Abstract
Cryptococcus neoformans is an important human, fungal pathogen that sheds polysaccharide (exo-PS) into host tissues. While shed exo-PS mediates numerous untoward effects (including promoting increased intracranial pressure), little is known about the regulation of this phenomenon. Since down-regulation of the Allergen 1 (ALL1) gene is associated with high ICP, we investigated the relationship between ALL1 expression and exo-PS structure using a variety of biophysical techniques. The Δall1 mutants of two serotypes produced a shorter exo-PS with less branching and structural complexity than the parental strains. Consistent with lower branching, these exo-PSs manifested higher intrinsic viscosity than the parental strains. The Δall1 mutant strains manifested differences in epitope expression and significant resistance to phagocytosis. Exo-PS of Δall1 mutant exhibited anti-phagocytic properties. Comparative transcriptome analysis of mutant and parental strainunder iron-deprived conditions indicateda role of ALL1 in iron homeostasis, characterized by differential regulation of genes that mediate iron reduction and transport. Together, our results demonstrate a role of ALL1 in regulating conformational aspects of PS structure and iron homeostasis. These findings provide a mechanism to explain how changes in ALL1 expression influence virulence of switch variants and suggest that structural changes and polymer length are epigenetically regulated.
Keywords: Cryptococcus neoformans, GXM, allergen 1, polysaccharide structure, branching
INTRODUCTION
C. neoformans is an important fungal pathogen in patients with AIDS. The most common clinical presentation of cryptococcosis is chronic meningoencephalitis (CME), a leading infectious cause of death in the world, particularly in Africa (Park et al., 2009), with 10-week mortality rates between 25–50% (Bicanic et al., 2007; Imwidthaya and Poungvarin, 2000; Kambugu et al., 2008). One of the unique features of C. neoformans is its PS capsule that is induced during infection (Zaragoza et al., 2009). The PS capsule is essential for virulence (Chang and Kwon-Chung, 1994; McClelland et al., 2005) and is predominantly composed of glucuronoxylomannan (GXM) (McFadden et al., 2006). This large polymer consists of structural reporter groups (SRGs), which are composed of an α-(1,3)-mannan main chain with β-(1,2)-glucuronic acid and a β-(1,2)- or β(1,4) xylosyl residues units (Cherniak et al., 1998).
C. neoformans induces its polysaccharide capsule in iron deprived conditions and releases substantial quantities of exo-PS into the cerebrospinal fluid (CSF) during chronic infection causing serious complications (Goldman et al., 1997; Graybill et al., 2000). The accumulation of shed exo-PS in host tissues interferes with CSF re-absorption at the level of the granulations in the arachnoid villi and thus is thought to play a key role in the development of elevated ICP, (Graybill et al., 2000). High ICP is difficult to treat, and is associated with significant morbidity and mortality in patients with CME (Dromer et al., 2007; Graybill et al., 2000; Seaton et al., 1996). In patients with CME (Bicanic et al., 2007; Bicanic et al., 2008) a high CSF fungal burden correlates with increased ICP before and after fungal therapy. Nonetheless, a high fungal burden does not appear to be sufficient to produce elevated ICP (Bicanic et al., 2009), suggesting that strain-related properties of exo-PS affect the development of high ICP.
Our previous studies using phenotypic switch variants support a role for exo-PS differences in promoting elevated ICP. In these studies we found that phenotypic switching from RC2-SM to a RC2-MC variant was associated with the shedding of an altered exo-PS, which caused elevated ICP and rapidly fatal CME in rats (Fries et al., 2005). NMR analysis confirmed serotype D predicted SRG composition in RC2-SM and RC2-MC derived exo-PS. Static light scattering (SLS) analysis, demonstrated that the radius of gyration (Rg) differed in RC2-SM and RC2-MC exo-PS despite comparable molecular mass (Mw) and thus suggested that yet undefined macromolecular properties of the exo-PS were altered by phenotypic switching (McFadden et al., 2007).
Phenotypic switching of RC2-SM to RC2-MC is associated with down-regulation of a defined set of genes, in particular Allergen1 (ALL1) (Jain et al., 2009b). ALL1 down-regulation is noteworthy as null mutant (Δall1) mimics the hypervirulence of the RC2-MC switch variant including the strain’s propensity to cause high ICP and premature death in rats (Jain et al., 2009b). This is analogous to the pathogenesis of human CME with poor outcome.
In the present study, we used viscosity, SLS and dynamic light scattering (DLS) analysis to investigate the macromolecular properties of exo-PS that are shed in Δall1 mutants of serotype A and D strains as well as their respective parental and reconstituted strains. The results obtained imply that the structure of shed exo-PS is greatly affected by the loss of ALL1 function and phenotypic switching. Analysis of mass density, shape factor and viscosity, consistently demonstrate that changes of RC2-MC exo-PS are mediated by loss of ALL1. Specifically, Δall1 strains produce a more viscous and smaller exo-PS with a lower degree of branching. In addition, microarray analysis of Δall1 mutant in serotype D strain RC2 (RC2-Δall1) versus wild type (wt) of RC2 in minimal medium identified up-regulation of genes encoding cell surface reductases, iron permease/ferroxidase, cytokine inducing proteins and siderophore transporters. The impact and correlations of this structural variation and its association with iron homeostasis and pathogenesis of CME are discussed. We propose that biophysical characteristics of the exo-PS constitute emerging properties that are regulated and can be changed by phenotypic switching.
RESULTS
Null Mutants of ALL1 (Δall1) in the Serotype A and D Strains Exhibited Impaired Capsule Induction and Shed a More Viscous PS
Previous work demonstrated that RC2-Δall1 exhibited subtle changes in capsule size and impaired capsule induction (Jain et al., 2009b). We sought to establish whether these changes were also present in the serotype A strain background and generated a Δall1 mutant in H99 reference strain (H99-Δall1). The baseline capsule diameter was not increased in H99-Δall1, but impaired capsule induction was confirmed in H99-Δall1 in vivo (Fig 1A) and in vitro under various capsule-inducing conditions (Table S1). Capsule induction defects were corrected in reconstituted strains (Δall1 + ALL1) and more pronounced induction was observed in the over expression mutant PCTR4-2-ALL1 (Fig S1, Table S2), when ALL1 was placed under a copper inducible promoter resulting in 23 fold over expression of ALL1 (data not shown).
Figure 1. Impaired capsule induction and differences in PS shedding and viscosity upon loss of ALL1.
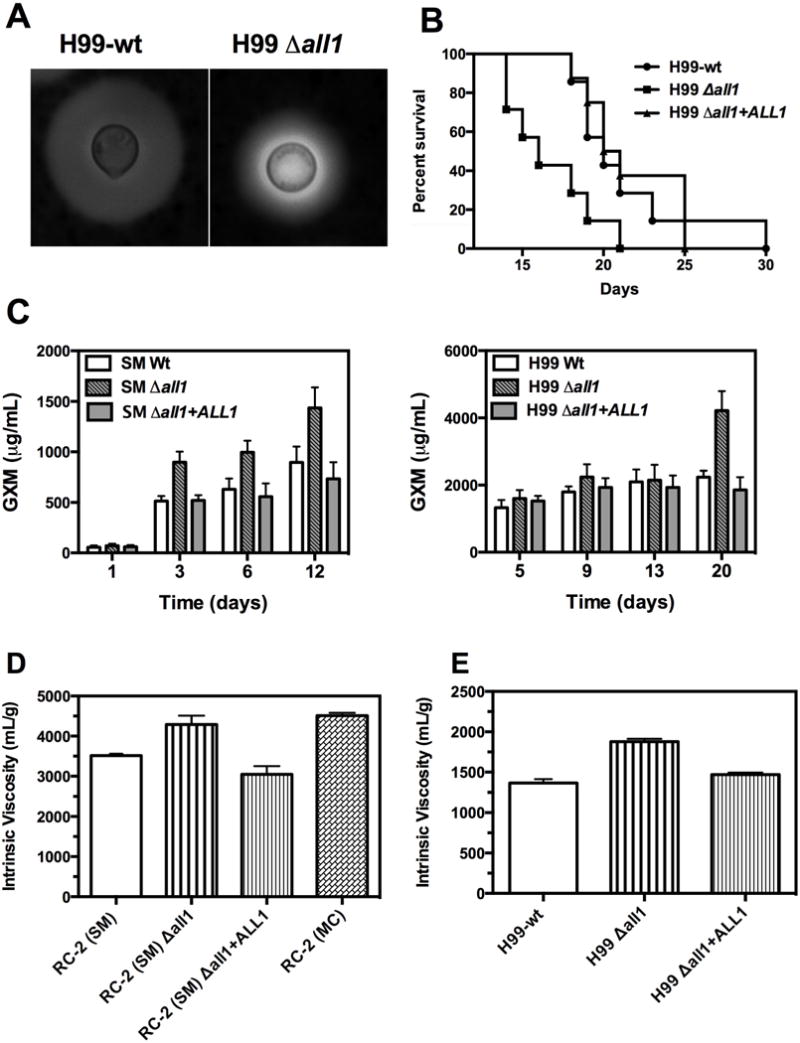
(A) H99-Δall1 mutant showedan impaired capsule induction defect after 24 h in vivo growth. Exo-PS concentrations from supernatants of in vitro cultures of RC2-wt (B) and H99-wt (C) was determined by ELISA after 1, 3, 6, 12 days, and 5, 9, 13, and 20 days of growth, respectively. (D) Intrinsic viscosities of exo-PSs from RC2-SM, RC2-Δall1, RC2-Δall1 + ALL1, RC2-MC and (E) of exo-PS from H99-wt, and H99-Δall1, H99-Δall1 + ALL1.
