Abstract
GCN5-related N-acetyltransferases (GNATs) are the most widely distributed acetyltransferase systems among all three domains of life. GNATs appear to be involved in several key processes, including microbial antibiotic resistance, compacting eukaryotic DNA, controlling gene expression, and protein synthesis. Here, we report the crystal structure of a putative GNAT Ta0374 from Thermoplasma acidophilum, a hyperacidophilic bacterium, that has been determined in an apo-form, in complex with its natural ligand (acetyl coenzyme A), and in complex with a product of reaction (coenzyme A) obtained by cocrystallization with spermidine. Sequence and structural analysis reveals that Ta0374 belongs to a novel protein family, PaiA, involved in the negative control of sporulation and degradative enzyme production. The crystal structure of Ta0374 confirms that it binds acetyl coenzyme A in a way similar to other GNATs and is capable of acetylating spermidine. Based on structural and docking analysis, it is expected that Glu53 and Tyr93 are key residues for recognizing spermidine. Additionally, we find that the purification His-Tag in the apo-form structure of Ta0374 prevents binding of acetyl coenzyme A in the crystal, though not in solution, and affects a chain-flip rotation of “motif A” which is the most conserved sequence among canonical acetyltransferases.
Keywords: GNAT acetyltransferase, pai operon, degradative enzyme production, sporulation, acetyl coenzyme A, spermidine/spermine, P-loop
INTRODUCTION
When the environment can no longer support exponential increases in a bacterial population, the constituent cells induce expression of many genes normally repressed during conditions conducive to rapid growth. In some bacteria, for example Bacillus subtilis, the response leads to production of dormant spores. Two genes (abrB, hpr) and two operons (sin, pai) from Bacillus subtilis that act directly to silence postexponential gene expression during vegetative growth have been identified.1 The regulatory proteins encoded by abrB, hpr, and sin are well characterized compared with pai.1
The pai operon was initially cloned as a fragment of the Bacillus subtilis chromosome, which, when on high-copy-number plasmid, caused decreased levels of extracellular degradative enzymes such as subtilisin, neutral protease, levansucrase, α-amylase, and alkaline phosphatases, and a greatly reduced frequency of sporulation.1 mRNA levels of the neutral protease gene (nprE) were examined and found to be reduced, implying that pai acts at the transcriptional level.2
The pai operon has been sequenced and consists of two open reading frames, ORF1 and ORF2, encoding proteins of 21 kDa (PaiA) and 24 kDa (PaiB), respectively.2 Strains bearing the multicopy pai plasmid overproduce two proteins of these sizes, and N-terminal analysis of partially purified preparations indicates that the proteins correspond to the two pai genes (paiA and paiB). The presence of these genes on the multicopy vector revealed that both are essential for the aforementioned phenotypic effects. Disruption of the pai gene at ORF1 in the genomic DNA resulted in sporulation resistance and protease secretion. The mutant carrying an insertional disruption at ORF2 could not be constructed, suggesting that ORF2 product, the PaiB protein, is essential for cell growth.2
Structure of PaiA from Bacillus subtilis in complex with coenzyme A (CoA) has been determined.3 Structural analysis demonstrates that PaiA belongs to the GNAT superfamily,4 which catalyzes the transfer of acyl groups from acetyl-CoA to the primary amines of a diverse set of substrates, ranging from small molecules to large proteins.5 Despite the low level of sequence homology, the GNAT proteins possess a similar fold with a mixed parallel/antiparallel β-sheet surrounded by a number of conserved α-helices and are similar in the mode of acetyl-CoA binding.4 The most conserved interactions between the protein and acetyl-CoA involves the P-loop which forms the conserved “motif A” among GNAT superfamily members.4
Various biochemical data indicate that GNAT proteins use a direct acetyl transfer mechanism, which requires the formation of a ternary complex among enzyme, acetyl-CoA, and the acceptor substrate.4 The primary amine of the acetyl acceptor can initiate the chemical reaction through nucleophilic attack upon the acyl carbon of the acetyl group of acetyl-CoA. The resulting bisubstrate intermediate can then decompose through proton transfer from a catalytic acid. Most members of the superfamily possess a tyrosine that is well positioned to act as the catalytic acid.4–7 The primary amine of an acceptor is positively charged at physiological pH, so deprotonation is required before it can function as a nucleophile. This deprotonation can be driven by an active site amino acid of GNAT proteins or associated water molecules in the cases that lack residues appropriate for this role.8,9
The biochemical data for PaiA from Bacillus subtilis reveals that it exhibits an N-acetyltransferase activity toward polyamines, including spermine and spermidine. The latter is suggested to be a physiological substrate for PaiA.3 PaiA from Bacillus subtilis has been proposed to be a novel nonmembrane-bound spermidine/spermine N1-acetyltransferase (SSAT)3 that catalyzes the transfer of acetyl groups from acetylcoenzyme A to spermidine and spermine, as part of a polyamine degradation pathway.7,9 However, the characterization of PaiA is not consistent with structural and catalytic details of the active site of the SSAT family proteins.10
In this study, we present crystal structures of GNAT acetyltransferase Ta0374 from Thermoplasma acidophilum. Sequence analysis of Ta0374 confirms that it is a member of the protease synthase and sporulation negative regulatory protein family (PaiA). The crystal structure of Ta0374 from Thermoplasma acidophilum is similar to PaiA from Bacillus subtilis.3 Structural analysis and proton chemical shift analysis by NMR spectroscopy of Ta0374 reveals that spermidine is a substrate and binds to the protein. Four crystal structures of Ta0374 are described: the apo-form, the complex with acetyl-CoA in two crystal forms and the complex with the product (CoA) prepared by cocrystallization with spermidine. Detailed analysis of the protein structure, conformational changes, spermidine binding site, and possible mechanism of pai regulation are discussed.
