Mesh Ewald method with an interpolation order of 4 and a maximum grid spacing of 0.12 nm.60,61 A time step of 2 fs was used, and pair lists were updated every 10 steps. The LINCS algorithm62 was employed to preserve bond lengths. The simple point charge water model63 was used in all simulations. The simulations were carried out with the GROMACS program, version 4.5.5, using the GROMOS96 53a6 force field.53,64,65
Acknowledgments
We thank Drs. Aneetha Halikhedkar, Christopher Morgan, Mark Williams, and Xiaoyu Tian for helpful discussion.
Glossary
- 2-AG
2-arachidonoylglycerol
- AHMMCE
arachidonoyl 7-hydroxy-6-methoxy-4-methylcoumarin ester
- CB1R
cannabinoid receptor 1
- CB2R
cannabinoid receptor 2
- FPLC
fast protein liquid chromatography
- hMGL
human monoacylglyceorl lipase
- HX
hydrogen exchange
- Km
apparent dissociation constant
- MALDI-TOF
matrix-assisted laser desorption ionization-time of flight
- MD
molecular dynamics
- MGL
monoacylglycerol lipase
- MS
mass spectrometry
- MSP
membrane scaffold protein
- MSP1D1
membrane scaffold protein 1D1
- PDB
protein data bank
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Vmax
apparent maximum velocity
Supplementary material
Additional Supporting Information may be found in the online version of this article.
Figure S1. Analysis of hMGL (green line), POPC/POPG phospholipid bilayer nanodiscs (red line), and hMGL-nanodisc mixture (black line) by (a) size-exclusion FPLC and (b) SDS-PAGE. The calibration scale for the size-exclusion column is shown and was derived from proteins of known Stokes hydrodynamic diameter: thyroglobulin (bovine thyroid) (17 nm), ferritin (horse spleen) (12.2 nm), catalase (bovine liver) (10.4 nm), albumin (bovine serum) (7.1 nm).
References
- 1.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinh TP, Kathuria S, Piomelli D. RNA interference suggests a primary role for monoacylglycerol lipase in the degradation of the endocannabinoid 2-arachidonoylglycerol. Mol Pharmacol. 2004;66:1260–1264. doi: 10.1124/mol.104.002071. [DOI] [PubMed] [Google Scholar]
- 3.Blankman JL, Simon GM, Cravatt BF. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savinainen JR, Saario SM, Laitinen JT. The serine hydrolases MAGL, ABHD6 and ABHD12 as guardians of 2-arachidonoylglycerol signaling through canmnabinoid receptors. Acta Physiol. 2012;204:267–276. doi: 10.1111/j.1748-1716.2011.02280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janero DR, Vadivel SK, Makriyannis A. Pharmacotherapeutic modulation of the endocannabinoid signaling system in psychiatric disorders: drug-discovery strategies. Int Rev Psychiatry. 2009;21:122–133. doi: 10.1080/09540260902782778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermudez-Silva FJ, Cardinal P, Cota D. The role of the endocannabinoid system in the neuroendocrine regulation of energy balance. J Psychopharmacol. 2012;26:114–124. doi: 10.1177/0269881111408458. [DOI] [PubMed] [Google Scholar]
- 7.Guindon J, Guijarro A, Piomelli D, Hohmann AG. Peripheral antinociceptive effects of inhibitors of monoacylglycerol lipase in a rat model of inflammatory pain. Br J Pharmacol. 2011;163:1464–1478. doi: 10.1111/j.1476-5381.2010.01192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gosh S, Wise LE, Chen Y, Guijar R, Mahadevan A, Cravatt BF, Lichtman AH. The monoacylglycerol lipase inhibitor JZL 184 suppresses inflammatory pain in the mouse carrageenan model. Life Sci. 2013;92:498–505. doi: 10.1016/j.lfs.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comelli F, Giagnoni G, Bettoni I, Colleoni M, Costa B. The inhibition of monoacylglycerol lipase by URB602 showed an anti-inflammatory and anti-nociceptive effect in a murine model of acute inflammation. Br J Pharmacol. 