Next, we quantified the shedding of exo-PS for both serotypes by ELISA. RC2 and H99 shed different amounts of GXM and alsoH99 sheds GXM at a later time. Therefore, experiments measuring GXM shedding were designed for H99 and RC2 and their respective mutants at different times. Regardless, the exo-PS shedding was found to be higher in liquid culture for Δall1 mutants of both serotypes (Fig 1C).
The intrinsic viscosities [η] of exo-PS shed by Δall1 mutants of H99 and RC2 were determined using a modified Ostwald-type capillary glass. Exo-PS derived from Δall1 mutants in both serotypes (A and D) consistently exhibited higher viscosity, when compared to exo-PS derived from the respective parental wt and reconstituted strain (Fig 1D). The increased viscosity of the RC2-Δall1 derived exo-PS was comparable to that of the RC2-MCderived exo-PS, which is the phenotypic switch variant of RC2-SM (RC2-wt) that exhibits down-regulation of ALL1 gene transcripts (Jain et al., 2009b).
Loss of ALL1 in H99 confers a hypervirulent phenotype similar to RC2-Δall1
In the murine intratracheal (i.t.) infection model, hypervirulence of H99-Δall1 relative to the H99-wt was documented analogous to the RC2-Δall1 mutant (Fig 1B). The median survival of animals infected with high inoculum (106) with H99-Δall1 and H99-wt was 15 d vs 21 d (p = 0.04). At lower infection inoculum (105), the H99-wt was successfully cleared from lung tissue at 10 days (< CFU log 2.3 ± 0.34 per lung) whereas the H99-Δall1 was not (log 6.4 ± 0.5 for, p = 0.01). Doubling times of mutant and wt were comparable confirming that enhanced virulence is not merely due to growth differences but rather altered host-pathogen interaction.
Loss of Allergen1 Altered PS Macro-structural Properties
Next, we pursued systematic analysis of the exo-PSs by both SLS and DLS analysis. All exo-PS solutions exhibited significant but comparable polydispersity, which was expected, based on prior studies (Cordero et al., 2011) (Fig 2). SLS yielded consistent differences in Mw and Rg (Table 1). A 2- to 5-fold decrease in PS Mw, Rg and hydrodynamic radius (Rh) was observed for Δall1 strains in both serotype backgrounds. Changes in Rh, which reflects the apparent size of the molecules in solution, were more pronounced than changes in Rg (Table 1), suggesting differences in PS conformation. Combining data from SLS and DLS further assessed structural conformation of exo-PS molecules in solution (Burchard et al., 1980). The mass density, which is the ratio between the Mw of exo-PS and their Rg, and the shape factor, which represents the ratio of Rg and Rh, were calculated for all exo-PS. These parameters indicated the occurrence of significant ALL1-dependent structural variation in shed exo-PS. A conserved decrease in mass density and increase in shape factor (1.6- and 4.6-fold, respectively) was evident upon loss of ALL1 (Table 1). Most importantly, most measured differences including the polymer size difference reverted to wt values in the reconstituted strains (Fig 2). In addition, differences were consistent for both Δall1 strains although the SRGs of serotype D and serotype A strains differ by a single xylose residue. In summary, DLS and SLS data confirm that Δall1 mutant exo-PS are less branched and have a lower mass density compared with exo-PS from the parental wt strains. These data were consistent with the observed increased viscosity in Δall1 mutants.
Figure 2. Hydrodynamic size distribution of PS samples determined by DLS.
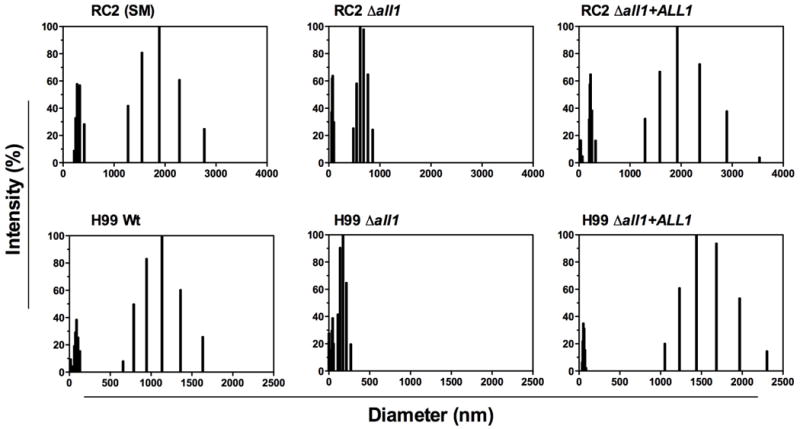
Hydrodynamic size distribution of exo-PS derived from wt, mutant and reconstituted strains was plotted as percentage of light scattering intensity versus effective diameter. Data are averages of 10 repeated measurements. All exo-PS solutions exhibited significant polydispersity. Note the polymer size difference reverted to wt in the reconstituted strains.
Table 1.
Macromolecular parameters of PS samples determined by static and dynamic light scattering analysis
| Strain | Mw (× 106) | Rg (nm) | Rh (nm) | PDI | Mass Density (× 104) | ρ |
|---|---|---|---|---|---|---|
| RC2 (SM) | 1.66 ± 0.24 | 116.0 ± 9.7 | 266.4 ± 17.3 | 0.434 ± 0.013 | 1.43 | 0.43 |
| RC2-Δall1 | 0.89 ± 0.07 | 104.0 ± 4.3 | 121.4 ± 5.3 | 0.464 0.015 | 0.83 | 0.86 |
| RC2-Δall1 + ALL1 | 0.54 ± 0.03 | 116.3 ± 6.6 | 240.3 ± 25.9 | 0.452 ± 0.022 | 0.46 | 0.48 |
| RC2 (MC) | 1.80 ± 0.90 | 182.0 ± 10.7 | 238.2 ± 6.4 | 0.381 ± 0.026 | 0.99 | 0.76 |
| H99-wt | 7.07 ± 0.65 | 170.2 ± 6.3 | 353.8 ± 23.7 | 0.445 ± 0.015 | 4.15 | 0.48 |
| H99-Δall1 | 1.26 ± 0.06 | 142.4 ± 3.3 | 162.9 ± 10.5 | 0.377 ± 0.009 | 0.88 | 0.87 |
| H99-Δall1 + ALL1 | 6.15 ± 0.58 | 168.0 ± 10.0 | 315.5 ± 15.7 | 0.461 ± 0.004 | 3.66 | 0.53 |
The average-molecular mass, Mw (g mol−1), radius of gyration, Rg (nm), hydrodynamic radius, Rh (nm), polydispersity index (PDI), average mass density (g mol−1 nm−1) and shape factor (ρ) of exo-PS samples. Mw, Rg data are represented as mean +/− SD of 2 measurements. Rh and polydispersity values are means +/− SE of 10 measurements.