MATERIALS AND METHODS
Protein cloning, expression, and purification
The recombinant Ta0374 from Thermoplasma acidophilum protein was cloned in the pMCSG7 vector developed at Midwest Center for Structural Genomics (MCSG) as a fusion protein containing a His-Tag with a Tobacco Etch Virus protease recognition site (MHHHHHHSSGVDLGT ENLYFQ↓SNA) at the N-terminus.11 Ta0374 was expressed in Escherichia coli BL21 magic cells by induction in Luria-Bertani medium (LB broth) for native Ta0374 protein and in High Yield M9 SeMet media kit (Medicilon) for selenomethionine-labeled Ta0374 protein. Purification was performed by Ni affinity chromatography.11
Crystallization
Both “Tag-on” (Ta0374) and “Tag-off” (Ta0374) proteins were crystallized by sitting-drop vapor-diffusion methods at room temperature. Drops were composed of one volume of the reservoir solution and one volume of the protein solution. The native and selenomethionine-labeled Ta0374 (8–10 mg/mL) protein solutions contained 500 mM sodium chloride, 5 mM β-mercaptoethanol in 10 mM Tris-HCl buffer (pH 8.3). The crystals of “Tag-on” selenomethionine-labeled apo-form of Ta0374 were grown in conditions containing 10% (w/v) PEG 8000, 10% ethylene glycol and 0.1M HEPES (pH 7.5). The crystals of the binary complex with acetyl-CoA of “Tag-off” native Ta0374 protein were obtained by cocrystallization with 5 mM acetyl-CoA in 30% (w/v) PEG6000 and 0.1M sodium acetate. The same tetragonal bipyramid crystals in complex with acetyl-CoA were also found in conditions with 0.2M sodium bromide, 20% (w/v) PEG 3350 and 0.1M Bis-Tris propane (pH 7.5). To obtain the ternary complex (dimeric complex with acetyl-CoA/CoA), these crystals were soaked with 2.5 mM spermidine. Crystals of the binary complex with CoA of “Tag-on” selenomethionine-labeled Ta0374 protein were obtained by cocrystallization with 2 mM spermidine in a condition containing 12% (v/v) glycerol, 1.5M ammonium sulfate, and 0.1M Tris (pH 8.5).
Data collection, structure determination, and refinement
Low-temperature (100 K) X-ray diffraction data sets were collected from single crystals of Ta0374 on beam-lines 21ID-D and 21ID-G (LS-CAT) at the Argonne National Laboratory.12 The diffraction data were processed with the HKL-2000 program suite.13 X-ray data-collection statistics are summarized in Table I.
Table I.
Crystallographic Parameters, Data-Collection and Refinement Statistics
| Ta0374 structures | Apo-form | Complex with acetyl-CoA | Complex with CoA | Dimeric complex with acetyl-CoA/CoA |
|---|---|---|---|---|
|
| ||||
| Number | I | II | III | IV |
| Data collection | ||||
| Resolution (Å) | 30-2.3 (2.34–2.30) | 54.5-2.5 (2.56–2.50) | 30-2.3 (2.34–2.30) | 55-2.3 (2.36–2.30) |
| Space group | P1 | P43212 | P43212 | P212121 |
| Unit cell parameters: | ||||
| a (Å) | 42.2 | 69.0 | 69.2 | 70.3 |
| b (Å) | 60.9 | 69.0 | 69.2 | 70.6 |
| c (Å) | 72.1 | 89.2 | 90.6 | 87.7 |
| α | 101.2 | |||
| β | 90.1 | |||
| γ | 90 | |||
| No of molecules per AU | 4 | 1 | 1 | 2 |
| Completeness (%) | 97.1 (91.2) | 99.87 (99.7) | 100 (100) | 98.7 (98.6) |
| Unique reflections | 28,727 | 7926 | 10,245 | 19,697 |
| I/σ(I) | 11.7 (2) | 19.2 (3.5) | 33.5 (4) | 17.5 (10.2) |
| Rmerge (%) | 5 (38) | 4.9 (56.5) | 15.1 (83.2) | 8.6 (18.5) |
| Wilson B-factor (Å2) | 67.7 | 53.2 | 46.3 | 46.2 |
| Refinement | ||||
| R (%)/Rfree (%)a | 22.9/27.5 | 17.7/22.9 | 18.7/23.1 | 19.2/24.7 |
| R.m.s. deviations from idealized geometry: | ||||
| Bond lengths (Å) | 0.022 | 0.012 | 0.017 | 0.015 |
| Bond angles (°) | 1.9 | 1.6 | 1.6 | 1.5 |
| Mean B value (Å2) | 40.2 | 26.2 | 18.9 | 21.3 |
| No of atoms | 5385 | 1453 | 1447 | 2658 |
| No of ligand atoms: | ||||
| Acetyl-CoA/CoA | 51/- | -/51 | 102/- | |
| Ni+/Cl/Br | 1/1/- | 1/-/- | 1/2/1 | |
| GOL/BME/SO4 | 6/8/5 | |||
| Mean B value of ligands (Å2): | ||||
| Acetyl-CoA/CoA | 29.2/- | -/23.1 | 23.1/- | |
| Ni+/Cl/Br | 23.6/37.7/- | 18.8/-/- | 21.2/46/33.5 | |
| GOL/BME/SO4 | 36.4/56.8/50.9 | |||
| Ramachandran analysis | ||||
| Favored (%) | 599 (94.2) | 151 (96.2) | 153 (97.5) | 308 (98.1) |
| Allowed (%) | 36 (5.7) | 6 (3.8) | 4 (2.5) | 5 (1.6) |
| Outlier (%) | 1 (0.2) | 1 (0.3) | ||
| PDB ID14 | 3FIX | 3F0A | 3NE7 | 3K9U |
Data for the highest resolution shell are given in parentheses.
R-factor = Σ (|Fobs|−k|Fcalc|)/Σ |Fobs|and R-free is the R value for a test set of reflections consisting of a random 5% of the diffraction data not used in refinement.
The selenomethionine-labeled “Tag-on” Ta0374 structure of apo-form was solved by the single-wavelength anomalous dispersion (SAD) method. Programs SHARP and HYSS were used to locate selenium atoms and calculate experimental phases to the limit of the data.15,16 SHELXD, SHELXE, RESOLVE, DM were used to perform density modification and improvement of the phases.17–21 The initial model of the apo-form structure was built by ARP/wARP.22 The structures of binary complexes of Ta0374 were solved by molecular replacement using MOLREP.23 The structure of the “Tag-on” apo-form Ta0374 was used as the starting model. Gaps, turns, and side chains for all solved structures were fitted manually using the program COOT.24 The structures were refined with REFMAC.25 Solvent atoms were initially built using the program ARP/wARP and later added or removed by manual inspection. The refinement statistics for four different structures are given in Table I.