2007;152:787–794. doi: 10.1038/sj.bjp.0707425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinsey SG, Nomura DK, O'Neal ST, Long JZ, Mahadevan A, Cravatt BF, Grider JR, Lichtman AH. Inhibition of monoaclglycerol lipase attenuates nonsteroidal anti-inflammatory drug-induced gastric hemorrhages in mice. J Pharmacol Exp Ther. 2011;338:795–802. doi: 10.1124/jpet.110.175778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melis M, Pillolla G, Bisogno T, Minassi A, Petrosino S, Perra S, Muntoni AL, Lutz B, Gessa GL, Marsicano G, Di Marzo V, Pistis M. Protective activation of the endocannabinoid system during ischemia in dopamine neurons. Neurobiol Dis. 2006;24:15–27. doi: 10.1016/j.nbd.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Sciolino NR, Zhou W, Hohmann AG. Enhancement of endocnnabinoid signaling with JZL184, an inhibitor of the 2-arachidonoylglycerol hydrolyzing enzyme monoacylglycerol lipase, produces anxiolytic effects under consitions of high environmental aversiveness in rats. Pharmacol Res. 2011;64:226–234. doi: 10.1016/j.phrs.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sticht MA, Long JZ, Rock EM, Limebeer CL, Mechoulam R, Cravatt BF, Parker LA. Inhibition of monoacylglycerol lipase attenuates vomiting in Suncus murinus and 2-arachdonoyl glycerol attenuates nausea in rats. Br J Pharmacol. 2012;165:2425–2435. doi: 10.1111/j.1476-5381.2011.01407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nithipatikom K, Endsley MP, Isbell MA, Wheelock CE, Hammock BD, Campbell WB. A new class of inhibitors of 2-arachidonoylglycerol hydrolysis and invasion of prostate cancer cells. Biochem Biophys Res Commun. 2005;332:1028–1033. doi: 10.1016/j.bbrc.2005.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen SG, Ramesh D, Booker L, Burston JJ, Thomas EA, Selley DE, Sim-Selley LJ, Liu QS, Lichtman AH, Cravatt BF. Chronic monoacylglyerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat Neurosci. 2010;13:1113–1119. doi: 10.1038/nn.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler CJ. Monoacyglycerol lipase—a taregt for drug development? Br J Pharmacol. 2012;166:1568–1585. doi: 10.1111/j.1476-5381.2012.01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulvihill MM, Nomura DK. Therapeutic potential of monoacylglycerol lipase inhibitors. Life Sci. 2013;92:492–497. doi: 10.1016/j.lfs.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aloulou A, Rodriguez JA, Fernandez S, van Oosterhout D, Puccinelli D, Carrière F. Exploring the specific features of interfacial enzymology based on lipase studies. Biochim Biophys Acta. 2006;1761:995–1013. doi: 10.1016/j.bbalip.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Rehm S, Trodler P, Pleiss J. Solvent-induced lid opening in lipases: a molecular dynamics study. Protein Sci. 2010;19:2122–2130. doi: 10.1002/pro.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derewenda ZS, Derewenda U, Dodson GG. The crystal and molecular structure of the Rhizomucor miehei triacylglyceride lipase at 1.9 Å resolution. J Mol Biol. 1992;227:818–839. doi: 10.1016/0022-2836(92)90225-9. [DOI] [PubMed] [Google Scholar]
- 22.Derewenda U, Swenson L, Wei Y, Green R, Kobos PM, Joerger R, Haas MJ, Derewenda ZS. Conformational lability of lipases observed in the absence of an oil–water interface: crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar. J Lipid Res. 1994;35:524–534. [PubMed] [Google Scholar]
- 23.Di Marzo V, Bisogno T, De Petrocellis L, Melck D, Orlando P, Wagner JA, Kunos G. Biosynthesis and inactivation of the endocannabinoid 2-arachidonoylglycerol in circulating and tumoral macrophages. Eur J Biochem. 1999;264:258–267. doi: 10.1046/j.1432-1327.1999.00631.x. [DOI] [PubMed] [Google Scholar]