Loss of ALL1 Produces PS Molecules with Differences in Antibody Reactivity
Given the conformational differences in the exo-PS, we studied the binding ability of a panel of anti-GXM monoclonal antibodies (mAb) that recognize distinct epitopes on the polysaccharide capsule. These mAbs yield defined staining patterns that correlate with specific serotype specific SRGs (Belay et al., 1997). Previous work documented that binding of mAbs can be greatly influenced by the degree of PS branching and conformation (Cordero et al., 2011). A decrease in mAb 13F1 and 21D2 binding was observed for exo-PS isolated from the H99-Δall1 (Fig 3A) by ELISA. Binding differences were also documented with mAb 12A1 for RC2-wt and RC2-Δall1 derived exo-PS by ELISA and also on capsules of live yeast cells as evident by capsular staining (Fig 3B).
Figure 3. Loss of Allergen1 produces PS with decreased mAb reactivity.
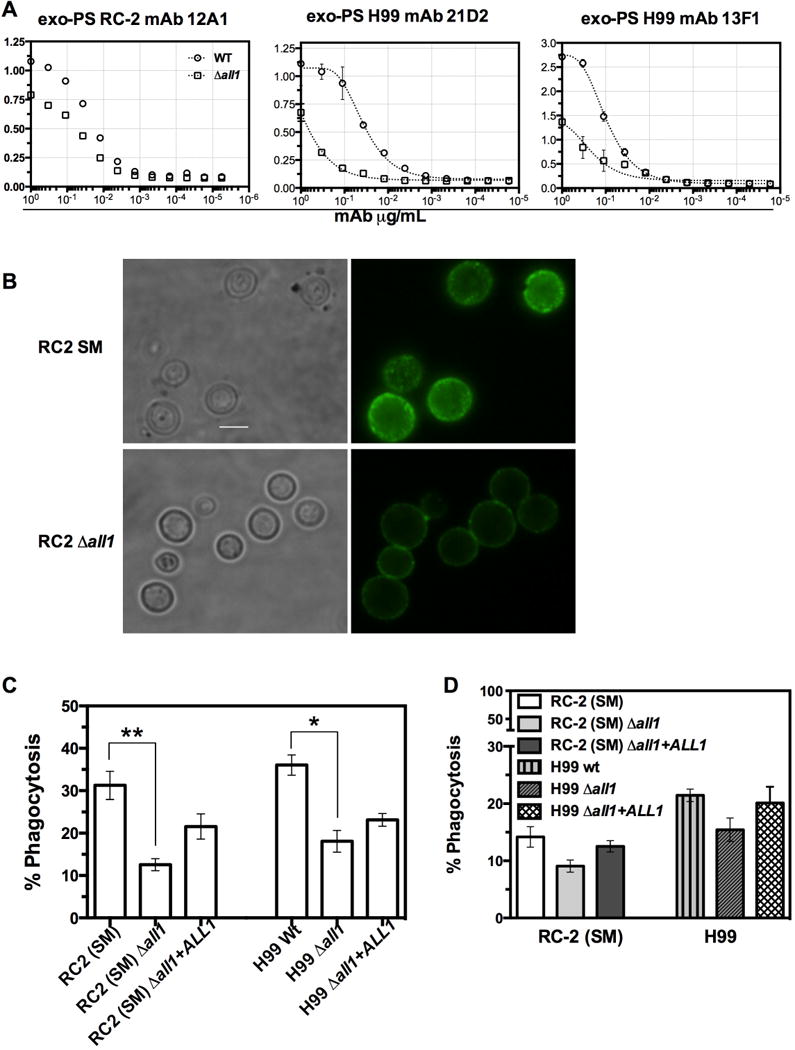
(A) ELISA of exo-PS isolated from wt and Δall1 of RC2 and H99 background strains using anti-GXM mAbs 12A1, 21D2, and 13F1. For all mAbs tested, Δall1 derived exo-PS showed lower reactivity. (B) Immunofluorescence micrograph of RC2-wt andRC2-Δall1 cells using mAb 12A1 directly conjugated to Alexa488. RC2-Δall1 cells showed a significant reduction in 12A1 binding to the capsule. Scale bar represents 10μm. (C) Complement-mediated phagocytosis of RC2-SM, RC2-Δall1, RC2-Δall1 + ALL1, H99-wt and H99-Δall1 and H99-Δall1 + ALL1. A significant reduction in percentage of phagocytosis was observed for Δall1 mutants relative to the parental and reconstituted strains in both serotypes. Data are mean +/− SE of 4 independent experiments of triplicate wells. (D) Exo-PS from wt, Δall1 and Δall1 + ALL1 strains were added to the wt wells and similar percent reduction of phagocytosis values were observed when exo-PS from, respective Δall1 mutant was added to SM-wt and H99-wtduring phagocytosis assay.
No differences were evident for binding of mAb 18B7, however, 18B7-mediated phagocytosis was reduced for mutants in both genetic backgrounds (Fig S2). Consistent with structural differences in the PS capsule, we also observed a significant decrease in complement-mediated phagocytosis of Δall1 cells by J774 macrophage-like cells (Fig 3C) in both serotypes. In addition, when exo-PS secreted by wt, Δall1 and Δall1 + ALL1 was added to respective wt cells in both serotypes, we documented that Δall1 exo-PS inhibited complement mediated phagocytosis more effectively than wt-exo-PS and Δall1 + ALL1 exo-PS (Fig 3D).
Gene regulation in Δall1 under different growth conditions
To further elicit gene function of ALL1, a transcriptome analysis of RC2-Δall1 vs. RC2-wtwas performed. For microarray experiments, RC2-wt and RC2-Δall1 cells were grown in minimal medium, and Yeast Extract Peptone Dextrose medium (YPD) overnight at 37°C, diluted 1:100 into fresh medium and grown overnight up to an OD600nm of 0.6 at 37°C with agitation at 150 rpm. Microarray analysis demonstrated differential regulation of genes with very little overlap between the two growth conditions (Table S3). A total of 15 and 271 genes were found to be down regulated, 2 fold or more in RC2-Δall1 when grown in minimal media and YPD, respectively. In minimal media up-regulation of 111 genes (≥ 2 fold) was observed in Δall1, which includes a set of genes related to iron transport and homeostasis [e.g. cell surface reductase (FRE1), a high affinity iron permeases gene (FTR1), a metalloreductase, and siderophore transporters (Jung et al., 2006) Table 2], The most abundantly up-regulated gene in the RC2-Δall1 cells was CIG1 (19.7 fold), which encodes a mannoprotein involved in the retention of iron at the cell surface and/or in the uptake of siderophore-bound iron. It affects growth in low-iron medium, and alters capsule response to iron-replete conditions (Lian et al., 2005). This up-regulation was only observed in minimal media, which was also the growth condition for exo-PS isolations. In YPD, loss of ALL1 resulted in the up-regulation of approximately 270 genes, which were related to multiple cellular pathways. We further analyzed the data for gene ontology (GO) categories for the biological processes of differentially expressed genes in minimal media. The analysis indicates that differentially expressed genes were significantly enriched with GO categories related to oxidoreductase activity, oxidation-reduction process and transmembrane transport after loss of ALL1 (Table 3). Differentially regulated genes with GO term related to oxidoreductase activity were genes related to reductive iron uptake systems such as FRE1, Ferric-chelate reductases (CNG00110), metalloreductase (CNG00950) and Cytochrome-b5 reductase (CNB02540). The other highly ranked group, transmembrane transport also included 4 iron related transmembrane transporters, namely FTR1, CNM02430, CNG00120 and CNB00400. In addition, the drug transporter (CNJ00070) was 7- fold up-regulated in Δall1. The hexose transport related protein (CNE02910), and the carboxylic-acid transport protein (CNJ01360) were also up-regulated whereas the sugar transporter (CNH00490) was down-regulated. Genes classified under “integral to membrane” were also ranked in the analysis but did not reach significance (Table S3). Differential expression of iron transporters and siderophores were further confirmed by Real time PCRonRC2 (Fig 4A) and H99 (Fig 4B). Real time PCR confirmed the signature of up-regulated iron-transport genes in Δall1 mutants in both serotypes, albeit difference were found among the serotypes and also minor differences in fold regulation with respect to the microarray analysis. It is noteworthy that these genes were not differentially regulated in microarrays done in YPD or sabouraud dextrose medium (Jain et al., 2009b), suggesting that ALL1 negatively controls iron homeostasis only under starvation conditions. A role of ALL1 in iron regulation was further supported, by demonstrating accumulation of intracellular iron under low-iron conditions (Fig 5A & 5B). Enhanced iron accumulation was not documented in the presence of iron (Fig 5C & 5D). Growth rate of wt and Δall1 was slow but comparable in low iron medium (data not shown). Additionally, sugar transporter (CNH00490) was down-regulated under all growth conditions. In summary, comparative transcriptional profiling supports a role of ALL1 in iron homeostasis under limited nutrition and iron conditions and a role in regulating sugar transport, which could affect production of capsular polysaccharide.
