Abstract
Regulator of G-protein signaling 4 (Rgs4) regulates the strength and duration of G-protein signaling, and plays an important role in cardiac development, smooth muscle contraction and psychiatric disorders. Rgs4 expression is regulated at both mRNA and protein levels. In order to examine the transcriptional mechanism of Rgs4 expression, we have cloned and characterized rabbit Rgs4 promoter. The coding sequence of rabbit Rgs4 was obtained by degenerative RT-PCR and used for Northern blot and 5′-RACE analysis. A single transcript was identified in rabbit colonic smooth muscle cells. The 5′-untranslated region (UTR) extended 120 bp nucleotides upstream of the Rgs4 start codon. A putative promoter sequence (1389 bp) showed a consensus TATA box and cis-acting binding sites for several potential transcriptional factors. Reporter gene assay identified strong promoter activity in various cell types. Further analysis by deletion mutagenesis suggested that the proximal region had a highest core promoter activity while the distal region is suppressive. IL-1β significantly increased the promoter activity. The in vitro and in vivo binding activities for NF-κB transcription factor were validated by electrophoretic mobility shift assay and chromatin immunoprecipitation assay respectively. Mutation of NF-κB site reduced the promoter activity. These data suggest that the cloned rabbit Rgs4 promoter is functionally active and NF-κB binding site possesses enhancer activity in regulating Rgs4 transcription. Our studies provide an important basis for further understanding of Rgs4 regulation and function in different diseases.
Keywords: Rgs4, Smooth muscle, Rabbit, Promoter, NF-κB
1. Introduction
Signal transduction is a key process of converting one signal to another, leading to a series of signaling reactions. One critical class of signal transduction pathways is the signaling controlled by the guanine-nucleotide-binding heterotrimeric proteins (Gαβ γ proteins). G-protein-coupled receptors (GPCRs) are involved in a vast array of physiological and pathological processes. Regulators of G-protein signaling (RGS) proteins belong to a large family of highly diverse, multifunctional signaling proteins, which share a conserved signature domain (RGS domain) that binds directly to the activated Gα subunits. RGS terminates G-protein signaling by accelerating intrinsic GTPase activity of Gα and thereby fostering re-association of the Gαβ γ trimer (Riddle et al., 2005). Over 30 mammalian RGS family members have been described so far, and classified into seven subfamilies based on sequence identity and functional similarities (Willars, 2006; Xie and Palmer, 2007). Rgs4 is one of the most extensively studied RGS proteins at the structural, biochemical, and cell biological levels (Riddle et al., 2005; Levitt et al., 2006; Willars, 2006; Roman et al., 2007; Xie and Palmer, 2007). Rgs4 regulates the strength and duration of the Gαi/o and Gαq/11 family members (Huang et al., 1997; Hao et al., 2006). However, the transcriptional and posttranscriptional regulations of RGS proteins are not well understood.
The expression and function of Rgs4 have been studied in cardiomyocyte (Mittmann et al., 2002; Lee et al., 2005; Hao et al., 2006), nerve tissue (Krumins et al., 2004) and cancers (Nikolova et al., 2008; Hurst et al., 2009; Xie et al., 2009), whereas little is known in smooth muscle cells. In cardiomyocyte, Rgs4 expression is induced by endotoxin and interleukin (IL)-1β (Patten et al., 2002; Patten et al., 2003) and may contribute to the loss of Gαq-mediated activation of phospholipase C by endothelin-1 (Mittmann et al., 2001). In the central nervous system, Rgs4 is linked to schizophrenia (Harrison and Weinberger, 2005; Erdely et al., 2006; Levitt et al., 2006; Lipska et al., 2006; Bowden et al., 2007), Alzheimer’s disease (Emilsson et al., 2006) and Huntington’s disease (Runne et al., 2008). However, the reports on the expression levels of Rgs4 under various neuropathologic conditions remain inconsistent (Guo et al., 2006; Li and He, 2006; Rizig et al., 2006; van Gemert et al., 2006a; Stuart Gibbons et al., 2008). In neuronal cell line, expression of Rgs4 is reduced after treatment with nerve growth factor (Krumins et al., 2004), cAMP (Pepperl et al., 1998) or camptothecin (Song and Jope, 2006), whereas opioid receptor agonists lead to an increase in Rgs4 mRNA expression (Zarnegar et al., 2006). Administration of corticosterone to adult rats decreased the level of Rgs4 mRNA in the paraventricular nucleus of the hypothalamus, and increased the levels in locus coeruleus (Ni et al., 1999), but had no effect in the hippocampus (van Gemert et al., 2006a; van Gemert et al., 2006b). Rapid kindling leads to an increase of Rgs4 mRNA in hippocampus and forebrain, but not in brainstem or cerebellum (Liang and Seyfried, 2001). Rgs4 expression is down-regulated in prefrontal cortex and striatum by neonatal status epilepticus (Lin et al., 2009). In rat adrenal glands, Rgs4 is up-regulated by aldosterone secretagogues, both in vivo and in vitro (Romero et al., 2007). Rgs4 mRNA is expressed only in glial cell line-derived neurotrophic factor (GDNF)-responsive neurons (Costigan et al., 2003). These studies suggest a complicate regulatory mechanism for Rgs4 expression and function.
In vascular smooth muscle cells, Rgs4 is highly expressed at mRNA level (Cho et al., 2003), which is passage-dependent (Hendriks-Balk et al., 2008). Activation of the nuclear hormone receptor peroxisome proliferator-activated receptor delta (PPARdelta) leads to the increase of vascular Rgs4 expression (Takata et al., 2008). In our previous studies, we demonstrate for the first time that Rgs4 expression is increased in rabbit intestinal smooth muscle cells after IL-1β treatment. These findings suggest that Rgs4 expression is regulated by inflammatory mediators and stress responses.
However, the molecular mechanisms and signaling pathways for Rgs4 regulation remain elusive. At the protein level, Rgs4 is regulated by the N-end rule pathway (Bodenstein et al., 2007). At the mRNA level, Rgs4 is regulated by a neural type-specific transcription factor Phox2b (Grillet et al., 2003). Our recent studies demonstrate that IL-1β-induced up-regulation of Rgs4 is transcription-dependent, mediated by the canonical IKK2/IκBα pathway of NF-κB activation (Hu et al., 2008), and differentially modulated by MAPK and PI3K/Akt/GSK3β pathways (Hu et al., 2009).
Genetic studies identified four common single-nucleotide polymorphisms (SNPs) for schizophrenia between the 5′ upstream sequence and the first intron of human RGS4 (Chowdari et al., 2008), suggesting that nucleotide change in the promoter region may affect Rgs4 promoter activity. Recent studies discovered 5 splice variants of human RGS4 and 3 splice variants of mouse Rgs4 (Ding et al., 2007), implying the complicate transcriptional regulation and promoter alternatives. However, the sequences of rabbit Rgs4 gene and promoter are not known. Here we report the cloning and characterization of rabbit Rgs4 gene and promoter. These studies provide an important basis to understand the transcriptional mechanism of Rgs4 regulation.