- 24.Duncan M, Thomas AD, Cluny NL, Patel A, Patel KD, Lutz B, Piomelli D, Alexander SPH, Sharkey KA. Distribution and function of monoacylglycerol lipase in the gastrointestinal tract. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1255–G1265. doi: 10.1152/ajpgi.90500.2008. [DOI] [PubMed] [Google Scholar]
- 25.Labar G, Bauvois C, Borel F, Ferrer JL, Wouters J, Lambert DM. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem. 2010;11:218–227. doi: 10.1002/cbic.200900621. [DOI] [PubMed] [Google Scholar]
- 26.Bertrand T, Auge F, Houtmann J, Rak A, Vallee F, Mikol V, Berne PF, Michot N, Cheuret D, Hoornaert C, Mathieu M. Structural basis for human monoglyceride lipase inhibition. J Mol Biol. 2010;396:663–673. doi: 10.1016/j.jmb.2009.11.060. [DOI] [PubMed] [Google Scholar]
- 27.Schalk-Hihi C, Schubert C, Alexander R, Bayoumy S, Clemente JC, Deckman I, DesJarlais RL, Dzordzorme KC, Flores CM, Grasberger B, Kranz JK, Lewandowski F, Liu L, Ma H, Maguire D, Macielag MJ, McDonnell ME, Mezzasalma Haarlander T, Miller R, Milligan C, Reynolds C, Kuo LC. Crystal structure of a soluble form of human monoglyceride lipase in complex with an inhibitor at 1.35 Å resolution. Protein Sci. 2011;20:670–683. doi: 10.1002/pro.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 29.Garavito RM, Ferguson-Miller S. Detergents as tools in membrane biochemistry. J Biol Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 30.Chou JJ, Kaufman JD, Stahl SJ, Wingfield PT, Bax A. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J Am Chem Soc. 2002;124:2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 31.Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho BK, Perahia D, Buckle AM. Hybrid approaches to molecular simulation. Curr Opin Struct Biol. 2012;22:386–393. doi: 10.1016/j.sbi.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol Chem. 2009;390:805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 34.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Recosntitution of membrane proteins in phospholipid bilayer nanodiscs. Meth Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L. Measurements and implications of the membrane dipole potential. Annu Rev Biochem. 2012;81:615–635. doi: 10.1146/annurev-biochem-070110-123033. [DOI] [PubMed] [Google Scholar]
- 36.Zvonok N, Williams J, Johnston M, Pandarinathan L, Janero DR, Li J, Krishnan SC, Makriyannis A. Full mass spectrometric characterization of human monoacylglycerol lipase generated by large-scale expression and single-step purification. J Proteome Res. 2008;7:2158–2164. doi: 10.1021/pr700839z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karageorgos I, Zvonok N, Janero DR, Vemuri VK, Shukla V, Wales TE, Engen JR, Makriyannis A. Endocannabinoid enzyme engineering: soluble human thio-monoacylglycerol lipase (sol-S-hMGL) ACS Chem Neurosci. 2012;3:393–399. doi: 10.1021/cn3000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diaz JCM, Cordova J, Baratti J, Carriere F, Abousalham A. Effect of nonionic surfactants on Rhizopus homothallicus lipase activity. Mol Biotechnol. 2007;35:205–214. doi: 10.1007/BF02686006. [DOI] [PubMed] [Google Scholar]
- 39.Reis P, Holmberg K, Watzke H, Leser ME, Miller R. Lipases at interfaces: a review. Adv Colloid Interface Sci. 2009;147-148:237–250. doi: 10.1016/j.cis.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Carr PD, Ollis DL. α/β-Hydrolase fold: an update. Protein Pept Lett. 2009;16:1137–1148. doi: 10.2174/092986609789071298. [DOI] [PubMed] [Google Scholar]