Table 2.
ALL1 regulates genes required for Iron transport
| Gene ID and Accession number | Fold Change |
|---|---|
| RC2-SM vs RC2-Δall1 | |
| CIG1, cytokine inducing-glycoprotein; CNC01660 | 19.7 |
| Ferric-chelate reductase activity: CNG00110 | 15.87 |
| High-affinity iron permease, FTR1: CNC05700 | 6.52 |
| FRE1, ferroxidase: CNC05690 | 5.94 |
| Hypothetical protein: CNK02840 | 3.94 |
| Metalloreductase, ferric-chelate reductase activity: CNG00950 | 2.72 |
| Zinc-Finger protein putative: CNI01980 | 2.65 |
| Iron ion transmembrane transporter activity: CNM02430 | 2.57 |
| Siderophore-iron uptake transmembrane transporter activity: CNG00120 | 2.48 |
| Cytochrome-b5 reductase: CNB02540 | 2.35 |
| Acidic laccase, putative: CNM02420 | 2.1 |
| Siderophore-iron transmembrane transporter activity: CNB00400 | 2.0 |
| Cytochrome b2, Mitochondrial Precursor: CNH01230 | 3.29 |
Table 3.
GO terms identified in the differentially expressed genes
| GO Term and Identification number | p-value |
|---|---|
| Oxidoreductase activity, GO:0016491 | 3.97 × 10−5 |
| Oxidation-reduction process, GO:0055114 | 2.86 × 10−4 |
| Transmembrane transport, GO:0055085 | 4.11 × 10−4 |
Figure 4. ALL1 indirectly regulate genes required for Iron-transport.
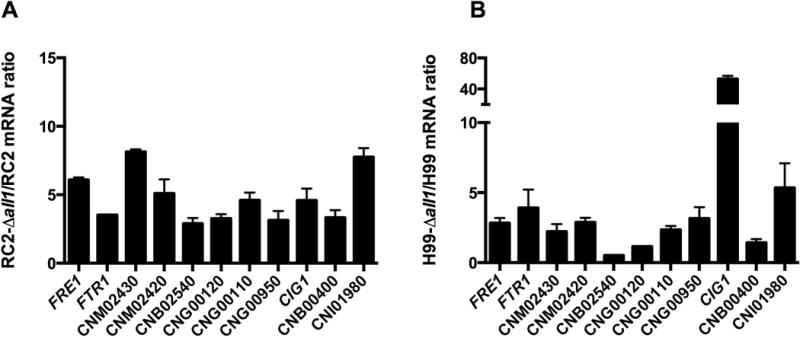
Differential gene expression in wild type (RC2-wt, H99-wt) and their respective Δall1 mutants (RC2-Δall1, H99-Δall1) were confirmed by real time PCR with selected iron-related genes, Real-time PCR results on RC2-wt, RC2-Δall1 (A) and H99-wt, H99-Δall1 (B) are shown. Black bars represents genes up-regulated in mutants. Bars represent average +/− SD mRNA ratios. The mRNA levels were normalized against the respective β-actin expression. Fold expression was calculated using the delta-delta CT method.
Figure 5. Mutant of ALL1 accumulates Intracellular iron and exo-PS affects the regulation of iron transport related genes.
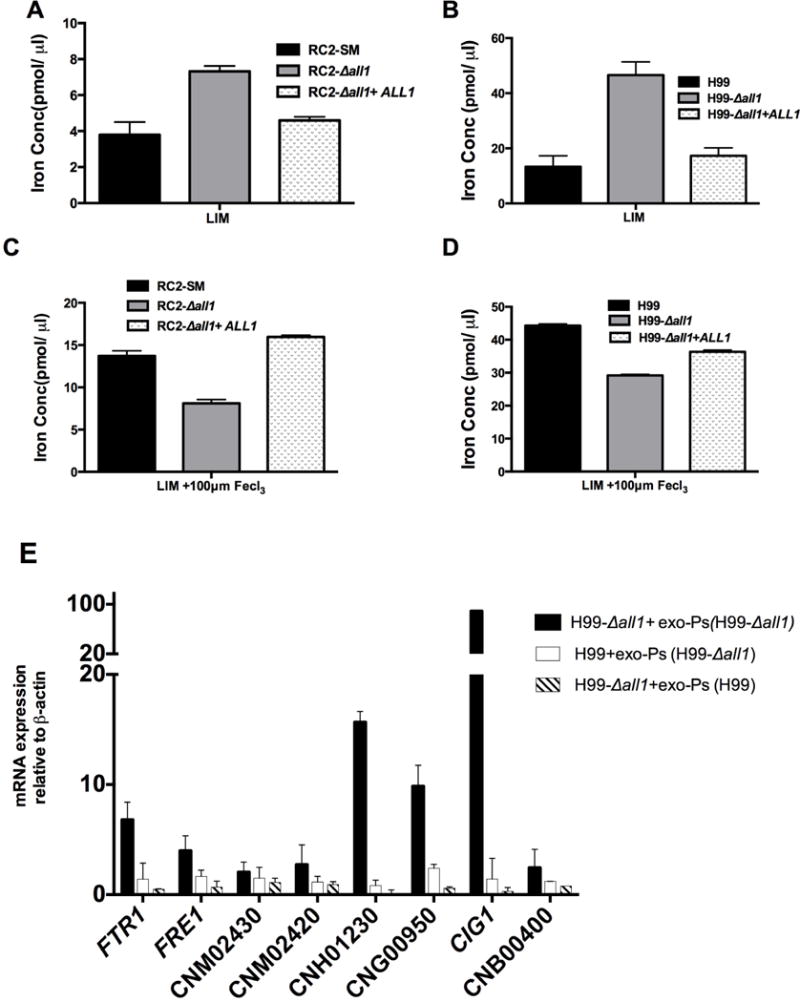
Iron concentration was altered for RC2-SM, RC2-Δall1, H99-wt, H99-Δall1 and reconstituted strains under low iron conditions (A, B) and in response to ferric chloride (C, D). Differences in exo-PS structure of H99-wt and H99-Δall1 mutant affect expression of iron transport related genes. FTR1, FRE1, CNM02430, CNM02420, CNH01230, CNG00950, CIG1, and CNB00400 were up-regulated in H99-Δall1 (black bars) and in H99-wt cells (white bars) by addition of 40μg/ml mutant exo-PS. In contrast, addition of H99-wt exo-PS to H99-Δall1 cells (striped bars) resulted in significant down-regulation of iron transport related genes (E).
Δall1 mutant exo-PS assists or activates the iron homeostasis related genes expression
Next, we tested the effect of exo-PS derived from the Δall1 mutant on expression of iron transport related genes. H99-wt and H99-Δall1 cells were grown in minimal media for 16h supplemented with 40μg/ml exo-PS of H99-Δall1 and H99-wt respectively at 37°C with agitation at 150 rpm. Transcript levels of selected genes related to iron homeostasis were quantified by real time PCR. Addition of H99-Δall1 exo-PS in H99-wt cells resulted in up-regulation of FTR1, FRE1, CNM02430, CNM02420, CNH01230, CNG00950, CIG1, and CNB00400 transcripts (Fig 5E, white bars). Up-regulation was also seen if the mutant exo-PS was added to mutant cells (Fig 5E, Black bars). Interestingly, addition of H99-wt exo-PStoH99-Δall1 cells resulted in down-regulation of iron transport associated genes (Fig 5E, crossed lines bars). Consistent with our expectation, transcript levels of the sugar transporter were not affected by the addition of exo-PS.
DISCUSSION
Here we report that structural characteristics of shed cryptococcal PS are regulated by ALL1 expression, a gene that is regulated in vivo during chronic infection. This result is important because it provides a mechanism by which ALL1 can modulate the virulence of C. neoformans. Biophysical alterations of the exo-PS affect clinically relevant characteristics of the polymers such as their viscosity, mAb reactivity, and complement-mediated phagocytosis. In vivo, these altered polymers affect capsule induction and the propensity of the mutants to cause high ICP in rats (Jain et al., 2009b). In addition, we demonstrate that loss of ALL1 up-regulates iron transport resulting in enhanced accumulation of intracellular iron. Our results link the presence of high ICP in Δall1 infected rats, to impaired capsule induction and in vivo shedding of shorter and less branched PS polymers that exhibit significantly higher viscosity when compared to the wt polymers. Hence, this work associates the modulation of a gene that is regulated in vivo with the most catastrophic complication of cryptococcal CME through the production of a structurally different exo-PS. These results also further support a link between exo-PS and iron regulation possibly through altered binding and sensing of extracellular iron level, which then indirectly regulates expression of iron transport and iron reduction pathway associated genes.