2. Materials and methods
2.1. Cell culture
Rabbit colonic smooth muscle cells were isolated and cultured as previously described (Murthy and Makhlouf, 1997; Hu et al., 2007). Briefly, distal colon from euthanized New Zealand White rabbit (2~2.5 kg) following IACUC guidance at Temple University was placed in HEPES-buffered smooth muscle media. The circular and longitudinal smooth muscle layers were dissected under stereo microscope and treated with 0.1% collagenase (type II) and 0.1% soybean trypsin inhibitor for 30 min at 31 °C. The isolated single muscle cells after several rounds of spontaneous dispersion were harvested by filtration through 500-μm Nitex and centrifuged twice at 350×g for 10 min. The isolated smooth muscle cells were plated in 10-cm dish with DMEM containing 10% fetal bovine serum and 1% antibiotics and antimycotics. After 10–14 days, the smooth muscle cells attained confluence and were then passaged once for use in various experiments.
HEK293T and Hela cells were cultured in DMEM containing 10% fetal bovine serum and 1% antibiotics.
2.2. Cloning of rabbit Rgs4 coding sequence by degenerative RT-PCR
Total RNA was extracted from cultured rabbit colonic smooth muscle cells using the Trizol reagent (Invitrogen, Carlsbad, CA). The potentially contaminated genomic DNA was removed by treating 10 μg of RNA sample at 37 °C for 30 min with 1 μl of TURBO DNase (Ambion, Austin, TX) followed by extraction with phenol/chloroform/isoamylalcohol (25:24:1). Two micrograms of RNA was used to synthesize cDNA using SuperScript II reverse transcriptase (Invitrogen) with random hexanucleotide primer.
Conventional PCR was performed on the cDNA using HotMaster™ Taq DNA polymerase (Eppendorf), which generates 3′-A overhang for T-A cloning. The degenerative primer sequences were obtained by comparing the coding sequence of Rgs4 from human, mouse and rat. The nucleotide sequences of the forward and reverse primers are shown in Table 1. The PCR reaction was heated at 94 °C for 2 min, and then cycled 30 times at 94 °C for 30 s, 55 °C for 45 s, and 72 °C for 1 min. Reaction products were analyzed by electrophoresis on 1% agarose gel. The band of predicted size was purified and cloned into pCRII-TOPO T-A vector (Invitrogen) for confirmation by sequencing using T7 and SP6 primers.
Table 1.
Primer sequences.
| Target | Forward (F) and reverse (R) nucleotide sequence |
|---|---|
| RGS4-cds | 5′-ATGTGCAAAGGRCTYGCWGGTC-3′(F) 5′-GTGAGAATTAGGCACACTGRG-3′(R) |
| RGS4-P1 | 5′-GGGAGTGGAGGCAGCTTGAGAT-3′ (F) 5′-CTCAAGCTTCGGTTTCACTCCTGC-3′ (R) |
| pMLuc3-sequence | 5′-AACTAGCAAAATAGGCTGTCCC-3′ (F) 5′-GTGCAAGACTTAGTGCAATGCA-3′ (R) |
| NF-κB mutation | 5′-GTCTATTTTGGTGCACTGCAACAGTCGCTTGTCCATTTTG-3′ (F) 5′-CAAAATGGACAAGCGACTGTTGCAGTGCACCAAAATAGAC-3′ (R) |
2.3. Northern blot
Ten micrograms of total RNA from cultured rabbit colonic smooth muscle cells was separated by 1.2% denaturing agarose gel electrophoresis and transferred to a positively charged nylon membrane. The PCR probe of the coding sequence of rabbit Rgs4 was labeled by random priming with [32P]dCTP. Hybridizations were conducted in the ExpressHyb hybridization solution (Clontech) at 65 °C for 2 h according to the manufacturer’s protocol.
2.4. Rapid amplification of cDNA ends (RACE)
The 5′ ends of rabbit Rgs4 transcripts were identified by using SMART RACE cDNA amplification kit (BD Biosciences, Clontech, Palo Alto, CA) following the manufacturer’s instructions. PCR was carried out using the reverse primer of rabbit Rgs4 as described above (Table 1) and the universal primer provided in the kit. The PCR products were gel-purified and cloned into the pCRII-TOPO T-A vector (Invitrogen), and the nucleotide sequence of independent clones was determined by sequencing.
2.5. Promoter cloning and vector construction
Genomic DNA was extracted from rabbit intestine using Wizard® SV genomic DNA purification system (Promega). The 5′-untranslated region (UTR) sequence of rabbit Rgs4 identified by 5′-RACE was used to blast the draft assembly of rabbit genome (http://genome-test.cse.ucsc.edu/). Various pairs of primers against the blasted sequence of Rgs4 5′-flanking region were designed for PCR cloning using rabbit genomic DNA as PCR template.
A fragment of −1389/+50 (from the putative transcription initial site) with single 3′-dA overhangs was generated by PCR using sense and antisense primers (Table 1) and non-proofreading thermostable DNA polymerase (HotMaster™ Taq). This fragment (designed as Rgs4-P1) was inserted by T-A cloning into the lineated promoter-less pMlu3 AccepTor vector at EcoRV of multiple cloning sties upstream of the secreted renilla luciferase reporter (EMD-Bioscience/Novagen). The direction and sequence of the insert were validated by sequencing with upstream and downstream vector primers (Table 1).
Various deletion constructs of pMluc3-Rgs4-P1 were generated through digestion, blunting and ligation by analyzing and combining the digestion sites within the insert and the backbone vector. The mutants pMluc3-Rgs4-P2 (−962 to +50) and pMluc-Rgs4-P3 (−1389 to −816) were generated by single digestion with PstI and HindIII respectively followed by ligation. The mutant pMluc-Rgs4-P4 (−247 to +50) was generated by double digestion with XhoI/XbaI followed by blunting and ligation, which does not affect the sequence of Rgs4-P4.
2.6. Site-directed mutagenesis
Potential transcription factor binding sites within Rgs4 promoter were identified by MatInspector (http://www.genomatix.de) and TFSEARCH (http://www.cbrc.jp). Mutation of the binding site for the transcription factors in the reporter vector construct was performed by site-directed mutagenesis using the QuikChange kit (Stratagene). Mutagenic primers as shown in Table 1 led to nucleotide change in entire binding site for transcriptional factor. Mutation was confirmed by nucleotide sequencing.
2.7. Cell transfection and reporter assays
All the vectors for mammalian expression were prepared with EndoFree Plasmid Maxi kit (Qiagen). Cells (2–4×104/well) cultured on 96-well plate were transfected with the renilla luciferase reporter constructs and 1:10 normalization firefly luciferase vector pGL4-CMV (Promega) using Lipofectamine-2000 kit (Invitrogen). The transfection efficiency of rabbit smooth muscle cells (~60%) is determined using pEGFP-N1 vector (BD Biosciences Clontech).
After incubation for the indicated time periods in the absence or presence of stimulators, the media were harvested for measurement of renilla luciferase activity and at the end the cell lysate was used for measurement of firefly luciferase activity. The renilla luciferase was determined with renilla luciferase assay kit (Promega). The firefly luciferase was determined with ONE-Glo luciferase assay system (Promega). Data are normalized by dividing renilla luciferase activity with that of the corresponding firefly luciferase activity. Four to eight separate experiments were conducted and, in each experiment, data are calculated as the average of 4–6 samples.