The model validation was performed with SFCHECK, PROCHECK, ADIT, MOLPROBITY, and KING.26–29 Figures were produced using PyMOL, CCP4MG, and LIGPLOT.30–32
1H NMR spectroscopy
A Varian Inova 600 MHz NMR spectrometer equipped with a triple resonance, z-gradient cold probe was used to monitor the depletion of acetyl-CoA or the generation of acetylspermidine by measuring the heights of 1H resonances near 2.3 or 1.9 ppm, respectively.9 The data were acquired at a temperature of 25°C in 90% H2O, 10% D2O, and 10 mM Tris buffer at pH 8.0. A stock solution of 10 mg/mL Ta0374 enzyme containing 100 mM sodium chloride, 3 mM sodium azide, and 20 mM phosphate buffer at pH 8.3 was diluted in 10 mM Tris buffer (pH 8.0). For substrate titration, 5 mM acetyl-CoA and 5 mM spermidine were added to the final protein solution that had a concentration of 0.01 mg/mL. NMR one-dimensional proton spectra were recorded for acetyl-CoA and spermidine with Ta0374 enzyme (see supplemental material). kcat rates were determined by measuring NMR peak heights as a function of time. NMR experiments were processed with Varian VNMRJ software.
Sequence and structural analysis
A search for amino acid sequence homologs of Ta0374 from Thermoplasma acidophilum was carried out with PSI--BLAST.33 Multiple sequence alignment was constructed with MUSLE for sequence homologs identified by the database search.34 To identify a group of more closely related homologous sequences, they were clustered using CLANS.35 CLANS uses the P-values of high-scoring segment pairs obtained from an N × N BLAST search, to compute attractive and repulsive forces between each sequence pair. The most related sequences of PaiA N-acetyltransferases were aligned using CLUSTALW formatted using ESPript.36,37
Search for structural neighbors of Ta0374 by direct comparison of three-dimensional protein structures was performed with DALI, VAST, and ProFunc web servers.38–40
Prediction of the spermidine-binding site
The MEDock web server was used for prediction of the spermidine binding site in Ta0374.41 A global search strategy that exploits the maximum entropy property of the Gaussian distribution was employed. The spermidine was docked to one molecule of the structure of Ta0374 in complex with acetyl-CoA and to both independent molecules of the dimer of Ta0374 in complex with acetyl-CoA/CoA. The best solution was found for molecule A of the dimer of Ta0374 in complex with acetyl-CoA/CoA. For the docking protocol, the maximum generation in each run was set to 1000. A grid of 0.375 Å spacing was used for the calculation. Five separate docking calculations were performed for the separate molecules of Ta0374. Each calculation was performed with a population size of 50, and a probability of 0.05 to invoke a local search.
RESULTS
Sequence analysis and overall structure suggest that the protease and sporulation negative regulatory Ta0374, is a N-acetyltransferase (PaiA)
Ta0374 from Thermoplasma acidophilum (GenBank: CAC11518.1) is a hypothetical protein that has 161 amino acid residues and molecular weight of 19 kDa. Sequence clustering analysis (described in Materials and Methods section) reveals that Ta0374 belongs to the protease synthase and sporulation negative regulatory proteins (PaiA) that are closely connected to a cluster comprising the GNAT acetyl-transferases. It is interesting to note that the closest homolog to Ta0374 (identity—63%; sequence similarity—83%) is another PaiA N-acetyltransferase encoded by the neighboring gene Ta0375. It is not clear how this associates to the physiological function of these proteins. Sequence alignment of PaiA related acetyltransferases are shown in Figure 1(a).
Figure 1.
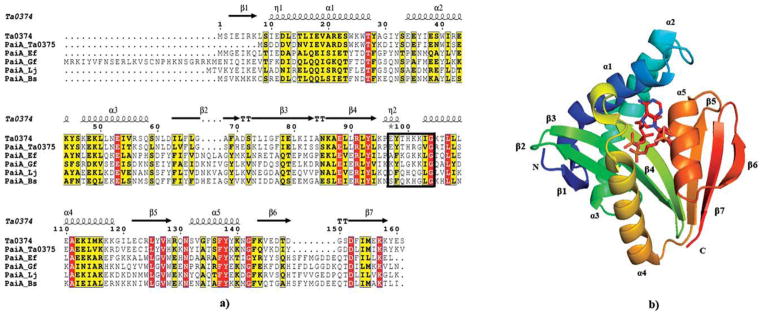
(a) Sequence alignment of PaiA from Thermoplasma acidophilum (Ta0374) and other PaiA family members: PaiA from Thermoplasma acidophilum (PaiA_Ta0375), PaiA from Enterococcus faecalis (PaiA_Ef), PaiA from Gramella forsetii (PaiA_Gf), PaiA from Lactobacillus johnsonii (PaiA_Lj), PaiA from Bacillus subtilis (PaiA_Bs). Secondary structure elements of Ta0374 are indicated above the sequence. P-loop is boxed. Sequence homologies are highlighted by red background (identities) and yellow (similarities); (b) Ribbon diagram of the Ta0374 molecule in complex with acetyl-CoA (the stick model). The secondary structure elements of the molecule are labeled.
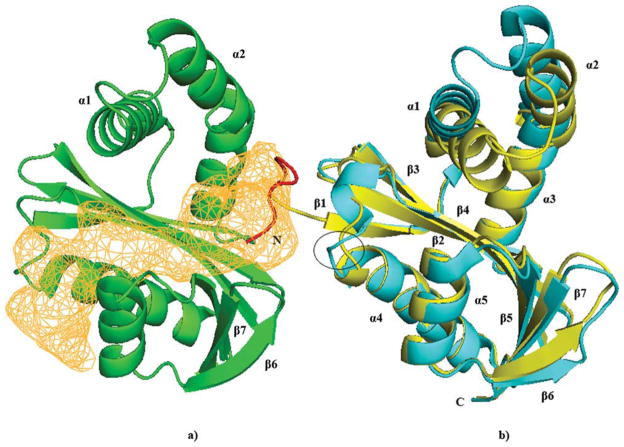
The crystal structure of the apo-form of Ta0374 was determined by the SAD method at 2.3 Å resolution (Table I, structure I). The asymmetric unit contains four protein molecules; each has a mixed α/β fold with a central seven-stranded β-sheet connected by five α-helixes [Figs. 1(b) and 2]. The central β-sheet is comprised mostly of antiparallel strands with the exception of strands 4 and 5, which are parallel. The order of the strands within the β-sheet is linear with respect to the sequence except at the C-terminus, where β7 is positioned between β5 and β6 [Fig. 1(a)]. The overall fold confirms that Ta0374 is a member of GNAT superfamily of acetyltransferases.
Figure 2.
Ribbon diagram of molecules A (C) (yellow) and B (D) (green) of the apo-form of Ta0374; (a) The acetyl-CoA and ligand-binding channel in Ta0374 that also found in known GNATs is presented in orange. The part of the His-Tag localized in the structure is shown in red. Helixes and β-strands that interact with “Tag” in the structure I crystal form and have different conformational states are labeled; (b) Superposition of A (C) (yellow) and B (D) (cyan) molecules of the apo-form Ta0374. The conformational differences of the P-loop are circled.