Cryptococcal PSs are complex polymers that have distinct physical and chemical properties reflected by their molecular mass, shape, and composition. Previous studies have shown that polymer diameter does not vary significantly as a function of SRG composition, but is more affected by secondary and tertiary structure of the molecule (McFadden et al., 2007). Consistent with this finding, SRG type does not correlate with clinical presentation or outcome of CME in a consistent matter (Cordero et al., 2011). Yet, macromolecular properties such as molecular mass and hydrodynamic size and/or shape have shown to underlie GXM biological functions or reactivity (Cordero et al., 2011; Fonseca et al., 2010). The view that GXM molecules are linear polymers (Doering, 2009) was recently revised and it was shown that degree of PS branching and higher order structural characteristics could be modulated by the dextrose concentration of the medium in which C. neoformans was cultured (Cordero et al., 2011; Guimaraes et al., 2010). We investigated exo-PS from mutants (RC2-Δall1 & H99-Δall1) and parental (RC2 & H99) strains that have previously been well characterized with respect to their virulence and relative ability to elicit high ICP in rats (Jain et al., 2009b). Raised ICP is a major and commonly fatal complication in patients with cryptococcal CME and is not necessarily responsive to antifungal therapy. Although the presence of high GXM titers are necessary to cause high ICP, they are not sufficient as there are patients with high crypt-Ag titers that do not develop high ICP (Bicanic et al., 2009).
GXM are large heterodisperse polymers (> 1 mDa) and cannot be studied by NMR or crystallography. However, LS provides the means to gain insight into their shape. LS analysis showed that exo-PS polymers from Δall1 mutants are shorter, and consequently exhibit less mass density. Exo-PS from mutants generated in both the serotype D and A strain backgrounds exhibited consistently these length and mass changes, although GXM polymers from serotype D and A strains are composed of distinct SRGs that can be distinguished by NMR. Shorter length of polymer is corrected to wt length in the reconstituted all1 + ALL1 mutants of RC2 and H99. In addition when the gene is over expressed capsule induction is even more pronounced. This finding further supports our conclusion that ALL1 regulates length of the GXM-polymer either directly or indirectly. The finding of shorter polymers with smaller masses was consistent with the impaired capsule induction of both Δall1 mutants in host tissue and in vitro (Jain et al., 2009b). Our data substantiate previous conclusions that the longest GXM polymers span the entire diameter of the capsule (Frases et al., 2009) and support the notion that larger PS capsules shed larger exo-PSs.
Transcriptome analysis in minimal medium conditions unexpectedly demonstrated up-regulation of genes involved in iron homeostasis including iron transporters and reductases. This signature was confirmed by real time PCR and is consistent with data from the GO analysis. CIG gene was the most abundantly up-regulated transcript by real time PCR in H99 and by microarray in RC2. Why it RT PCR yielded a lower up regulation in RC2 is not clear because the growth conditions were exactly the same for both assays. Consistent with the observed transcript profile, both Δall1 mutants accumulate more iron intracellularly under low-iron conditions. Co-cultivation of wt and mutant cells with wt and mutant exo-PS indicates that 9 of 13 iron related genes tested are up-regulated, when mutant-exo-PS is added to wt cells and down-regulated when wt-exo-PS is added to mutant cells. This observation further indicates that there is indeed a link between exo-PS and iron transport. One possible explanation is that mutant exo-PS binds more iron resulting in up-regulation of genes associated with iron sensing and transport. In the CSF, iron levels are low and the pathogen must compete with the host for iron and thus enhanced iron availability can exacerbate the outcome of cryptococcal CME (Barluzzi et al., 2002). Based on our findings we hypothesize that down-regulation ofALL1 does not only affect the polysaccharide capsule but may also render the pathogen more fit in an iron depleted host environment.
A complex regulatory network composed of distinct but partially overlapping pathways, regulates capsule induction. Others have shown that 316 genes are positively regulated during capsule induction, including ALL1 (Haynes et al., 2011). This network includes the cAMP pathway, which activates Pka1 and then Nrg1, which regulates genes directly involved in capsule assembly. An important pathway of capsule regulation involves iron homeostasis Hap3, Hap5, and Hapx (Jung et al., 2010) as well as the iron sensing transcription factor CIR1 (Jung et al., 2006). Review of previously published microarray data revealed that ALL1 was not differentially regulated under low iron conditions, however 5 and 3 fold down-regulated in Δcir1 mutant under low and high iron conditions, (Jung et al., 2006). ALL1 was also significantly up-regulated in Δhapx under low and high iron conditions (FeCl3 and transferrin) and was also up-regulated in Δhap3 mutantin response to transferrin (Jung et al., 2010). One gene that was not related to iron transport but nevertheless highly regulated in the mutant was a gene that encodes a multidrug resistance transporter (DHA1 family) (CNJ00070). Interestingly, in Saccharomyces cerevisiae this transporter is up-regulated in cells exhibiting reduced susceptibility to azoles (Barker et al., 2003).
Importantly, deletion of ALL1 gene affects baseline capsule size in RC2. So far, the phenotype of impaired capsule induction has only been described for Δall1 mutants. Here we document that loss of ALL1 results in the secretion of more exo-PS molecules with higher viscosity and smaller size and lower degree of branching. Several studies have indicated in the past that GXM polymer length is dynamic. Yoneda et al. showed that exo-PS shed early in the course of in vitro growth exhibited smaller molecular sizes when compared to exo-PS shed after 72 h (Yoneda and Doering, 2008). Frases et al. demonstrated that the capsule associated GXM was significantly different from shed exo-PS and that the diameter of the capsular PS and the exo-PS correlated (Frases et al., 2008). Finally, Cordero et al. showed that increasing dextrose in culture medium decreased polymer length and branching, which again resulted in structural differences (Cordero et al., 2011).
How and in what sub-cellular compartment the length of PS polymers are determined is not understood. It is also not clear if iron is specifically required in that process or important as a sensor only. ALL1 is regulated by SP1 in stress-associated pathways (Adler et al., 2011) as well as during capsule induction (Haynes et al., 2011). In contrast to bacterial PSs, GXM is a heteropolymer and magnitude larger in size, thus its assembly process is very different (Zaragoza et al., 2009). Current thinking is that the cryptococcal GXM polymers are pre-assembled in lipid associated cytoplasmic structures (Oliveira et al., 2009) and exported across the cell wall in vesicles (Rodrigues et al., 2007). All1p is a cytoplasmic protein, which is not present in vesicles (Jain et al., 2009b; Rodrigues et al., 2008) but loss of this protein down-regulates a sugar transporter (CNH00490). We therefore hypothesize that All1p is indirectly involved in GXM assembly, as its only homologues are found in non-encapsulated fungi that infect plants.
In the phenotypic switch variant RC2-MC hydrodynamic diameter and Rg differ from that of the RC2-Δall1 mutant, however shape factor, and viscosity are similarly altered consistent with the presence of high ICP in RC2-MC infected rats. Several studies demonstrate that structural changes alter the biological effects of GXM, which is a potent immunomodulator. For instance, the hydrodynamic effective diameter of the GXM molecules correlated with their ability to stimulate cellular responses (Fonseca et al., 2010) and affects the binding of monoclonal antibodies (Frases et al., 2008; Nimrichter et al., 2007).
Studies on in vivo derived C. neoformans cells further supported the notion that PS structure affects Ab and complement binding and demonstrated that the spatial deposition of C3 within the capsular matrix was influenced by the density of the capsular matrix (Gates and Kozel, 2006). Charlier et al. showed that brain invasion by C. neoformans was associated with changes in cryptococcal capsule structure and altered binding of PS specific mAbs (Charlier et al., 2005). Our data now links structural changes to loss of ALL1, which is down-regulated in the setting of phenotypic switching during chronic infection.