2.8. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed according to the manufacturer’s protocol (Upstate Biotechnology Inc., Lake Placid, NY). Smooth muscle cells were cultured in 10-cm dishes until full confluence and serum-starved overnight. IL-1β (Alexis Biochemicals) was added at 10 ng/ml and incubated for 3 h. The DNA-chromatin of cells were cross-linked by the addition of 280 μl of 37% formaldehyde to 10 ml of culture medium for 10 min at room temperature and stopped with 0.125 M glycine. Cells were washed twice with PBS and harvested with 1 ml of SDS lysis buffer containing protease inhibitors. After sonication and centrifugation, the supernatants were used for standard immunoprecipitation with anti-p65 antibody or control IgG. The immune complexes were eluted, reverse cross-linked using 5 M NaCl, and purified by phenol/chloroform extraction. Ethanol-precipitated DNA pellets were redissolved in Tris–EDTA buffer. The supernatant of an immunoprecipitation reaction carried out in the absence of antibody was purified and diluted 1:100 as total input DNA control. PCR was carried out on 1 μl of each sample using sense and anti-sense primers against the promoter of interest. PCR products were analyzed on 1 % agarose gels and images were analyzed with NIH ImageJ densitometric measurements. Relative changes were calculated with the mean density after background subtraction.
3. Results
3.1. Single transcript of rabbit Rgs4 in gut smooth muscle cells
Since rabbit Rgs4 sequence is not available, we performed degenerative PCR to obtain the coding sequence of rabbit Rgs4. The coding sequence was selected because of its high homology between human, mouse and rat. A pair of degenerative primers was designed based on the available Rgs4 coding sequence. RT-PCR using total RNA from cultured rabbit colonic smooth muscle cells generated a single product of predicted size. T-A cloning and sequencing of this PCR product confirmed the coding sequence of rabbit Rgs4 with a correct open-reading frame (Genebank access number NM_001082214), which shares 92%, 90% and 89% identity with coding sequence of human, rat and mouse Rgs4 respectively. Northern blot analysis of Rgs4 mRNA expression in rabbit colonic smooth muscle cells was performed using this coding sequence as a probe. As shown in Fig. 1A, one single band of around 2.8 kb transcript was detected in both circular and longitudinal colonic smooth muscle cells after treatment with IL-1β for 3 days. However, Rgs4 transcript was undetectable by Northern blot analysis in cultured colonic smooth muscle cells at full confluence after serum starvation. These data suggest that only one transcript of Rgs4 is present in rabbit gut smooth muscle cells. The inducible expression of Rgs4 is consistent with our previous reports as determined by real time PCR and Western blot analysis (Hu et al., 2007).
Fig. 1.
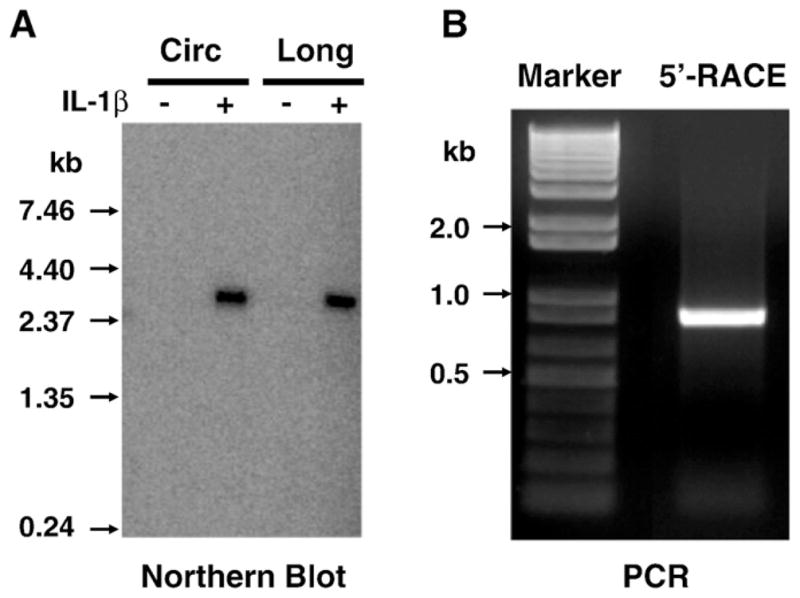
Single transcript of Rgs4 gene in rabbit colonic smooth muscle. Rabbit circular (Circ) and longitudinal (Long) colonic smooth muscle cells were serum-starved for 24 h and treated with IL-1β (10 ng/ml) for 3 days. Total RNA was extracted for Northern blot (A) and SMART 5′-RACE (B).
3.2. Cloning of rabbit Rgs4 5′-untranslated region (UTR) in gut smooth muscle cells
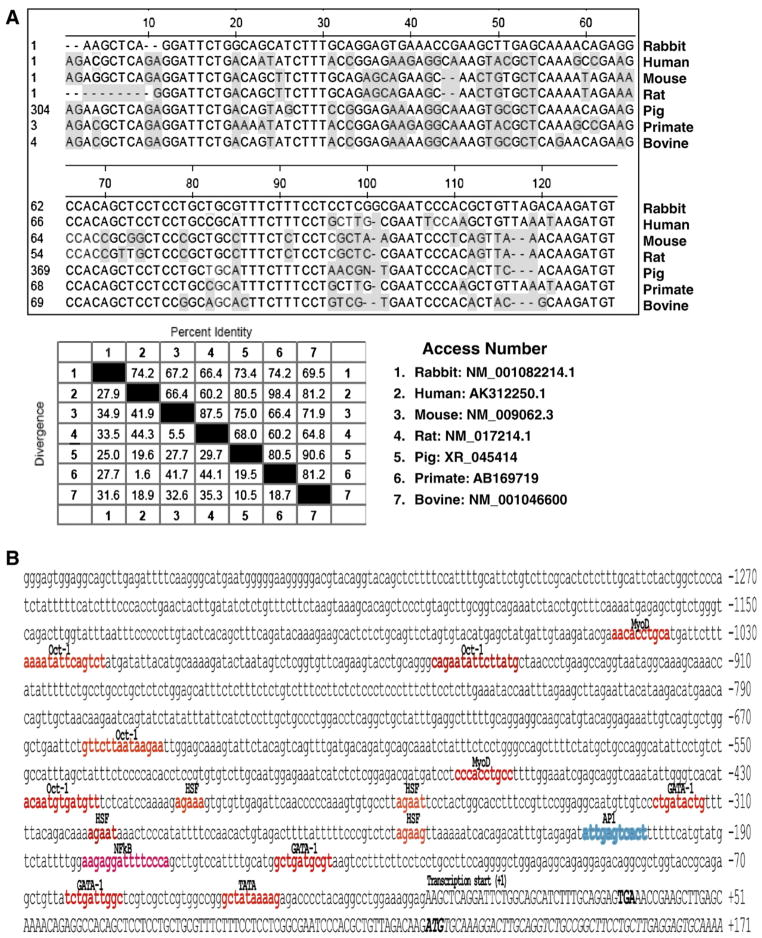
To characterize the 5′-UTR and identify the 5′-flanked sequence (promoter), we performed 5′-RACE on the cDNA from cultured rabbit colonic smooth muscle cells using the primers from rabbit Rgs4 coding sequence. As shown in Fig. 1B, a single 5′-RACE product of around 800 bp was observed, supporting the conclusion of single transcript in gut smooth muscle cells as determined by Northern blot. After confirmation by sequencing, the partial sequence of Rgs4 transcript containing the 5′-UTR and coding sequence was deposited in GeneBank with access number NM_001082214. BLAST search analysis showed that the 5′-UTR of rabbit Rgs4 shared high identity with the corresponding 5′-UTR of RGS4 from human isoform A (74%), mouse (67%), rat (66%), bovine (70%), primate (Macaca fascicularis and Pan troglodytes, 74%), pig (Sus scrofa, 73%) (Fig. 2A). Analysis of the open-reading frame for rabbit Rgs4 transcript identified a typical kozak translation start site and an in-frame stop codon upstream of the start codon. The sequence of 1–51 bp of rabbit Rgs4 was less conserved than the other region when compared with corresponding region of Rgs4 from other species (Fig. 2A).