GNAT acetyltransferases with oligomeric structures are typically dimers constructed by joining the central β-sheet of each molecule into one continuous β-sheet.4,5 Often the active site contains residues from both molecules of the dimer. Examination of the protein-protein interface in the crystal form of the apo-form Ta0374 structure having multiple polypeptides in the asymmetric unit reveals that it is different from those found in other known GNAT members. Additionally, analysis by dynamic light scattering indicates that Ta0374 is monomeric in solution (data not shown).
Structural homology searches produced a long list of GNAT acetyltransferases including ribosomal-protein N1-acetyltransferases,42,43 phosphinothricin N-acetyl-transferase,44 bleomycin N-acetyltransferase,45 SSAT,10 etc. All share a conserved region involved in acetyl-CoA binding. However, the other parts of the structures are more variable, and most probably this is related to binding of their specific substrates.
The highest ranked protein based on Z-score, the root mean square deviation (RMSD 1.5 Å), the number of aligned residues (154), and the sequence identity (27%) is an acetyltransferase (PaiA) in complex with CoA and DTT from Bacillus subtilis (PDB ID 1TIQ).3 Despite the high resemblance between the structures, Ta0374 possesses several unique structural features compared with PaiA from Bacillus subtilis. The loops connecting strands β3–β4 and β6–β7 show significant structural differences. In the PaiA structure from Bacillus subtilis, these loops bind the second CoA molecule of the oxidized dimer CoA2 and DTT,3 leading to the suggestion that these loops in PaiA acetyltransferases may be involved in binding of the substrate. However, as is discussed by Dyda et al.,4 the upstream end of α2 and loop between α1 and α2 play an important role in the binding of the acceptor molecule in GNAT proteins.
His-Tag interactions in the structure of apo-form
In most cases, the His-Tags used to purify proteins are disordered when present in crystal structures and appear to be innocuous in functional studies of the proteins.46 However, in our case, the His-Tag residues bind to the adjacent molecule and interact at the active site of Ta0374 (Fig. 2). Six of the 22 N-terminal amino acid residues from the purification His-Tag attached to the protein are found in the electron density map in two molecules (A and C) of the apo-form structure. This “Tag” region localizes in the interface between molecules (A) and (B) or (C) and (D) and interacts with residues of α1, α2, β6, β7 of molecule (B), and with the P-loop and residues of α4 in molecule (A) (Fig. 2).
Superposition of (A) and (B) molecules of the apo-form structure reveals conformational changes (RMSD >1.5 Å), which occur in the regions involved in interactions with the “Tag” mentioned earlier [Fig. 2(b)]. Additionally, we find that the P-loop residues in molecule (A) have a “flip-flop” rotation of the backbone chains (His100-Lys101-Lys102) compared with their position in molecule (B) [Fig. 2(b)]. In this orientation, the side chain of the Lys101 moves to a position corresponding to the pyrophosphate-binding mode of acetyl-CoA observed in the structures of known GNAT. The “Tag” appears to prevent acetyl-CoA binding only in the structure I crystal lattice. We found that the Ta0374/acetyl-CoA complex is formed in the lysate and remains throughout purification. The structure of “Tag-on” Ta0374 obtained by cocrystallization with spermidine in different crystallization conditions (Table I, structure III) demonstrates the presence of the cofactor in the active site of the protein.
Acetyl-CoA binding site
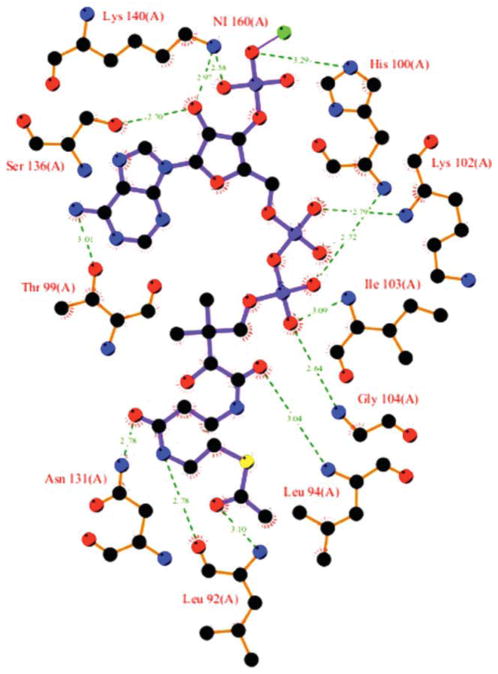
The acetyl-CoA molecule in the structure of the binary complex of Ta0374 (Table I, structure II) is well defined in the electron density map and found in the active site cleft between helix α4, the end of strand β4, and beginning of helix α3 [Fig. 1(b)]. This is the same coenzyme-binding mode that is typical for most GNAT acetyl-transferase superfamily members.4,5 A schematic diagram of the interaction with acetyl-CoA in the active site of the Ta0374 structure is presented in Figure 3.
Figure 3.
Schematic diagram of the acetyl-CoA binding site in Ta0374 structure of the binary complex. The nitrogen atoms are drawn in blue, oxygen in red, carbon in black, sulfur in yellow. Ni2+ ion and hydrogen bonds are shown in green.
The most conserved interactions between the protein and acetyl-CoA involve the P-loop, consisting of residues 97–104 between β4 and α4 [Fig. 1(a)]. This includes the conserved GNAT acetyltransferase “motif A. ”4 The backbones of residues Thr99, His100, Lys102, Ile103, and Gly104 along with water molecules form a network of hydrogen bonds with the pyrophosphate group of acetyl-CoA. This motif appears to be crucial for the recognition of the pyrophosphates of acetyl-CoA in the N-acetyl-transferases.4
The adenine portion of the acetyl-CoA is located on the Ta0374 surface. There is a significant variation in the position and conformation of the ribose possessing the 3′-phosphate and the base among structures of complexes of GNAT acetyltransferases.4 In fact, the position of this part of the acetyl-CoA in our structure is different from that in the structure of PaiA from Bacillus subtilis,3 and this is likely caused by strong crystal contacts. In our structure, the adenine group is located close to the conserved P-loop and interacts with the side chain of Thr99 (Fig. 3). In the structure of PaiA from Bacillus subtilis, the adenine group is located close to helix α6,3 where in our case the 3′-phosphate and ribose have hydrogen bonds with the side chains of the residues Ser136 and Lys140 of helix α6 and His100 of the P-loop (Fig. 3). Furthermore, a Ni2+ ion is found in structure of the binary complex, which coordinates two 3′-phosphates of acetyl-CoA from the symmetry related protein molecules.