Older conclusions that impaired capsule induction is associated with loss of virulence (Granger et al., 1985) were not confirmed. In the rat model the altered exo-PS elicits high ICP resulting in rapid death and in the pulmonary model the altered exo-PS constitutes a potent immunomodulator and has dramatic effects on the host response especially macrophage activation and T-cell response (Guerrero and Fries, 2008; Guerrero et al., 2010; Jain et al., 2009a). Most importantly, the mutant elicits high ICP at a comparable fungal burden. Hence, hypervirulence is the consequence of altered host pathogen response to altered exo-PS, which results in increased fungal burden as the host response becomes ineffective.
We propose a model whereby loss of ALL1 function in clinical C. neoformans strains will result in altered iron homeostasis that results in an ICP inducing hypervirulent strain, because GXM polymers with altered length and shape that are more viscous and cannot be cleared by the host are produced. The ability to alter polymer length is a trait that was most likely selected for survival in the environment where the fungus encounters amoeboid predators (Pirofski and Casadevall, 2008) and extreme climate conditions. This observation underscores the notion that pathoadaptation of C. neoformans to the mammalian host is not driven by a single virulence factor that is shut on or off but rather constitutes fine epigenetic regulation of sophisticated pathways.
EXPERIMENTAL PROCEDURES
Yeast strains and culture conditions
C. neoformans strains were cultivated in chemically defined minimal medium (10 mM MgSO4, 29.3 mM KH2PO4, 13 mM glycine, 3 μM thiamine-HCl, 15 mM glucose; pH 5.5) for exo-PS isolation, microarray and Real time PCR. For exo-PS isolation, microarray and Real time PCR, cells were grown overnight at 37°C, diluted 1:100 into fresh medium and grown overnight up to an OD600nm of 0.6 at 37°C with agitation at 150 rpm. Asparagine salt medium contains 1 g L−1 asparagine, 10 mM sodium phosphate (pH 6.5), and 0.25 g L−1 MgSO4 and was used to grow ALL1 overexpression strain, PCTR4-2-ALL1. Low iron medium was prepared as described elsewhere (Nyhus et al., 1997). For GXM isolation, RC2-wt and H99-wt and their respective mutants were cultured for 14 days and 21 days, respectively at 37°C under shaking in minimal medium. For screening mutants YPD agar supplemented with 100mg L−1 nourseothricin (NAT) or 200mg L−1 neomycin G418 (NEO) was used. The C. neoformans strains used in the study are listed in Table 4.
Table 4.
Strains used in this study
| Strain | Additional information | Reference |
|---|---|---|
| RC2-SM | Serotype D, Switch variant Mat α | (Fries et al., 2001) |
| RC2-MC | Serotype D, Switch variant Mat α | (Fries et al., 2001) |
| RC2-Δall1 | Δall1: :NEO | (Jain et al., 2009b) |
| RC2-Δall1 + ALL1 | ALL1-NAT inserted to Δall1: :NEO | (Jain et al., 2009b) |
| H99 | Serotype A, Mat α | (Perfect et al., 1980) |
| H99-Δall1 | Δall1: :NEO in H99 | This study |
| H99-Δall1 + ALL1 | ALL1-NAT inserted to Δall1: :NEO | This study |
| PCTR4-2-ALL1 | ALL1 under CTR4 copper transport promoter | This study |
Generation of ALL1 mutant in H99
H99 reference sequence was accessed through Fungal Genome Initiative database at the Broad Institute (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html). To generate gene disruption mutants H99-wtgenomic DNA was isolated to amplify upstream and downstream regions of ALL1 using primers listed in Table S4. The entire coding region of ALL1 was replaced with a neomycin resistance marker (2098bp) by homologous recombination using standard procedure described earlier (Jain et al., 2009b). Homologous recombination was confirmed by PCR, southern blot analysis and transcription level of ALL1 by real time PCR.
Complementation of ALL1 gene in H99 Δall1
The wt ALL1 gene was amplified with the native promoter from the H99-wt genomic DNA with primers H99ALL1-RecF and H99ALL1-RecR (Table S4) and cloned into plasmid pJAF13. The plasmid was linearized using ApaI and introduced into H99-Δall1 cells by biolistic transformation. The ALL1-positive clones were selected on YPD agar containing 100mg L−1 NAT (Werner Bioagents, Germany). Gene complementation was confirmed by PCR and transcription level of ALL1 by real time PCR.
Construction of ALL1 Overexpression strain, PCTR4-2-ALL1
To construct RC2-SM strain overexpressing ALL1, PCTR4-2 promoter was amplified from plasmid pCTR4-2/Gust (Ory et al., 2004) and fused with ALL1 gene amplified from RC2-SM genomic DNA, in frame using restriction site NdeI. Flanking region of 1000bp upstream of ALL1 including 5′UTR, Ori/Ampr and NEO cassette was excised from previously generated TA clones using van91I digestion to generate plasmid 5′UTR/NEO/PCTR4-2-ALL1/Amp. The plasmid was used to amplify 5′UTR-NEO-PCTR4-2-ALL1 using primers stated in Table S4 and biolistically transformed in RC2-SM. Standard procedure was further adapted to screen and confirm clones as mentioned above. To overexpress ALL1, the cells were grown in minimal medium or asparagine salt medium with 200μM of bathocuproinedisulfonic acid (BCS), copper chelator. Overexpression was confirmed via checking transcription levels by real time PCR.
GXM isolation
Culture supernatants were separated from yeast cells by centrifugation at 6,000 g (20 min, 4°C). Supernatants were successively centrifuged and filtered through 0.45μm membranes. Supernatants were concentrated ≅ 20-fold using an Amicon ultrafiltration cell and regenerated cellulose 100 KDa, 76 mm ultrafiltration membrane (Millipore, Billerica, MA) with stirring. After concentration the fluid phase was discarded and the viscous film over the membrane was scraped carefully and lyophilized.
Light Scattering Analysis
Average-molecular mass (Mw) and radius of gyration (Rg) were determined by multi-angle laser (static) light scattering (BI-MwA; Brookhaven Instruments Corp.), as described (Cordero et al., 2011). Each PS sample was measured twice with good reproducibility. Average hydrodynamic radius (Rh) of PS samples was determined by Dynamic light scattering using a 90 Plus Particle Sizing analyzer (Brookhaven Instruments Corp.), as described (Cordero et al., 2011). Average Rh and multimodal size distributions were calculated from ten individual measurements.
Viscosity
Intrinsic viscosity of the exo-PS samples isolated from H99 and RC2 and their respective mutants cultures supernatants were measured using a modified Ostwald-type capillary glass viscometer (Cannon-Manning Semi-Micro, Technical Glass Co. Dover, NJ) in a temperature-controlled water bath at 25°C, as previously described (Cordero et al., 2011). Exo-PS samples were dissolved in ultrapure water and temperature-stabilized before measurements. Measurements were done in triplicate.
GXM ELISA
ELISA were done as described previously (Casadevall et al., 1992). Briefly, a 96-well polystyrene plate was coated with IgM (1μg/ml) at 4°C overnight. Unbound sites are blocked with 1% BSA for 2hrs at room temperature (RT). Anti-GXM mAb 2D10, 13F1 or 12A1 (100μl of 1μg/ml) was added to the plate and incubated for 1hr at RT. Wells were washed with TBS-T 3 times (3X) and incubated with isolated exo-PS [10–0.740nM, diluted in PBS (obtained by 1:3 serial dilutions vertically)] either from wt (SM, H99) or their respective Δall1 mutant. After washed with TBS-T (3X), wells were incubated with 100μl of 1μg/ml mAb18B7 diluted in PBS ranging from 1–0.0000016 μg/ml (obtained by 1:3 serial dilutions horizontally), followed by detection with 1:1000 dilution of anti-IgG1-AP antibody. All samples were done in triplicates and all incubations were done for 1h at 37°C. For GXM shed experiment, the supernatants were collected at days 1, 3, 6, 12, 21 of cultivation and ELISA was performed using above-mentioned procedure (used mAb 2D10) to quantitate GXM concentration.
Complement mediated Phagocytosis Assay
Approximately 2.5 × 104 J774 macrophage-like cells/well (ATCC) were plated in 96-well plates containing Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS) at 37°C in a 5% CO2 atmosphere and under the stimulation of recombinant murine γ-interferon and LPS; 100U/mL and 0.5 μg/ml, respectively. C. neoformans cells were washed and suspended in DMEM supplemented with 20% mouse serum (Pel-Freez Biologicals, Rogers, AR) and added to macrophages to generate a ratio of 5 yeast cells per macrophage. For some experiments, 5 μg of exo-PS from wt, Δall1 and Δall1 + ALL1 strains were added to the wt wells to test the inhibitory capacity of different PSs. Mixture was incubated at 37°C and 5% CO2 for 2h. After incubation, wells were washed with PBS to remove non-adherent yeast cells. Cells were then fixed with ice-cold methanol for 30 min and stained with 1:5 solution of Giemsa stain in distilled water. The percentage of phagocytosis was determined by microscopic examination of macrophages with internalized C. neoformans cells divided by total macrophages. At least 300 macrophages cells were examined per well and each condition was done in triplicates.