- 41.Bowman AL, Makriyannis A. Refined homology model of monoacylglycerol lipase: toward a selective inhibitor. J Comput Aided Mol Des. 2009;23:799–806. doi: 10.1007/s10822-009-9289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zvonok N, Pandarinathan L, Williams J, Johnston M, Karageorgos I, Janero DR, Krishnan SC, Makriyannis A. Covalent inhibitors of human monoacylglycerol lipase: ligand-assisted characterization of the catalytic site by mass spectrometry and mutational anlaysis. Chem Biol. 2008;25:854–862. doi: 10.1016/j.chembiol.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindhal E, Sansom MSP. Membrane proteins: molecular dynamics simulations. Curr Opin Struct Biol. 2008;18:425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Xie X-Q, Melvin LS, Makriyannis A. The conformational properties of the highly selective cannabinoid receptor ligand CP-55,940. J Biol Chem. 1996;271:10640–10647. doi: 10.1074/jbc.271.18.10640. [DOI] [PubMed] [Google Scholar]
- 45.Kalgutkar AS, Dalvie DK. Drug discovery for a new generation of covalent drugs. Expert Opin Drug Disc. 2012;7:561–581. doi: 10.1517/17460441.2012.688744. [DOI] [PubMed] [Google Scholar]
- 46.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 48.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hebling CM, Morgan CR, Stafford DW, Jorgenson JW, Rand KD, Engen JR. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal Chem. 2010;82:5415–5419. doi: 10.1021/ac100962c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marcsisin SR, Narute PS, Emert-Sedlak LA, Kloczewiak M, Smithgall TE, Engen JR. On the solution conformation and dynamics of the HIV-1 viral infectivity factor. J Mol Biol. 2011;410:1008–1022. doi: 10.1016/j.jmb.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kukol A. Lipid models for united-atom molecular dynamics simulations of proteins. J Chem Theory Comput. 2009;5:615–626. doi: 10.1021/ct8003468. [DOI] [PubMed] [Google Scholar]
- 54.Janosi L, Gorfe AA. Simulating POPC and POPC/POPG bilayers: conserved packing and altered surface reactivity. J Chem Theory Comput. 2010;6:3267–3273. doi: 10.1021/ct100381g. [DOI] [PubMed] [Google Scholar]
- 55.Trotter BW, Nanda KK, Burgey CS, Potteiger CM, Deng JZ, Green AI, Hartnett JC, Kett NR, Wu Z, Henze DA, Della Penna K, Desai R, Leitl MD, Lemaire W, White RB, Yeh S, Urban MO, Kane SA, Hartman GD, Bilodeau MT. Imidazopyridine CB2 agonists: optimization of CB2/CB1 selectivity and implications for in vivo analgesic efficacy. Bioorg Med Chem Lett. 2011;21:2354–2358. doi: 10.1016/j.bmcl.2011.02.082. [DOI] [PubMed] [Google Scholar]
- 56.Malde AK, Zuo L, Breeze M, Stroet M, Poger D, Nair PC, Oostenbrink C, Mark AE. An automated force field topology builder (ATB) and repository: version 1.0. J Chem Theory Comput. 2011;7:4026–4037. doi: 10.1021/ct200196m. [DOI] [PubMed] [Google Scholar]
- 57.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 58.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 60.Darden T, York D, Pedersen L. Particle mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 61.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle-Mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 62.Hess B, Bekker H, Berendsen HJC, Fraaije J. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 63.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular forces. Dordrecht, Netherlands: Reidel; 1981. pp. 331–342. [Google Scholar]
- 64.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 65.van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG. Biomolecular simulation: the GROMOS96 manual and user guide. Zürich, Switzerland: vdf Hochschulverlag AG; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.