The sulfhydryl and acetyl groups of acetyl-CoA are fitted into the splayed opening between β4 and β5. The side chain of Asn131, which is conserved among GNAT acetyltransferases, is hydrogen-bonded to the carbonyl oxygen of the pantothenate moiety closest to its sulfhydryl group (Fig. 3). The acetyl group of the cofactor is hydrogen-bonded to the main chain of Leu92 (Fig. 3). This mode of interaction, where the carbonyl of the acetyl group is hydrogen bonded to the main-chain amide nitrogen of a residue just downstream of the βbulge on β4, has been observed in complexes of other GNAT proteins.4 The splay between β4 and β5 is a distinctive characteristic of GNAT proteins and is an essential structural feature in promoting acetyl transfer.5
Ta0374 structure of the binary complex with acetyl-CoA/CoA obtained by soak with spermidine
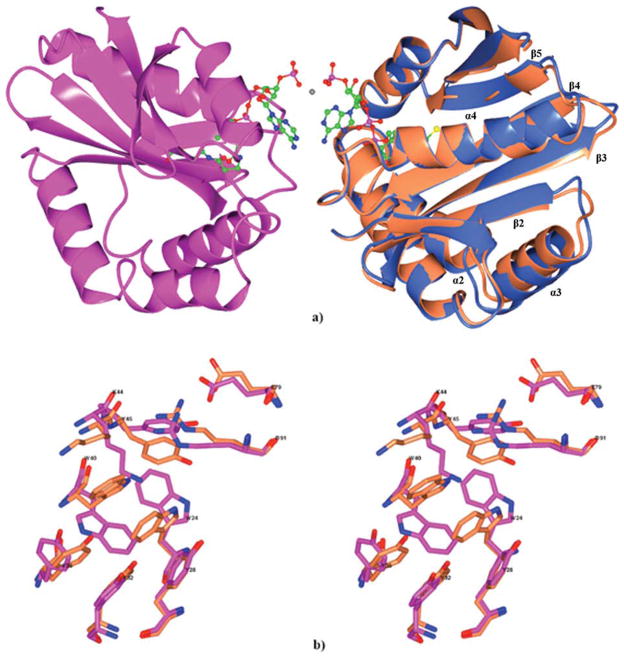
The binary complex with acetyl-CoA that was determined following an attempt to obtain a complex with spermidine by the soaking technique (Table I, structure IV) contains a dimer in asymmetric part of the unit cell [Fig. 4(a)]. The dimer is composed of molecules related by a noncrystallographic two-fold axis that runs along the diagonal of the crystallographic cell. Each channel of the dimer contains a well-ordered acetyl-CoA in the position observed in the structure of complex with acetyl-CoA (Table I, structure II). The dimer interface involves the cofactor molecules and residues from helix α1 (22–26) and conserved “motif A” (97–104). In structure IV, the 3′-phosphates of the cofactor from each molecule of the dimer are coordinated through strong interactions with a Ni2+ ion [Fig. 4(a)]. A similar dimer is found in the structure II, but in this case, the protein molecules are related by a crystallographic twofold rotation axis. The light scattering analysis and the structure of the apo-form of Ta0374 in a different crystal form (Table I, structure I) indicate that the observed dimer is a strong crystal contact but probably not physiologically relevant. In one molecule (A) of the noncrystallographic dimer, the acetyl group of the cofactor has an estimated occupancy of 70%. Because this does not occur in the absence of spermidine, this indicates that the crystallized enzyme can transfer the acetyl group to spermidine.
Figure 4.
(a) Ribbon diagram of the Ta0374 dimeric complex with acetyl-CoA/CoA is shown in light blue (molecule A) and magenta (molecule B). The acetyl-CoA/CoA is shown as a ball-and-stick model (nitrogen in blue, oxygen in red, carbon in green, sulfur in yellow). Ni2+ is shown as a grey dot between 3′-phosphate of acetyl-CoA/CoA molecules. Superposition of (A) molecule of the Ta0374 dimeric complex with acetyl-CoA/CoA (light blue) with molecule of the Ta0374 in complex with acetyl-CoA (coral); (b) Stereo view of the superposition of side chain residues that have different positions between (B) molecule (magenta) of Ta0374 dimeric complex with acetyl-CoA/CoA and the molecule (coral) of Ta0374 in complex with acetyl-CoA.
The molecules in the noncrystallographic dimer of Ta0374 (structure IV) were compared with the one molecule of structure II by LSQKAB.47 The RMSD between the coordinates of the Cα atoms are 0.9 and 0.3 Å, respectively for molecules (A) and (B) in structure IV. Deviations were observed in helixes α2, α3, α4, and loops connecting elements α3-β2, β3-β4, and α4-β5 of molecule A of structure IV compared with those positions in structure II [Fig. 4(a)]. Furthermore, analysis has shown differences in the position of the side chains of Trp24, Tyr28, Tyr32, Tyr36, Trp40, Lys44, Tyr45, Glu79, and Arg91 residues in molecule B of structure IV [Fig. 4(b)]. Crystal-packing analysis using the CONTACT program48 has shown that in both structures of complexes Ta0374 with acetyl-CoA (II and IV) of the molecules have similar contact networks with symmetry related molecules, and this seems unlikely to be responsible for the structural changes. We assume that such differences together with the partial occupancy of acetyl-CoA can be explained by the presence of spermidine; however, spermidine was not found in the structure of the “attempted” complex.
1H NMR spectroscopy activity assay
Analysis of product formation by 1H NMR spectroscopy demonstrated the spermidine dependent acetyltransferase activity of Ta0374. This NMR-based assay was advantageous in that it could be used to simultaneously monitor the appearance of products and disappearance of reactants, while identifying the products formed. The ability of Ta0374 to acetylate spermidine was tested at pH 8.3, using 5 mM acetyl-CoA and 5 mM spermidine. Catalytic rates (kcat) were determined by measuring NMR peak heights as a function of time (Supporting Information Fig. 1S) and were found to be 31 and 42.6 min−1, respectively, for acetyl-CoA and acetylspermidine. PaiA from Bacillus subtilis has been found to have a similar value.3 Taken together the above structural data, these findings support the concept that Ta0374 is capable of acetylating spermidine.