Animal Studies
Balb/c mice (Female, 6–8 weeks) were obtained from the National Cancer Institute (Bethesda, MD). Mice (n = 10 per group) were infected intratracheal (i.t) with 106 or 105 C. neoformans H99-wt, H99-Δall1 and H99 Δall1 + ALL1 cells that were grown overnight in SAB medium. Mice were observed on a daily basis for survival and sacrificed if they were moribund in accordance with IACUC regulations.
RNA Purification and quantitative Real Time PCR Assays
For microarray, RC2-SM and RC2-Δall1 were grown in two different culture medium conditions including minimal media (same media which was used for exo-PS isolation) and YPD broth. Cells were grown for overnight, diluted 1:100 and then again grown overnight or up to an OD of 0.6–0.7 with agitation (150 rpm) at 37°C. Approximately, 108 cells were collected and mechanically disrupted by bead beating in RLT buffer (Qiagen) using 0.5mm zirconia beads for 1 min, for a total of 4 cycles with 2 min rest on ice in between cycles. Following lysis, total RNA was isolated using QIAGEN® RN easy mini kit, according to the manufacture’s instructions. For real time PCR, RNA was further cleaned up for residual DNA using MessageClean Kit (genHunter corp.) and cDNA was synthesized using First strand superscript II RT kit (Life technologies) with 1μg total RNA according to manufacturer’s instruction. RNA was also isolated from H99 and H99-Δall1. Relative expression of selective genes was measured by real time PCR using power SYBR green (applied biosystem) in ABI 384 system and primers listed in Table S4. Gene expression levels were normalized using the control gene β-actin. Primer efficiency was determined by serially diluting the cDNA and monitoring DNA amplification by real-time PCR. The relative transcript levels were determined using the delta-delta CT method.
To investigate the link between exo-PS structure and regulation of Iron- related genes, H99-wt and H99-Δall1 cells were grown for 16h in minimal media supplemented with 40μg/ml exo-PS of H99-Δall1 and H99-wt respectively. Specifically for control, H99-wt cells were grown with 40μg/ml exo-PS of H99-wtand H99-Δall1 cells were grown with H99-Δall1 exo-PS for 16h with shaking at, 37°C. Total RNA was isolated and real time PCR was done on 13 genes differentially expressed in microarray analysis related to iron-transport i.e., FTR1, FRE1, CNM02430, CNM02420, CNB02540, CNG00120, CNH01230, CNG00110, CNG00950, CNC01660, CNK02840, CNB00400, CNI01980 and β-actin as control. The mRNA levels were normalized against their respective β-actin and expression was calculated using the delta-delta CT method. The fold change was calculated as mRNA ratio.
Microarray Hybridization
Experiments were performed in duplicate (Minimal Media arrays) ortriplicate (YPD arrays). Total RNA hybridization and data acquisition were performed by the Genome technology access center, Washington University (http://gtac.wustl.edu/services/microarray/rna-analysis/cryptococcus-neoformans.php) according to their established protocols for C. neoformans microarrays. For analysis, raw data was imported in Partek genomic suite and intensity-dependent (LOESS) normalization was implemented in the software. Values for replicate probes on the array were averaged to represent the expression of the associated gene. Genes were considered for further evaluation if they showed a fold change of > 2.0.
Gene Ontology (GO) Enrichment Analysis
For gene ontology enrichment, each gene was assigned GO category according to the Broad Institute’s PFAM annotations using the mapping provided by the Gene Ontology project (http://www.geneontology.org/external2go/pfam2go). A hypergeometric test was applied for each GO category, the resulting p-values were corrected for multiple hypothesis testing, and a cutoff of 0.05 was used to determine significance.
Determination of Cellular Iron Concentration
To determine intracellular iron concentration, the wt and the Δall1 mutants were grown in minimal media overnight and subcultured in minimal media +100μM bathophenanthrolinedisulfonic acid (BPS), iron chelator for 16h to deplete intracellular iron stores. Cells were washed twice with iron free water and grown with or without 100μM FeCl3 in low iron media for 16h. Cell cultures were washed three times with low-iron water and mixed with lysis buffer (50 mM Hepes-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA and 1% Triton × −100) as described (Choi et al., 2012). Cells were disrupted and homogenized using glass-beads in a bead beater for a total of 4 cycles with 2 min rest on ice in between cycles. Iron concentration of lysate was measured using the Abnova Iron assay kit according to the manufacturer’s instruction. Tests were run in triplicates and each sample iron concentration was normalized against total protein concentration.
Supplementary Material
Acknowledgments
This work was supported by NIH awards AI059681, AI033774, HL059842, and AI033142. RJBC was supported by the Training Program in Cellular and Molecular Biology and Genetics, T32 GM007491. We thank Dr. Goldman for critical reading of the manuscript.
References
- Adler A, Park YD, Larsen P, Nagarajan V, Wollenberg K, Qiu J, Myers TG, Williamson PR. A novel specificity protein 1 (SP1)-like gene regulating protein kinase C-1 (Pkc1)-dependent cell wall integrity and virulence factors in Cryptococcus neoformans. The Journal of biological chemistry. 2011;286:20977–20990. doi: 10.1074/jbc.M111.230268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker KS, Pearson MM, Rogers PD. Identification of genes differentially expressed in association with reduced azole susceptibility in Saccharomyces cerevisiae. J Antimicrob Chemother. 2003;51:1131–1140. doi: 10.1093/jac/dkg217. [DOI] [PubMed] [Google Scholar]
- Barluzzi R, Saleppico S, Nocentini A, Boelaert JR, Neglia R, Bistoni F, Blasi E. Iron overload exacerbates experimental meningoencephalitis by Cryptococcus neoformans. J Neuroimmunol. 2002;132:140–146. doi: 10.1016/s0165-5728(02)00324-7. [DOI] [PubMed] [Google Scholar]
- Belay T, Cherniak R, Kozel TR, Casadevall A. Reactivity patterns and epitope specificities of anti-Cryptococcus neoformans monoclonal antibodies by enzyme-linked immunosorbent assay and dot enzyme assay. Infect Immun. 1997;65:718–728. doi: 10.1128/iai.65.2.718-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicanic T, Meintjes G, Wood R, Hayes M, Rebe K, Bekker LG, Harrison T. Fungal burden, early fungicidal activity, and outcome in cryptococcal meningitis in antiretroviral-naive or antiretroviral-experienced patients treated with amphotericin B or fluconazole. Clin Infect Dis. 2007;45:76–80. doi: 10.1086/518607. [DOI] [PubMed] [Google Scholar]
- Bicanic T, Wood R, Meintjes G, Rebe K, Brouwer A, Loyse A, Bekker LG, Jaffar S, Harrison T. High-dose amphotericin B with flucytosine for the treatment of cryptococcal meningitis in HIV-infected patients: a randomized trial. Clin Infect Dis. 2008;47:123–130. doi: 10.1086/588792. [DOI] [PubMed] [Google Scholar]
- Bicanic T, Brouwer AE, Meintjes G, Rebe K, Limmathurotsakul D, Chierakul W, Teparrakkul P, Loyse A, White NJ, Wood R, Jaffar S, Harrison T. Relationship of cerebrospinal fluid pressure, fungal burden and outcome in patients with cryptococcal meningitis undergoing serial lumbar punctures. Aids. 2009;23:701–706. doi: 10.1097/QAD.0b013e32832605fe. [DOI] [PubMed] [Google Scholar]
- Burchard W, Schmidt M, Stockmayer WH. Information on Polydispersity and Branching from Combined Quasi-Elastic and Intergrated Scattering. Macromolecules. 1980;13:1265–1272. [Google Scholar]
- Casadevall A, Mukherjee J, Scharff MD. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J Immunol Methods. 1992;154:27–35. doi: 10.1016/0022-1759(92)90209-c. [DOI] [PubMed] [Google Scholar]