Fig. 2.

Homology analysis of Rgs4 5′-UTR and putative promoter. (A) Highly conserved 5′-UTR. Multiple alignment from various species by Clustal W method showing the shaded residues different from rabbit Rgs4 (upper panel) and the identity percentage between species (lower panel). (B) Sequence of rabbit Rgs4 promoter and partial exon-1. Putative transcription factor binding sites analyzed by MatInspector are highlighted with color. Sequence from the transcription start site is in capital case. NF-κB: nuclear factor kappa B; AP-1: activator protein 1; HSF: heat shock factor. (C) Evolutionary analysis of Rgs4 promoter. Nucleotide sequence alignment of rabbit Rgs4 promoter (−257 to −1) with human, mouse and rat demonstrating highly conserved region at the proximal promoter. The shaded residues exactly match rabbit Rgs4 promoter sequence.
3.3. Cloning of Rgs4 promoter and comparative analysis
Using the unique 5′-UTR of rabbit Rgs4 obtained above to blast the draft assembly of rabbit genome (http://genome-test.cse.ucsc.edu/), we obtained potential promoter sequence of rabbit Rgs4. The 5′-upstream regulatory region of 1389 bp plus 50 bp of partial exon-1 was cloned by PCR from genomic DNA isolated from rabbit intestine. The cloned sequence was validated by T-A cloning and sequencing (Fig. 2B), and deposited into GeneBank with access number (GQ848293). Comparative analysis with the potential 5′-flanking segments of human, mouse and rat Rgs4 genes showed that only the proximal region of rabbit Rgs4 promoter shares a highly conserved homology with that from the other species (Fig. 2C).
3.4. Promoter activity of 5′-flanked region of rabbit Rgs4 in different cells
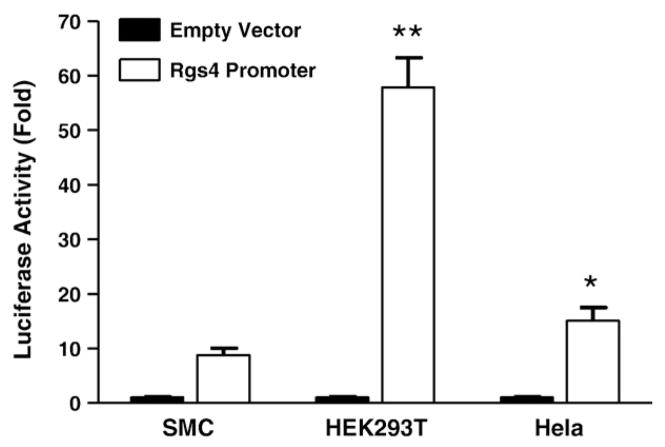
To examine the promoter activity of the cloned promoter region of rabbit Rgs4, a fragment of −1389/+50 (the initial transcription site was defined as +1, Fig. 2B) was cloned into pMluc3 secreted renilla luciferase vector. This fragment (designed P1) showed strong constitutive activity in rabbit smooth muscle cell (Fig. 3). This region also showed strong activity in smooth muscle cells of other species (data not shown), as well as other cell line such as HEK293T and Hela cells (Fig. 3). The highest promoter activity in HEK293T cells may reflect the highest transfection efficiency but also possibly the better transcription machinery. These data suggest that this cloned fragmental region of Rgs4 displays promoter activity and is shared by different species and different cell types. This observation is supported by the bioinformatics analysis showing high homology of the Rgs4 promoter region between different species (Fig. 2C).
Fig. 3.
Promoter activity of rabbit Rgs4 5′-flanked region (1.4 kb) in various cells. Cultured cells were cotransfected with promoter-less pMlu3 vector or Rgs4 promoter vector carrying secreted renilla luciferase and pGL4-CMV vector carrying firefly luciferase. After 24 h, renilla and firefly luciferases were measured separately. Relative fold changes in renilla luciferase activity after normalization by firefly luciferase were expressed as compared with the promoter-less empty vector. Data represent the mean±SEM of 4 experiments. **P<0.01 and *P<0.05 indicate statistically significant increase by Student’s t test compared with the fold change in smooth muscle cells (SMC).
3.5. Identification of core promoter activity
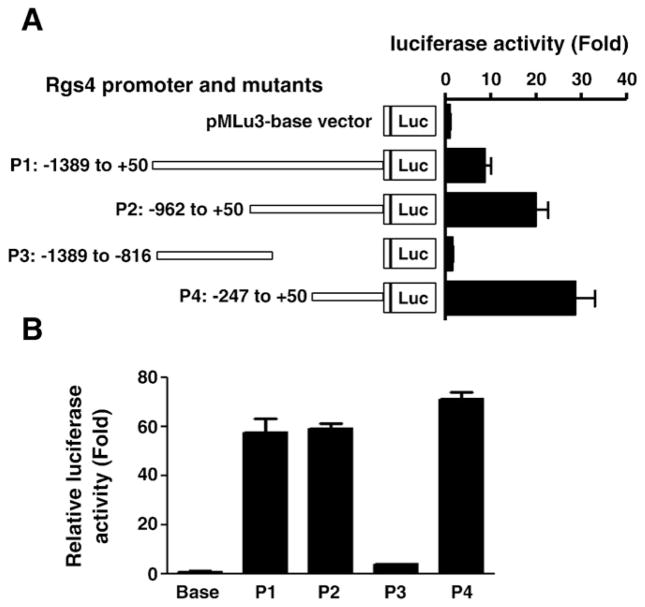
To characterize the promoter responsive elements, deletion mutants of rabbit Rgs4 1.4 kb promoter (P1) were prepared. As shown in Fig. 4, mutant P2 containing −962/+50 showed stronger activity than original promoter P1 (−1389/+50), suggesting that the region between −1389/−962 contains regulatory elements negatively modulating the promoter activity of Rgs4. Mutant P3 containing −1389/−816 had no significant effect on the basal reporter activity, implying that the proximal region −816/+50 of the promoter P1 holds the main promoter activity and the inhibitory function of distal region −1389/−816 depends upon the presence of the proximal cis-elements. Deletion of −1389/−247 (mutant P4) further increased the promoter activity, suggesting that the most proximal region −247/+50 holds a maximal core promoter activity and the region −1389/−247 inhibits the core promoter activity.
Fig. 4.
Deletion analysis of Rgs4 promoter in rabbit colonic smooth muscle cells (A) and human HEK293T cells (B) showing higher promoter activity in the proximal region and repressive activity in the distal region. Cultured cells were cotransfected with indicated Rgs4 promoter and various mutant vectors plus normalization vector. After 24 h, renilla and firefly luciferases were measured separately. Relative fold changes in renilla luciferase activity after normalization by firefly luciferase were expressed as compared with the promoter-less empty vector. Values represent the mean±SEM of 4 individual experiments.