Structure of Ta0374 in complex with CoA
Cocrystallization of the enzyme with spermidine yielded the crystal structure of Ta0374 in complex with one of the products of the reaction (CoA) (Table I, structure III), confirming that the enzyme catalyzes the transfer of the acetyl group in the presence of spermidine. The asymmetric unit contains one molecule similar to that observed in structure II. The RMSD between α-carbons of these two structures is 0.2 Å. The CoA molecule has well-defined electron density and is localized in the same position as acetyl-CoA. The sulfhydryl group of CoA is found in two conformations (Fig. 5). In one position, the conformation places the sulfur atom of CoA close to the conserved Tyr138 that serves as the general acid that protonates the thiolate anion of CoA.4 This is the major conformation for the sulfur atom bound to the acetyl group of acetyl-CoA in structures of GNAT members and supports a mechanism of direct nucleophilic attack by the nearby N1 atom of the substrate on acetyl-CoA. In the other position, the sulfur atom of the CoA is in an oxidized state. It makes a disulfide bridge with β-mercaptoethanol (BME) and locates within hydrogen bonding distance of residues Tyr28 and the conserved Tyr93 (Fig. 5). The role of Tyr93 is uncertain, however, its position within the structure suggests that this residue could be involved in catalysis and participate in substrate binding.
Figure 5.
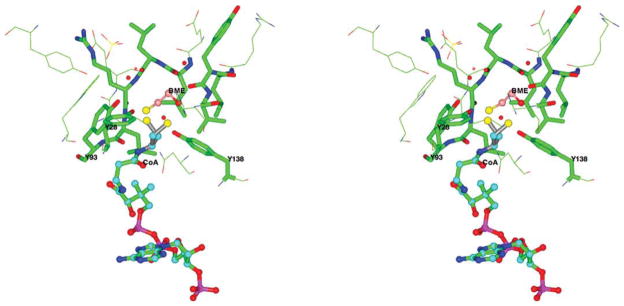
Stereo view of the sulfhydryl group of CoA in the binary complex of Ta0374 with CoA. BME and CoA are shown as ball-and-stick models with different color carbon atoms, nearby (less then 5 Å) protein residues as cylinder and line models. The nitrogen atoms of protein residues are drawn in blue, oxygen in red, carbon in green. In one orientation the sulfur atom of CoA is located close to Tyr138; in the other position the sulfur atom of CoA makes a disulfide bond with BME and is located within hydrogen bonding distance of Tyr28 and Tyr93.
Additionally, in the active site of the structure of the complex with CoA, a sulfate ion and some uncharacterized density for another small molecule are found. The sulfate ion makes hydrogen bonds with residues Trp40, Tyr45, and the conserved Arg91, and via water molecules with Tyr93 and Leu90. The unknown molecule is localized near conserved residues Glu53, Arg56, and Lys81. These interactions give us additional evidence to suggest that residue Trp40, Tyr45, Glu53, Arg91, and Tyr93 could be involved in the binding of the spermidine.
Spermidine binding site
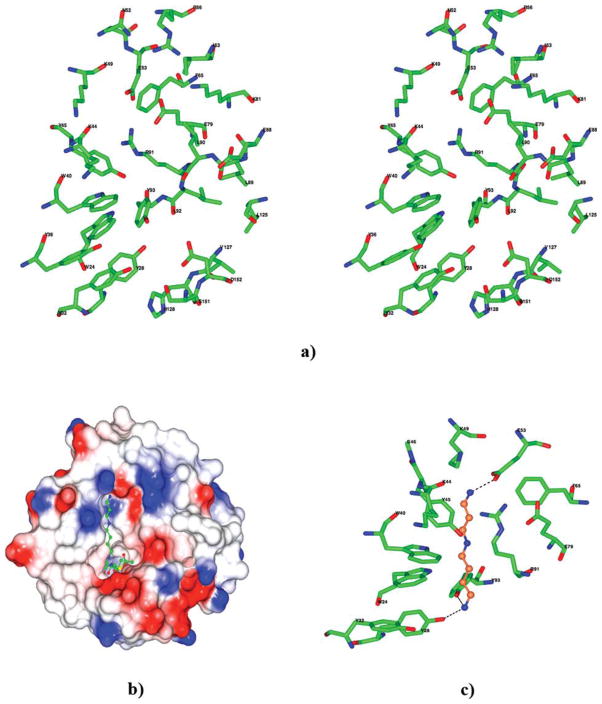
In the Ta0374 crystal structure, a large channel is present [Fig. 2(a)]. A similar cleft is found in the structure of PaiA from Bacillus subtilis.3 The acetyl-CoA occupies one half of the channel; the other end is presumably where the other substrate, spermidine, is bound for catalysis. The amino acid residues composing the surface of the predicted binding pocket for spermidine in the Ta0374 structure is shown in Figure 6(a). Most of the residues that line the binding pocket (Trp24, Tyr28, Tyr32, Tyr36, Trp40, Lys44, Tyr45, Glu79, Arg91) coincide with residues that could be involved in binding spermidine and have differences in the positions of the side chains observed between the two crystal structures of complexes [Fig. 4(b)]. Predominantly, hydrophobic residues (Trp24, Tyr28, Tyr32, Tyr36, Trp40, Tyr45, Phe65, Leu89, Leu90, Leu92, Tyr93, Leu125, Val127, His128) form the bottom of the pocket. The inner surface at the opposite ends of the pocket display a hydrophilic nature. There are patches of negative electrostatic potential, created by the residues Asp152, Glu53, and Glu79 from both sides of the pocket that could potentially be areas for interacting with the elongated spermidine and its positively charged nitrogen atoms [Fig. 6(b)]. Furthermore, sequence alignment shows that residues Glu53, Glu88, Arg91, Tyr93, Leu125, Val127, and Asp152 are very conserved among related PaiA proteins suggesting that the substrate-binding pocket is designed to recognize the specific and common ligand among this class of the N-acetyltransferases [Fig. 1(a)]. Additionally, there are two conserved ion pairs in the region between Glu53-Arg91 and Glu79-Arg91, which were also found in the structure of PaiA from Bacillus subtilis.3
Figure 6.
(a) Stereo view of the amino acid residues near the predicted spermidine-binding pocket in the Ta0374 structure. (b) Electrostatic surface of the Ta0374 molecule with spermidine (ball-and-stick model) docked in the binding pocket of the protein. (c) Predicted spermidine position in the active site. Possible hydrogen bonds to nitrogen atoms of spermidine are shown as dashed lines. Carbon and nitrogen atoms of the spermidine are colored in coral and blue, respectively. (a, c) In the model oxygen atoms of the surrounding residues are colored in red, nitrogen in blue, and carbon in green.