- Chang Y, Kwon-Chung K. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores virulence. MCB. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Chretien F, Baudrimont M, Mordelet E, Lortholary O, Dromer F. Capsule structure changes associated with Cryptococcus neoformans crossing of the blood-brain barrier. Am J Pathol. 2005;166:421–432. doi: 10.1016/S0002-9440(10)62265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniak R, Valafar H, Morris L, Valafar F. Cryptococcus neoformans Chemotyping by quantitative analysis of 1H nuclear magnetic resonance spectra of glucuronxylomannans with a computer-simulated artificial neural network. Clinical and Diagnostic Laboratory Immunology. 1998;5:146–159. doi: 10.1128/cdli.5.2.146-159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JN, Kim J, Kim J, Jung WH, Lee CH. Influence of Iron Regulation on the Metabolome of Cryptococcus neoformans. PLoS One. 2012;7:e41654. doi: 10.1371/journal.pone.0041654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero RJ, Frases S, Guimaraes AJ, Rivera J, Casadevall A. Evidence for branching in cryptococcal capsular polysaccharides and consequences on its biological activity. Mol Microbiol. 2011;79:1101–1117. doi: 10.1111/j.1365-2958.2010.07511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering TL. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu Rev Microbiol. 2009;63:223–247. doi: 10.1146/annurev.micro.62.081307.162753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromer F, Mathoulin-Pelissier S, Launay O, Lortholary O. Determinants of disease presentation and outcome during cryptococcosis: the CryptoA/D study. PLoS Med. 2007;4:e21. doi: 10.1371/journal.pmed.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca FL, Nohara LL, Cordero RJ, Frases S, Casadevall A, Almeida IC, Nimrichter L, Rodrigues ML. Immunomodulatory effects of serotype B glucuronoxylomannan from Cryptococcus gattii correlate with polysaccharide diameter. Infect Immun. 2010;78:3861–3870. doi: 10.1128/IAI.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frases S, Nimrichter L, Viana NB, Nakouzi A, Casadevall A. Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical, and antigenic differences. Eukaryot Cell. 2008;7:319–327. doi: 10.1128/EC.00378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frases S, Pontes B, Nimrichter L, Viana NB, Rodrigues ML, Casadevall A. Capsule of Cryptococcus neoformans grows by enlargement of polysaccharide molecules. Proc Natl Acad Sci U S A. 2009;106:1228–1233. doi: 10.1073/pnas.0808995106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries BC, Taborda CP, Serfass E, Casadevall A. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J Clin Invest. 2001;108:1639–1648. doi: 10.1172/JCI13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries BC, Lee SC, Kennan R, Zhao W, Casadevall A, Goldman DL. Phenotypic switching of Cryptococcus neoformans can produce variants that elicit increased intracranial pressure in a rat model of cryptococcal meningoencephalitis. Infect Immun. 2005;73:1779–1787. doi: 10.1128/IAI.73.3.1779-1787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates MA, Kozel TR. Differential localization of complement component 3 within the capsular matrix of Cryptococcus neoformans. Infect Immun. 2006;74:3096–3106. doi: 10.1128/IAI.01213-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DL, Casadevall A, Zuckier LS. Pharmacokinetics and biodistribution of a monoclonal antibody to Cryptococcus neoformans capsular polysaccharide antigen in a rat model of cryptococcal meningitis: implications for passive immunotherapy. J Med Vet Mycol. 1997;35:271–278. doi: 10.1080/02681219780001261. [DOI] [PubMed] [Google Scholar]
- Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J Clin Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill JR, Sobel J, Saag M, van Der Horst C, Powderly W, Cloud G, Riser L, Hamill R, Dismukes W. Diagnosis and management of increased intracranial pressure in patients with AIDS and cryptococcal meningitis. The NIAID Mycoses Study Group and AIDS Cooperative Treatment Groups. Clin Infect Dis. 2000;30:47–54. doi: 10.1086/313603. [DOI] [PubMed] [Google Scholar]
- Guerrero A, Fries BC. Phenotypic switching in Cryptococcus neoformans contributes to virulence by changing the immunological host response. Infect Immun. 2008;76:4322–4331. doi: 10.1128/IAI.00529-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero A, Jain N, Wang X, Fries BC. Cryptococcus neoformans variants generated by phenotypic switching differ in virulence through effects on macrophage activation. Infect Immun. 2010;78:1049–1057. doi: 10.1128/IAI.01049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes AJ, Frases S, Cordero RJ, Nimrichter L, Casadevall A, Nosanchuk JD. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell Microbiol. 2010;12:740–753. doi: 10.1111/j.1462-5822.2010.01430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BC, Skowyra ML, Spencer SJ, Gish SR, Williams M, Held EP, Brent MR, Doering TL. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 2011;7:e1002411. doi: 10.1371/journal.ppat.1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwidthaya P, Poungvarin N. Cryptococcosis in AIDS. Postgrad Med J. 2000;76:85–88. doi: 10.1136/pmj.76.892.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Cook E, Xess I, Hasan F, Fries D, Fries BC. Isolation and characterization of senescent C. neoformans and its implications for phenotypic switching and the pathogenesis of chronic cryptococcosis. Eukaryot Cell. 2009a;8:858–866. doi: 10.1128/EC.00017-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Li L, Hsueh YP, Guerrero A, Heitman J, Goldman DL, Fries BC. Loss of allergen 1 confers a hypervirulent phenotype that resembles mucoid switch variants of Cryptococcus neoformans. Infect Immun. 2009b;77:128–140. doi: 10.1128/IAI.01079-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, Kronstad JW. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambugu A, Meya DB, Rhein J, O’Brien M, Janoff EN, Ronald AR, Kamya MR, Mayanja-Kizza H, Sande MA, Bohjanen PR, Boulware DR. Outcomes of cryptococcal meningitis in Uganda before and after the availability of highly active antiretroviral therapy. Clin Infect Dis. 2008;46:1694–1701. doi: 10.1086/587667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian T, Simmer MI, D’Souza CA, Steen BR, Zuyderduyn SD, Jones SJ, Marra MA, Kronstad JW. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2005;55:1452–1472. doi: 10.1111/j.1365-2958.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- McClelland EE, Bernhardt P, Casadevall A. Coping with multiple virulence factors: which is most important? PLoS Pathog. 2005;1:e40. doi: 10.1371/journal.ppat.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DC, De Jesus M, Casadevall A. The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J Biol Chem. 2006;281:1868–1875. doi: 10.1074/jbc.M509465200. [DOI] [PubMed] [Google Scholar]
- McFadden DC, Fries BC, Wang F, Casadevall A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell. 2007;6:1464–1473. doi: 10.1128/EC.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimrichter L, Frases S, Cinelli LP, Viana NB, Nakouzi A, Travassos LR, Casadevall A, Rodrigues ML. Self-aggregation of Cryptococcus neoformans capsular glucuronoxylomannan is dependent on divalent cations. Eukaryot Cell. 2007;6:1400–1410. doi: 10.1128/EC.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyhus KJ, Wilborn AT, Jacobson ES. Ferric iron reduction by Cryptococcus neoformans. Infect Immun. 1997;65:434–438. doi: 10.1128/iai.65.2.434-438.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DL, Nimrichter L, Miranda K, Frases S, Faull KF, Casadevall A, Rodrigues ML. Cryptococcus neoformans cryoultramicrotomy and vesicle fractionation reveals an intimate association between membrane lipids and glucuronoxylomannan. Fungal genetics and biology : FG & B. 2009;46:956–963. doi: 10.1016/j.fgb.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory JJ, Griffith CL, Doering TL. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast. 2004;21:919–926. doi: 10.1002/yea.1139. [DOI] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am J Pathol. 1980;101:177–194. [PMC free article] [PubMed] [Google Scholar]
- Pirofski LA, Casadevall A. The damage-response framework of microbial pathogenesis and infectious diseases. Adv Exp Med Biol. 2008;635:135–146. doi: 10.1007/978-0-387-09550-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, Alvarez M, Nakouzi A, Feldmesser M, Casadevall A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, Almeida IC, Casadevall A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton RA, Naraqi S, Wembri JP, Warrell DA. Predictors of outcome in Cryptococcus neoformans var. gattii meningitis. QJM. 1996;89:423–428. doi: 10.1093/qjmed/89.6.423. [DOI] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. Regulation of Cryptococcus neoformans capsule size is mediated at the polymer level. Eukaryot Cell. 2008;7:546–549. doi: 10.1128/EC.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol. 2009;68:133–216. doi: 10.1016/S0065-2164(09)01204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