3.6. IL-1β up-regulates Rgs4 promoter activity
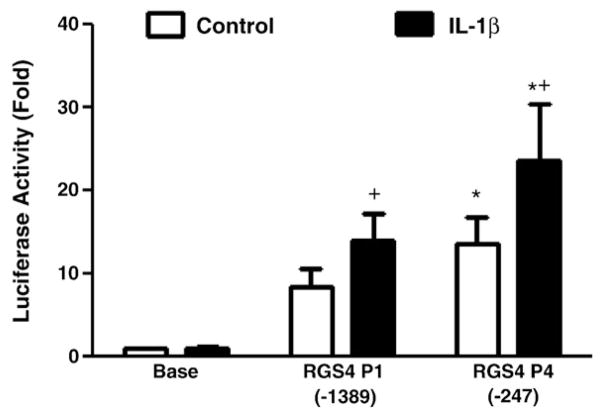
We have shown that IL-1β up-regulates Rgs4 mRNA expression in colonic smooth muscle cells (Hu et al., 2007), and this up-regulation is inhibited by a transcription inhibitor actinomycin D (Hu et al., 2008). To further investigate whether the transcriptional mechanism is involved in IL-1β-induced up-regulation of Rgs4 expression, we examined the effect of IL-1β on the reporter activity of Rgs4 promoter. As shown in Fig. 5, IL-1β treatment significantly increased the promoter activity of both Rgs4-P1 and Rgs4-P4 by around 50%.
Fig. 5.
Induction of Rgs4 promoter activity by IL-1β in rabbit colonic smooth muscle cells. Cultured cells were cotransfected with Rgs4 promoter P1 or P4 plus normalization vector for 24 h, serum-starved for 24 h and treated with IL-1β (10 ng/ml) for 24 h. The renilla and firefly luciferases were measured separately. Fold changes in renilla luciferase activity after normalization were expressed as compared with the promoter-less empty vector. Values represent the mean±SEM of 8 individual experiments with quadruplicate each. *P<0.05 indicates statistical significance by Student’s t test compared with corresponding Rgs4-P1 vector; +P<0.05 indicates a significant increase by IL-1β treatment in promoter activity compared with corresponding vehicle control.
3.7. Analysis of Rgs4 promoter binding sites for potential transcription factors
On-line program analysis of the 5′-flanking region of rabbit Rgs4 (−1389) identified the presence of a typical TATA box at -35, and multiple putative binding sites for various transcription factors such as NF-κB, AP-1, GATA, heat shock transcription factor (HSF), MyoD, Oct-1, etc. (Fig. 2B).
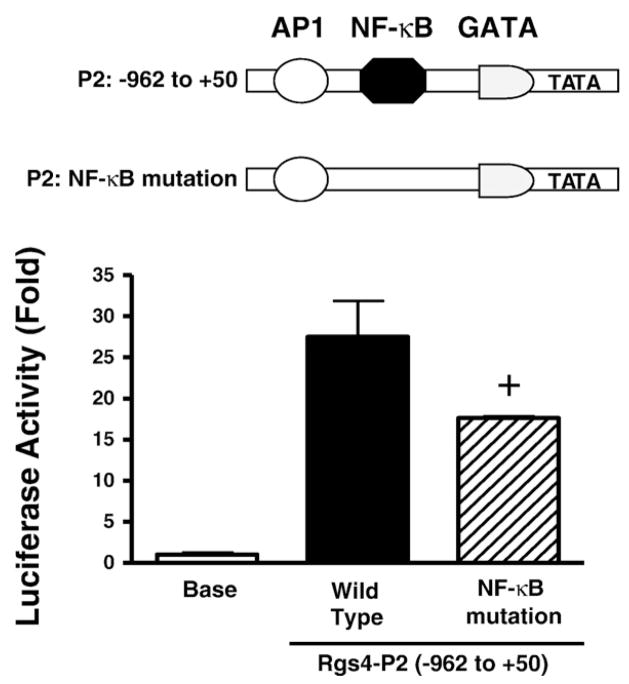
Since NF-κB transcription factor is the major target for IL-1β signaling, which up-regulates Rgs4 mRNA expression (Hu et al., 2007; Hu et al., 2008), we have focused our interest on studying the role of NF-κB binding sites in regulating Rgs4 promoter activity. One predicted NF-κB binding site within the cloned promoter region of Rgs4 was verified by gel shift and supper-shift assays in our previous studies (Hu et al., 2008). To address whether this NF-κB site is functionally active in regulating the promoter activity, we performed site-directed mutagenesis studies using Rgs4 P2 promoter reporter assay. As shown in Fig. 6, mutation of NF-κB site within P2 promoter reduced the promoter activity by 30%, suggesting that NF-κB binding site is involved positively and partially in the promoter activity of Rgs4.
Fig. 6.
Stimulatory function of NF-κB binding site within Rgs4 promoter for activation of reporter gene. Site-directed mutant of NF-κB site from Rgs4 promoter P2 was cotransfected with normalization vector into cultured smooth muscle cells. After 24 h, relative luciferase activity was determined. +P<0.05 indicates a significant decrease by NF-κB site mutation in promoter activity compared with the wild-type Rgs4-P2 promoter. Values represent the mean±SEM of 4 individual experiments and are analyzed by Student’s t test.
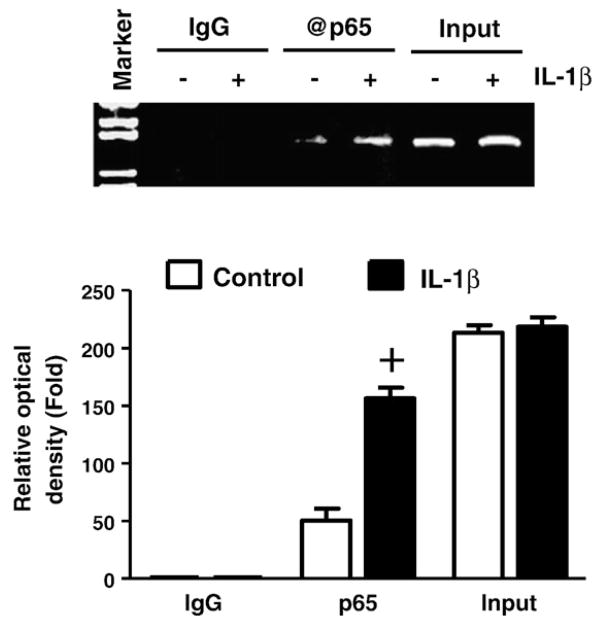
In order to determine the binding activity of endogenous NF-κB factors within the cloned Rgs4 promoter under physiological condition, we performed ChIP assay using cultured colonic smooth muscle cells. As shown in Fig. 7, the Rgs4 promoter region 1.4 kb was immunoprecipitated from the chromatin extracts by the antibody against p65 but not by the control IgG, indicating that endogenous p65 binds to Rgs4 promoter 1.4 kb. In addition, IL-1β treatment increased the endogenous NF-κB binding activity (Fig. 7).
Fig. 7.
Induction of endogenous NF-κB (p65)-DNA binding activity within Rgs4 promoter by IL-1β. Cultured rabbit colonic smooth muscle cells were treated with or without IL-1β for 3 h and chromatin immunoprecipitation assay was performed. The relative optical density on the bottom was compared to IgG control. +P<0.05 indicates a significant increase by IL-1β treatment through Student’s t test compared with corresponding vehicle control. Values represent the mean±SEM of 3 individual experiments. Input indicates the DNA from supernatant after precipitation without IgG.
4. Discussion
Rgs4 accelerates the GTPase activity of the Gαi/o and Gαq/11 family members and regulates the strength and duration of G-protein signaling in neurons, cardiomyocytes, smooth muscle cells and other cells. Thus, Rgs4 is implicated in psychiatric disorders, cardiovascular diseases and intestinal inflammation. However, the regulatory mechanism of Rgs4 expression remains elusive. Our previous studies demonstrate that IL-1β-induced up-regulation of Rgs4 mRNA in rabbit colonic smooth muscle cells is transcription-dependent because inhibition of transcription blocked IL-1β-induced up-regulation of Rgs4 (Hu et al., 2008). The transcriptional machinery involves the epigenic coordination among various transcription factors, promoters and chromatin. In the present study, we cloned and characterized for the first time the potential promoter region of rabbit Rgs4. This promoter contains a canonical TATA box, is evolutionally conserved, has highly constitutive activity and can be up-regulated by IL-1β treatment. The proximal region of rabbit Rgs4 promoter has a highest core promoter activity while the distal region holds repressive activity.