A docking study using the program MEDock (see Materials and Methods section) shows that this binding pocket can accommodate the spermidine [Fig. 6(b)]. In the model, spermidine is localized between two negatively charged areas of the pocket. The docking study suggests that the N1 atom of spermidine is likely to have hydrogen bonds with the OH atoms of the side chains Tyr28 and Tyr93 [Fig. 6(c)]. The N8 atom of spermidine would have hydrogen bonds with the O atom of the main chain of Lys44 and OE2 atom of the side chain of Glu53 [Fig. 6(c)]. The distinguishing feature of such binding interactions in the docking model is that the positively charged ends of spermidine bind to conserved polar residues such as Tyr93 and the acidic Glu53 in the binding site.
DISCUSSION
Based on our studies, the crystal structure of Ta0374 from Thermoplasma acidophilum confirms that it belongs to the PaiA N-acetyltransferase subfamily of the GNAT family and is capable of acetylating spermidine. The original proposal that PaiA may be a novel nonmembrane-bound SSAT is unlikely to be correct,3 as the structural and catalytic details of the active site differ considerably between these two protein families.10 The structures of the two groups have some relatively well-conserved residues and similar topology in the core region involved in acetyl-CoA binding; however, they are highly divergent in sequence and topology in the region involved in dimerization and polyamine binding in SSAT. The PaiA protein is a monomer and differs at most of the SSAT positions of the polyamine active site.
The polyamines such as spermidine, spermine, and putrescine are important in many biological processes, including DNA/RNA binding and stability, chromatin condensation, mRNA translation, protein binding, etc.49–52 They are abundant and ubiquitous components of all cells and can protect from reactive oxygen species.53 Acetylation of polyamines appears to be necessary for their breakdown and export from the cell.54 Mammalian SSAT is a well-characterized cytoplasmic enzyme that catalyzes the rate-limiting step in polyamine catabolism by transferring the acetyl group from acetyl-CoA to the N1 position of spermidine or spermine.7,9,10,55–57 Acetylated spermidine or spermine are excreted from the cells or metabolized by the constitutive flavin adenine dinucleotide (FAD)—requiring polyamine oxidase.58–60 As an alternative to SSAT, PaiA N-acetyltransferase could acetylate the N8-nitrogen or both the N1 and N8 positions of spermidine. This suggestion could be confirmed by 1H NMR spectroscopy where we used N8-acetylspermidine for the comparison of chemical shifts associated with the incubation of acetyl-CoA, spermidine, and Ta0374.
It is observed that in Bacillus subtilis spermidine is the major polyamine and is essential for sporulation, which requires compaction of the genomic DNA into a small-specialized cell, the spore.61 A decrease in spermidine content during the logarithmic phase of cell growth delays spore formation of Bacillus subtilis.61 It is also found that the pai operon from Bacillus subtilis includes regulatory genes that are involved in sporulation and degradative enzyme production.1,2 The pai operon contains two genes paiA and paiB. The characterization of these genes on a multicopy vector reveals that both a greatly reduced frequency of sporulation and decreased levels of extracellular degradative enzymes such as subtilisin, neutral protease, levansucrase, α-amylase, and alkaline phosphatases.1
In comparison with PaiA, even less is known about the PaiB protein. Based on the study by Strauch (1993), the paiB gene appears to encode a regulator belonging in a class with AbrB, Hpr, and Sin family regulators that control synthesis of degradative enzymes during the transition between vegetative growth and the onset of the stationary phase and sporulation in Bacillus subtilis.1 During vegetative growth and in the early stages of sporulation, these regulators prevent the inappropriate expression of various spo genes.1 Structural and bioinformatic analysis confirms that PaiB is a transcriptional regulator with a dimeric architecture (Filippova et al, accompanying structure note). Structure analysis reveals that PaiB has a flavin mononucleotide (FMN)—binding cavity similar to the pyridoxine 5′-phosphate oxidases.62 Superposition of the ligand binding domains of these two proteins demonstrates the conservation of the FMN-binding core suggesting that PaiB can accommodate flavin ligands. This is consistent with evidence that after acetylation, the acetylated spermidine/spermine is subsequently metabolized by FAD-requiring proteins.
In conclusion, as a regulator of sporulation and degradative enzymes,2 we hypothesize that the pai proteins may be important for the regulation of expression of the spo genes by controlling spermidine levels. Confirmation of this assessment, and elucidation of where pai fits into the scheme of preventing inappropriate transition-state gene expression, obviously awaits further biochemical experimentation.
Supplementary Material
Acknowledgments
The results are based on work performed at Argonne National Laboratory, at the Advanced Photon Source (APS); University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. The authors thank members of the Midwest Center for Structural Genomics and Life Sciences Collaborative Access Team at the APS for their help in the development of the project.
Grant sponsor: NIH PSI; Grant number: GM074942
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Strauch MA. Regulation of Bacillus subtilis gene expression during the transition from exponential growth to stationary phase. Prog Nucleic Acid Res Mol Biol. 1993;46:121–153. doi: 10.1016/s0079-6603(08)61020-x. [DOI] [PubMed] [Google Scholar]
- 2.Honjo M, Nakayama A, Fukazawa K, Kawamura K, Ando K, Hori M, Furutani Y. A novel Bacillus subtilis gene involved in negative control of sporulation and degradative-enzyme production. J Bacteriol. 1990;172:1783–1790. doi: 10.1128/jb.172.4.1783-1790.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forouhar F, Lee I-S, Vujcic J, Vujcic S, Shen J, Vorobiev SM, Xiao R, Acton TB, Montelione GT, Porter CW, Tong L. Structural and functional evidence for Bacillus subtilis PaiA as a novel N-spermidine/spermine acetyltransferase. J Biol Chem. 2005;280:40328–40336. doi: 10.1074/jbc.M505332200. [DOI] [PubMed] [Google Scholar]
- 4.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vetting MW, de Carvalho LPS, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Scheibner KA, De Angelis J, Burley SK, Cole PA. Investigation of the roles of catalytic residuies in serotonin N-acetyltransferase. J Biol Chem. 2002;277:18118–18126. doi: 10.1074/jbc.M200595200. [DOI] [PubMed] [Google Scholar]
- 7.Hegde SS, Chandler J, Vetting MW, Yu M, Blanchard JS. Mechanistic and structural analysis of human spermidine/spermine N1-acetyltransferase [hSSAT] Biochem. 2007;46:7187–7195. doi: 10.1021/bi700256z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung MN, Rangarajan E, Munger C, Nadeau G, Sulea T, Matte A. Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis. J Bacteriol. 2006;188:5606–5617. doi: 10.1128/JB.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montemayor EJ, Hoffman DW. The crystal structure of spermidine/spermine N1-acetyltransferase in complex with spermine provides insights into substrate binding and catalysis. Biochemistry. 2008;47:9145–9153. doi: 10.1021/bi8009357. [DOI] [PubMed] [Google Scholar]
- 10.Bewley MC, Graziano V, Jiang J, Matz E, Studier FW, Pegg AE, Coleman CS, Flanagan JM. Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target. Proc Natl Acad Sci USA. 2006;103:2063–2068. doi: 10.1073/pnas.0511008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LS-CAT: Life Sciences Collaborative Access Team. 2011 Available at: http://ls-cat.org/index.html.
- 13.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystal. 1997;A276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 16.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr. 2003;D59:1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 17.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 18.Sheldrick GM. Macromolecular phasing with SHELXE. Kristallogr. 2002;217:644–650. [Google Scholar]
- 19.Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr. 2002;D58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 20.Cowtan KD, Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr. 1993;D49:148–157. doi: 10.1107/S0907444992007698. [DOI] [PubMed] [Google Scholar]
- 21.Cowtan KD, Zhang KYJ. Density modification for macromolecular phase improvement. Progr Biophys Mol Biol. 1999;72:245–270. doi: 10.1016/s0079-6107(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 22.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 23.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. 1999;D55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 28.Yang H, Guranovic V, Dutta S, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr. 2004;D60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 29.Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 30.Delano WL. The pymol molecular graphics system. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 31.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. 2004;D60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 32.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Prot Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and 23 rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holm L, Sander C. 3-D lookup: fast protein structure database searches at 90% reliability. ISMB. 1995;3:179–187. [PubMed] [Google Scholar]
- 39.Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:89–93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang DT, Oyang YJ, Lin JH. MEDock: a web server for efficient prediction of ligand binding sites based on a novel optimization algorithm. Nucleic Acids Res. 2005;33:233–238. doi: 10.1093/nar/gki586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuff ME, Li H, Moy S, Watson J, Cipriani A, Joachimiak A. Crystal structure of an acetyltransferase protein from Vibrio cholerae strain N16961. Proteins. 2007;69:422–427. doi: 10.1002/prot.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunzelle JS, Wu R, Korolev SV, Collart FR, Joachimiak A, Anderson WF. Crystal structure of Bacillus subtilis YdaF protein: a putative ribosomal N-acetyltransferase. Proteins. 2004;57:850–853. doi: 10.1002/prot.10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJ. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 45.Oda K, Matoba Y, Noda M, Kumagai K, Sugiyama M. Catalytic mechanism of bleomycin N-acetyltransferase proposed on the basis of its crystal structure. J Biol Chem. 2010;285:1446–1456. doi: 10.1074/jbc.M109.022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carson M, Johnson DH, McDonald H, Brouillette C, DeLucas LJ. His-tag impact on structure. Acta Crystallogr. 2007;D63:295–301. doi: 10.1107/S0907444906052024. [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. 1976;A32:922–923. [Google Scholar]
- 48.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Tabor CW, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Igarashi K, Kashiwagi K. Polyamines: mysterious modulators of cellular functions. Biochem Biophys Res Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 51.Childs AC, Mehta DJ, Gerner EW. Polyamine-dependent gene expression. Cell Mol Life Sci. 2003;60:1394–1406. doi: 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW. Genomic identification and biochemical characterization of a second spermidine/spermine N1-aceyltransferase. Biochem J. 2003;373:661–667. doi: 10.1042/BJ20030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rider JE, Hacker A, Mackintosh CA, Pegg AE, Woster PM, Casero RA., Jr Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids. 2007;33:231–240. doi: 10.1007/s00726-007-0513-4. [DOI] [PubMed] [Google Scholar]
- 54.Shappell NW, Fogel-Petrovic MF, Porter CW. Regulation of spermidine/spermine N1-acetyltransferase by intracellular polyamine pools. Evidence for a functional role in polyamine homeostasis. FEBS Lett. 1993;321:179–183. doi: 10.1016/0014-5793(93)80103-2. [DOI] [PubMed] [Google Scholar]
- 55.Yarlett N, Wu G, Waters WR, Harp JA, Wannemuehler MJ, Morada M, Athanasopoulos D, Martinez MP, Upon SJ, Marton LJ, Frydman BJ. Cryptosporidium parvum spermidine/spermine N1-acetyltransferase exhibits different characteristics from the host enzyme. Mol Biochem Paras. 2007;152:170–180. doi: 10.1016/j.molbiopara.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Liu B, Sutton A, Sternglanz R. A yeast polyamine acetyltransferase. J Biol Chem. 2005;280:16659–16664. doi: 10.1074/jbc.M414008200. [DOI] [PubMed] [Google Scholar]
- 57.Zhu Y-Q, Zhu D-Y, Zhang Y, Vonrhein C, Wang D-C. Crystal structure of human spermidine/spermine N1-acetyltransferase (hSSAT): The first structure of a new sequence family of transferase homologousd superfamily. Proteins. 2006;63:1127–1131. doi: 10.1002/prot.20965. [DOI] [PubMed] [Google Scholar]
- 58.Cook T, Roos D, Morada M, Zhu G, Keithly JS, Feagin JE, Wu G, Yarlett N. Divergent polyamine metabolism in the Apicomplexa. Microbiology. 2007;153:1123–1130. doi: 10.1099/mic.0.2006/001768-0. [DOI] [PubMed] [Google Scholar]
- 59.Casero RA, Pegg AE. Spermidine/spermine N1-acetyltransferase—the turning point in polyamine metabolism. FASEB J. 1993;7:653–661. [PubMed] [Google Scholar]
- 60.Pegg AE. Spermidine/spermine-N1-acetyltransferase: a key metabolic regulator. Am J Physiol Endocrinol Metab. 2008;294:995–1010. doi: 10.1152/ajpendo.90217.2008. [DOI] [PubMed] [Google Scholar]
- 61.Ishii I, Takada H, Terao K, Kakegawa T, Igarashi K, Hirose S. Decrease in spermidine content during logarithmic phase of cell growth delays spore formation of Bacillus subtilis. Cell Mol Biol. 1994;40:925–931. [PubMed] [Google Scholar]
- 62.Biswal BK, Cherney MM, Wang M, Garen C, James MNG. Structures of Mycobacterium tuberculosis pyridoxine 5′-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5′-phosphate. Acta Crystallogr. 2005;D61:1492–1499. doi: 10.1107/S0907444905026673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.