The characteristic of Rgs4 promoter has not yet been well established. Potential and alternative promoters have been described through the predictive analysis of the reported transcripts (Aceview). Only human and mouse Rgs4 promoters have been corroborated experimentally (Ding et al., 2007; Chowdari et al., 2008). We have identified rabbit Rgs4 promoter with high promoter activity. The reasons to use rabbit as a target are two fold: (i) Rabbit shares closer gene homology and functional correlation with human in several disease models (Percy et al., 1990; Depoortere et al., 2001; Woodruff-Pak et al., 2007); (ii) we have accomplished a plenty of signal transduction and molecular studies on rabbit gastrointestinal smooth muscle cells (Murthy, 2006; Hu et al., 2007). The identification of rabbit Rgs4 gene and regulatory mechanisms will be helpful to understand the biological relevance of Rgs4 in human diseases.
We have shown that the induction by IL-1β treatment in the mRNA expression of endogenous Rgs4 in rabbit colonic smooth muscle cells is 8–10 fold as compared with vehicle treatment (Hu et al., 2007; Hu et al., 2008). To obtain the direct evidence for IL-1β-induced up-regulation of Rgs4, we cloned the promoter sequence into luciferase reporter vector and examine whether IL-1β increases the promoter activity. Unexpectedly, the induction of Rgs4 promoter activity by IL-1β treatment (measured at 30 min till 3 d after treatment) was marginal though statistically significant with 8 individual experiments. The weak induction in the reporter gene assay has been reported in many genes (Nie et al., 2003; Liang et al., 2007; Mungunsukh et al., 2008). The possible interpretations for this phenomenon are given as follows: (i) IL-1β-induced up-regulation of Rgs4 mRNA level involves not only the transcriptional mechanism but also other mechanisms such as mRNA stability, which is currently under investigation; (ii) the constitutive promoter activity without IL-1β treatment is already high, which may limit further induction; and (iii) the complicate chromatin remodeling occurs to the endogenous Rgs4 transcription but not applied to the transient transfection of the reporter plasmid. To seek for the possibility of chromatin remodeling in regulating Rgs4 promoter activity, we established stable cell line incorporating Rgs4 promoter by cotransfection of hygromycin selection vector in Hela cells. Again, the induction by IL-1β remained marginal. Another possibility is that we have not yet identified the true full-length functional promoter of Rgs4, which may be much larger than the size we cloned. We tested the inducible effect of cytokines on the luciferase reporter activity of human RGS4 promoter (6.7 kb, gift from Nimgaonkar’s Lab in University of Pittsburg) (Chowdari et al., 2008). Similarly, there was only a marginal increase in human RGS4 promoter activity after IL-1β treatment. Since the distal regulatory elements are frequently dispersed in the regions far from the core promoter, even exceeding 100 kb (Wang et al., 2009), we are currently making BAC reporter construct by inserting secreted renilla luciferase reporter into Rgs4 BAC vector. This BAC reporter system will reflect endogenous chromatin contexts and identify potential elements of Rgs4 promoter in response to cytokine treatment.
The cloned Rgs4 promoter contains predicted binding sites for several transcription factors such as NF-κB, AP-1, GATA, MyoD, etc. We have identified the important role of NF-κB signaling in mediating IL-1β-induced up-regulation of Rgs4 mRNA expression in rabbit colonic smooth muscle cells (Hu et al., 2008). Here we provide additional evidence that NF-κB signaling is involved in Rgs4 transcriptional regulation. The family of NF-κB contains 5 members forming various patterns of dimmers. The best-studied heterodimer is p65/p50, which is translocated from the cytoplasm to nucleus upon cellular stimulation and bind to the consensus NF-κB sites to initiate the transcription of target genes. Our studies added Rgs4 as a new member of NF-κB target genes. Super gel shift and ChIP assay validated the binding activity of p65 within Rgs4 promoter. Site-directed mutation of NF-κB site within rabbit Rgs4 promoter reduced the promoter activity, suggesting that NF-κB binding site is functionally active in regulating the promoter activity of Rgs4. The transcriptional machinery is regulated by various transcription factors (enhancer and repressors). The functional regulations of other transcription factors on Rgs4 promoter need further investigation. The distal region (−1389 to −816) of Rgs4-P1 promoter by itself did not display any promoter activity. In contrast, deletion of the distal region led to higher promoter activity of the proximal region (−247). The proximal region comprises of an essential core TATA box and the cis-elements for GATA, NF-κB and AP-1. The distal region contains the binding site for Oct-1, MyoD and HSF. Although the outcome effects of these factors on Rgs4 promoter remain unknown, our data imply that the combined effects of these distal transcription factors may repress the transcription of Rgs4. This is supported by the fact that Oct-1 and MyoD have been shown to act as transcription suppressor in some genes (Chu et al., 1997; Schwachtgen et al., 1998; Cheng et al., 2002; Hitomi et al., 2007).
There are 5 splice variants of human RGS4 and 3 splice variants of mouse Rgs4 experimentally validated (Ding et al., 2007). According to ACEVIEW, human RGS4 gene produces 13 different transcripts including 11 alternatively spliced variants and 2 unspliced forms containing a single exon 5313 or 574 bp. There are 6 probable alternative promoters and 6 validated alternative polyadenylation sites. Mouse Rgs4 gene produces 2 alternatively spliced variants and 1 unspliced transcript with a single exon of 1068 bp (ACEVIEW). There is no information about rabbit Rgs4 in the GeneBank database. In the present study, only one transcript of rabbit Rgs4 in colonic smooth muscle cells was identified by Northern blot and 5′-RACE. However, the possibility of different variants for rabbit Rgs4 present in other cell types and/or tissues cannot be ruled out.
In conclusion, the present study has identified the functional promoter region of rabbit Rgs4 which contains a typical TATA box and various cis-acting elements for transcription factors. The NF-κB binding site plays an important role in mediating the constitutive and IL-1β-inducible promoter activity. Further characterization of Rgs4 promoter regulation will help understanding the molecular mechanisms and signaling pathways for Rgs4 expression and function.
Acknowledgments
This work is supported by Grant DK075964 and DK015564 from the National Institutes of Diabetes, and Kidney and Digestive Diseases.
Abbreviations
- AP-1
activator protein 1
- IL
interleukin
- NF-κB
nuclear factor κB
- RACE
rapid amplification of cDNA ends
- RGS
regulator of G-protein signaling
- RT-PCR
reverse transcription-polymerase chain reaction
- SMC
smooth muscle cells
- UTR
untranslated region
References
- Bodenstein J, Sunahara RK, Neubig RR. N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol Pharmacol. 2007;71:1040–1050. doi: 10.1124/mol.106.029397. [DOI] [PubMed] [Google Scholar]
- Bowden NA, Scott RJ, Tooney PA. Altered expression of regulator of G-protein signalling 4 (RGS4) mRNA in the superior temporal gyrus in schizophrenia. Schizophr Res. 2007;89:165–168. doi: 10.1016/j.schres.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Cheng CK, Yeung CM, Hoo RL, Chow BK, Leung PC. Oct-1 is involved in the transcriptional repression of the gonadotropin-releasing hormone receptor gene. Endocrinology. 2002;143:4693–4701. doi: 10.1210/en.2002-220576. [DOI] [PubMed] [Google Scholar]
- Cho H, Harrison K, Schwartz O, Kehrl JH. The aorta and heart differentially express RGS (regulators of G-protein signalling) proteins that selectively regulate sphingosine 1-phosphate, angiotensin II and endothelin-1 signalling. Biochem J. 2003;371:973–980. doi: 10.1042/BJ20021769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdari KV, et al. Linkage disequilibrium patterns and functional analysis of RGS4 polymorphisms in relation to schizophrenia. Schizophr Bull. 2008;34:118–126. doi: 10.1093/schbul/sbm042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Cogswell J, Kohtz DS. MyoD functions as a transcriptional repressor in proliferating myoblasts. J Biol Chem. 1997;272:3145–3148. doi: 10.1074/jbc.272.6.3145. [DOI] [PubMed] [Google Scholar]
- Costigan M, Samad TA, Allchorne A, Lanoue C, Tate S, Woolf CJ. High basal expression and injury-induced down regulation of two regulator of G-protein signaling transcripts, RGS3 and RGS4 in primary sensory neurons. Mol Cell Neurosci. 2003;24:106–116. doi: 10.1016/s1044-7431(03)00135-0. [DOI] [PubMed] [Google Scholar]
- Depoortere I, Van Assche G, Peeters TL. Motilin receptor density in inflamed and noninflamed tissue in rabbit TNBS-induced colitis. Neurogastroenterol Motil. 2001;13:55–63. doi: 10.1046/j.1365-2982.2001.00240.x. [DOI] [PubMed] [Google Scholar]
- Ding L, Mychaleckyj JC, Hegde AN. Full length cloning and expression analysis of splice variants of regulator of G-protein signaling RGS4 in human and murine brain. Gene. 2007;401:46–60. doi: 10.1016/j.gene.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Emilsson L, Saetre P, Jazin E. Low mRNA levels of RGS4 splice variants in Alzheimer’s disease: association between a rare haplotype and decreased mRNA expression. Synapse. 2006;59:173–176. doi: 10.1002/syn.20226. [DOI] [PubMed] [Google Scholar]
- Erdely HA, Tamminga CA, Roberts RC, Vogel MW. Regional alterations in RGS4 protein in schizophrenia. Synapse. 2006;59:472–479. doi: 10.1002/syn.20265. [DOI] [PubMed] [Google Scholar]
- Grillet N, Dubreuil V, Dufour HD, Brunet JF. Dynamic expression of RGS4 in the developing nervous system and regulation by the neural type-specific transcription factor Phox2b. J Neurosci. 2003;23:10613–10621. doi: 10.1523/JNEUROSCI.23-33-10613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, et al. RGS4 polymorphisms and risk of schizophrenia: an association study in Han Chinese plus meta-analysis. Neurosci Lett. 2006;406:122–127. doi: 10.1016/j.neulet.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Hao J, Michalek C, Zhang W, Zhu M, Xu X, Mende U. Regulation of cardiomyocyte signaling by RGS proteins: differential selectivity towards G proteins and susceptibility to regulation. J Mol Cell Cardiol. 2006;41:51–61. doi: 10.1016/j.yjmcc.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. image 5. [DOI] [PubMed] [Google Scholar]
- Hendriks-Balk MC, van Loenen PB, Hajji N, Michel MC, Peters SL, Alewijnse AE. S1P receptor signalling and RGS proteins; expression and function in vascular smooth muscle cells and transfected CHO cells. Eur J Pharmacol. 2008;600:1–9. doi: 10.1016/j.ejphar.2008.09.041. [DOI] [PubMed] [Google Scholar]
- Hitomi T, et al. Oct-1 is involved in the transcriptional repression of the p15 (INK4b) gene. FEBS Lett. 2007;581:1087–1092. doi: 10.1016/j.febslet.2007.01.092. [DOI] [PubMed] [Google Scholar]
- Hu W, Mahavadi S, Li F, Murthy KS. Upregulation of RGS4 and down-regulation of CPI-17 mediate inhibition of colonic muscle contraction by interleukin-1beta. Am J Physiol Cell Physiol. 2007;293:C1991–C2000. doi: 10.1152/ajpcell.00300.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Li F, Mahavadi S, Murthy KS. Interleukin-1beta up-regulates RGS4 through the canonical IKK2/IkappaBalpha/NF-kappaB pathway in rabbit colonic smooth muscle. Biochem J. 2008;412:35–43. doi: 10.1042/BJ20080042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Li F, Mahavadi S, Murthy KS. Upregulation of RGS4 expression by IL-1beta in colonic smooth muscle is enhanced by ERK1/2 and p38 MAPK and inhibited by the PI3K/Akt/GSK3beta pathway. Am J Physiol Cell Physiol. 2009;296:C1310–1320. doi: 10.1152/ajpcell.00573.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Hepler JR, Gilman AG, Mumby SM. Attenuation of Gi- and Gq-mediated signaling by expression of RGS4 or GAIP in mammalian cells. Proc Natl Acad Sci USA. 1997;94:6159–6163. doi: 10.1073/pnas.94.12.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst JH, Mendpara N, Hooks SB. Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell Mol Biol Lett. 2009;14:153–174. doi: 10.2478/s11658-008-0040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumins AM, et al. Differentially regulated expression of endogenous RGS4 and RGS7. J Biol Chem. 2004;279:2593–2599. doi: 10.1074/jbc.M311600200. [DOI] [PubMed] [Google Scholar]
- Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA. Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4) Biol Psychiatry. 2006;60:534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Li D, He L. Association study of the G-protein signaling 4 (RGS4) and proline dehydrogenase (PRODH) genes with schizophrenia: a meta-analysis. Eur J Hum Genet. 2006;14:1130–1135. doi: 10.1038/sj.ejhg.5201680. [DOI] [PubMed] [Google Scholar]
- Liang D, Seyfried TN. Genes differentially expressed in the kindled mouse brain. Brain Res Mol Brain Res. 2001;96:94–102. doi: 10.1016/s0169-328x(01)00287-x. [DOI] [PubMed] [Google Scholar]
- Liang KC, et al. Interleukin-1beta induces MMP-9 expression via p42/p44 MAPK, p38 MAPK, JNK, and nuclear factor-kappaB signaling pathways in human tracheal smooth muscle cells. J Cell Physiol. 2007;211:759–770. doi: 10.1002/jcp.20992. [DOI] [PubMed] [Google Scholar]
- Lin TC, Huang LT, Huang YN, Chen GS, Wang JY. Neonatal status epilepticus alters prefrontal–striatal circuitry and enhances methamphetamine-induced behavioral sensitization in adolescence. Epilepsy Behav. 2009;14:316–323. doi: 10.1016/j.yebeh.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Lipska BK, et al. RGS4 mRNA expression in postmortem human cortex is associated with COMT Val158Met genotype and COMT enzyme activity. Hum Mol Genet. 2006;15:2804–2812. doi: 10.1093/hmg/ddl222. [DOI] [PubMed] [Google Scholar]
- Mittmann C, et al. Evidence for a short form of RGS3 preferentially expressed in the human heart. Naunyn-Schmiedebergs Arch Pharmacol. 2001;363:456–463. doi: 10.1007/s002100000376. [DOI] [PubMed] [Google Scholar]
- Mittmann C, et al. Expression of ten RGS proteins in human myocardium: functional characterization of an upregulation of RGS4 in heart failure. Cardiovasc Res. 2002;55:778–786. doi: 10.1016/s0008-6363(02)00459-5. [DOI] [PubMed] [Google Scholar]
- Mungunsukh O, Marquez AP, Lee YH, Thiel G, Day RM. Characterization of the bovine angiotensin converting enzyme promoter: essential roles of Egr-1, ATF-2 and Ets-1 in the regulation by phorbol ester. Gene. 2008;421:81–88. doi: 10.1016/j.gene.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Murthy KS. Signaling for contraction and relaxation in smooth muscle of the gut. Annu Rev Physiol. 2006;68:345–374. doi: 10.1146/annurev.physiol.68.040504.094707. [DOI] [PubMed] [Google Scholar]
- Murthy KS, Makhlouf GM. Differential coupling of muscarinic m2 and m3 receptors to adenylyl cyclases V/VI in smooth muscle. Concurrent M2-mediated inhibition via Galphai3 and m3-mediated stimulation via Gbetagammaq. J Biol Chem. 1997;272:21317–21324. doi: 10.1074/jbc.272.34.21317. [DOI] [PubMed] [Google Scholar]
- Ni YG, Gold SJ, Iredale PA, Terwilliger RZ, Duman RS, Nestler EJ. Region-specific regulation of RGS4 (regulator of G-protein-signaling protein type 4) in brain by stress and glucocorticoids: in vivo and in vitro studies. J Neurosci. 1999;19:3674–3680. doi: 10.1523/JNEUROSCI.19-10-03674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie M, Pang L, Inoue H, Knox AJ. Transcriptional regulation of cyclooxygenase 2 by bradykinin and interleukin-1beta in human airway smooth muscle cells: involvement of different promoter elements, transcription factors, and histone h4 acetylation. Mol Cell Biol. 2003;23:9233–9244. doi: 10.1128/MCB.23.24.9233-9244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova DN, et al. Genome-wide gene expression profiles of thyroid carcinoma: identification of molecular targets for treatment of thyroid carcinoma. Oncol Rep. 2008;20:105–121. [PubMed] [Google Scholar]
- Patten M, et al. Endotoxin induces desensitization of cardiac endothelin-1 receptor signaling by increased expression of RGS4 and RGS16. Cardiovasc Res. 2002;53:156–164. doi: 10.1016/s0008-6363(01)00443-6. [DOI] [PubMed] [Google Scholar]
- Patten M, Stube S, Thoma B, Wieland T. Interleukin-1beta mediates endotoxin- and tumor necrosis factor alpha-induced RGS16 protein expression in cultured cardiac myocytes. Naunyn-Schmiedebergs Arch Pharmacol. 2003;368:360–365. doi: 10.1007/s00210-003-0798-0. [DOI] [PubMed] [Google Scholar]
- Pepperl DJ, Shah-Basu S, VanLeeuwen D, Granneman JG, MacKenzie RG. Regulation of RGS mRNAs by cAMP in PC12 cells. Biochem Biophys Res Commun. 1998;243:52–55. doi: 10.1006/bbrc.1997.8056. [DOI] [PubMed] [Google Scholar]
- Percy WH, Burton MB, Fallick F, Burakoff R. A comparison in vitro of human and rabbit distal colonic muscle responses to inflammatory mediators. Gastroenterology. 1990;99:1324–1332. doi: 10.1016/0016-5085(90)91157-2. [DOI] [PubMed] [Google Scholar]
- Riddle EL, Schwartzman RA, Bond M, Insel PA. Multi-tasking RGS proteins in the heart: the next therapeutic target? Circ Res. 2005;96:401–411. doi: 10.1161/01.RES.0000158287.49872.4e. [DOI] [PubMed] [Google Scholar]
- Rizig MA, et al. Failure to confirm genetic association between schizophrenia and markers on chromosome 1q23.3 in the region of the gene encoding the regulator of G-protein signaling 4 protein (RGS4) Am J Med Genet B Neuropsychiatr Genet. 2006;141B:296–300. doi: 10.1002/ajmg.b.30288. [DOI] [PubMed] [Google Scholar]
- Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- Romero DG, et al. Regulators of G-protein signaling 4 in adrenal gland: localization, regulation, and role in aldosterone secretion. J Endocrinol. 2007;194:429–440. doi: 10.1677/JOE-07-0153. [DOI] [PubMed] [Google Scholar]
- Runne H, et al. Dysregulation of gene expression in primary neuron models of Huntington’s disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J Neurosci. 2008;28:9723–9731. doi: 10.1523/JNEUROSCI.3044-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtgen JL, et al. Oct-1 is involved in the transcriptional repression of the von Willebrand factor gene promoter. Blood. 1998;92:1247–1258. [PubMed] [Google Scholar]
- Song L, Jope RS. Cellular stress increases RGS2 mRNA and decreases RGS4 mRNA levels in SH-SY5Y cells. Neurosci Lett. 2006;402:205–209. doi: 10.1016/j.neulet.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart Gibbons A, Scarr E, McOmish CE, Hannan AJ, Thomas EA, Dean B. Regulator of G-protein signalling 4 expression is not altered in the prefrontal cortex in schizophrenia. Aust N Z J Psychiatry. 2008;42:740–745. doi: 10.1080/00048670802206338. [DOI] [PubMed] [Google Scholar]
- Takata Y, et al. PPARdelta-mediated antiinflammatory mechanisms inhibit angiotensin II-accelerated atherosclerosis. Proc Natl Acad Sci USA. 2008;105:4277–4282. doi: 10.1073/pnas.0708647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gemert NG, Meijer OC, Morsink MC, Joels M. Effect of brief corticosterone administration on SGK1 and RGS4 mRNA expression in rat hippocampus. Stress. 2006a;9:165–170. doi: 10.1080/10253890600966169. [DOI] [PubMed] [Google Scholar]
- van Gemert NG, van Riel E, Meijer OC, Fehr S, Schachner M, Joels M. No effect of prolonged corticosterone over-exposure on NCAM, SGK1, and RGS4 mRNA expression in rat hippocampus. Brain Res. 2006b;1093:161–166. doi: 10.1016/j.brainres.2006.03.083. [DOI] [PubMed] [Google Scholar]
- Wang S, Zhao Y, Leiby MA, Zhu J. Studying human telomerase gene transcription by a chromatinized reporter generated by recombinase-mediated targeting of a bacterial artificial chromosome. Nucleic Acids Res. 2009;37:e111. doi: 10.1093/nar/gkp511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willars GB. Mammalian RGS proteins: multifunctional regulators of cellular signalling. Semin Cell Dev Biol. 2006;17:363–376. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Woodruff-Pak DS, Agelan A, Del Valle L. A rabbit model of Alzheimer’s disease: valid at neuropathological, cognitive, and therapeutic levels. J Alzheimers Dis. 2007;11:371–383. doi: 10.3233/jad-2007-11313. [DOI] [PubMed] [Google Scholar]
- Xie GX, Palmer PP. How regulators of G protein signaling achieve selective regulation. J Mol Biol. 2007;366:349–365. doi: 10.1016/j.jmb.2006.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, et al. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69:5743–5751. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar P, Persson AI, Ming Y, Terenius L. Opioid-induced regulation of gene expression in PC12 cells stably transfected with mu-opioid receptor. Neurosci Lett. 2006;396:197–201. doi: 10.1016/j.neulet.2005.11.040. [DOI] [PubMed] [Google Scholar]