Abstract
Fibroblast growth factor (FGF) signaling plays an important role in embryonic stem cells and adult tissue homeostasis, but the function of FGFs in mammary gland stem cells is less well defined. Both FGFR1 and FGFR2 are expressed in basal and luminal mammary epithelial cells (MECs), suggesting that together they might play a role in mammary gland development and stem cell dynamics. Previous studies have demonstrated that the deletion of FGFR2 resulted only in transient developmental defects in branching morphogenesis. Using a conditional deletion strategy, we investigated the consequences of FGFR1 deletion alone and then the simultaneous deletion of both FGFR1 and FGFR2 in the mammary epithelium. FGFR1 deletion using a keratin 14 promoter-driven Cre-recombinase resulted in an early, yet transient delay in development. However, no reduction in functional outgrowth potential was observed following limiting dilution transplantation analysis. In contrast, a significant reduction in outgrowth potential was observed upon the deletion of both FGFR1 and FGFR2 in MECs using adenovirus-Cre. Additionally, using a fluorescent reporter mouse model to monitor Cre-mediated recombination, we observed a competitive disadvantage following transplantation of both FGFR1/R2-null MECs, most prominently in the basal epithelial cells. This correlated with the complete loss of the mammary stem cell repopulating population in the FGFR1/R2-attenuated epithelium. FGFR1/R2-null MECs were partially rescued in chimeric outgrowths containing wild-type MECs, suggesting the potential importance of paracrine mechanisms involved in the maintenance of the basal epithelial stem cells. These studies document the requirement for functional FGFR signaling in mammary stem cells during development.
Keywords: Fibroblast growth factor receptors, Mammary gland, Stem cells, Limiting dilution analysis
Introduction
The fibroblast growth factor (FGF) signaling pathway consists of a highly conserved family of four single-pass membrane receptor tyrosine kinases (RTK), FGFR1–4, and at least 22 secreted FGF ligands. Each FGF ligand can selectively bind to and activate one or more of the four FGF receptors, making the pathway highly complex and often redundant [1]. Both FGFR1 and FGFR2 are expressed in the developing mammary bud and are the primary FGF receptors expressed in the mouse mammary epithelium during ductal morphogenesis [2, 3].
The importance of intact FGFR2 signaling during mammary placode and bud formation has been demonstrated in FGFR2iiib null mice. These mice fail to develop mammary placodes 1, 2, and 3 [4, 5]. Interestingly, deletion of FGFR2iiib-activating ligands FGF7 and FGF10 mirrors many of the same effects of FGFR2iiib loss. A critical role for FGFR signaling in the induction of mammary bud formation through FGF-dependent activation of Tbx3 and Lef1 expression has also been reported. Thus, FGF-Tbx3 and Wnt pathway cooperation are required for embryonic mammary gland development, suggesting a potential role for FGF signaling in mammary stem-progenitor cell functionality [6].
Postnatal deletion of FGFR2 has also recently been observed to transiently attenuate mammary ductal morphogenesis. Postnatal conditional deletion of FGFR2iiib resulted in a partial reduction in mammary outgrowth [7] and led to the complete loss of terminal end buds (TEBs) in the developing gland as well as an increase in apoptosis. Similar results were reported using a genetic mosaic analysis approach [3]. A competitive outgrowth of a minority of unrecombined cells with intact FGFR2 as compared to FGFR2− null mammary epithelial cells (MECs) was observed. These results demonstrated the selective proliferative advantage of intact FGFR2 signaling within the developing epithelium.
While both FGFR1 and FGFR2 are expressed in the TEBs during branching morphogenesis [3], the role of FGFR1 signaling in the developing mammary gland is not well understood. Because of the lack of appropriate immunological reagents, it is unknown whether these receptors are expressed in the same cells. The only study on the developmental effects of FGFR1 ablation on mammary development used a dominant negative isoform of FGFR1iiic driven by the MMTV promoter [8]. Dominant negative MMTV-(DN)FGFR1iiic mice did not display any detectable differences in lobuloalveolar development during pregnancy and lactation in contrast to mice expressing a dominant negative MMTV-(DN)FGFR2iiib construct that displayed impaired lobuloalveolar development.
In order to investigate the role of FGFR1 in normal mammary gland development, we have used a conditional deletion strategy. FGFR1 deletion, prenatally, resulted in a delay of mammary gland development, including a transient reduction in cellular proliferation. Additionally, while limiting dilution transplantation analysis did not reveal a requirement for functional FGFR1 in mammary fat pad reconstitution, simultaneous deletion of FGFR1 and FGFR2 led to a marked attenuation of MEC engraftment and outgrowth potential. Interestingly, this reduction in outgrowth potential also correlated with the loss of the mammary stem cell (MaSC) population. These studies demonstrate the requirement for functional FGFR signaling for the maintenance of mammary stem cells and for normal mammary gland development.
Materials and methods
Animal Breeding and Maintenance
Previously characterized, FGFR1 floxed mice were back-crossed to a C57BL/6 background expressing the Rosa 26 LacZ Reporter (R26R) construct [9–11] and then bred with mice expressing Cre-recombinase under the Keratin 14 (K14) promoter [12]. FGFR1/R2 double floxed mice maintained on an FVB/C57BL/6 background [9, 13] were generated by crossing previously generated FGFR1 and FGFR2 floxed mice [10, 14]. Both FGFR1 floxed and FGFR1/R2 double floxed mice were also crossed to Rosa26 fluorescent reporter mT-mG reporter line (mT-mG) [14]. All animals were maintained in accordance with the NIH Guide for the Care and Use of Experimental Animals with approval from the Baylor College of Medicine Institutional Animal Care and Use Committee.
Immunohistochemistry and Quantification
Immunohistochemistry was conducted according to manufacturers’ protocol using the antibodies listed in Supporting Information Table S1 as previously described [15]. Brightness and contrast of images were adjusted equally across both control and experimental groups using Adobe Photoshop CS3 Extended software version 10.0, San Jose, CA.
Quantification of BrdU incorporation was obtained through the acquisition of three random images of mammary epithelium, specifically in TEBs for each individual genotype/time point (anti-BrdU, BD Pharmingen, Franklin Lakes, NJ) (3 weeks, n = 3 for each genotype, 5 weeks, n = 3 for each genotype, 7 weeks, n = 3 for each genotype). Positive nuclear staining was then quantified as described previously [16].
RNA Isolation and Quantitative Reverse Transcription-PCR
Ad-Cre-transduced primary MECs were grown in two-dimensional culture for 10 days in order to determine the extent of recombination and deletion of FGFR1 and FGFR2. Cells for transplantation were never cultured on plastic to prevent loss of MaSCs and subsequent differentiation. RNA was collected through extraction with Trizol reagent (Invitrogen, Life Technologies, Carlsbad, CA) and cDNA templates were generated using a SuperScript III kit and 1 μg RNA per sample according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). qPCR reactions were run using SYBR Green reagent (Applied Biosystems, Carlsbad, CA) on a StepOnePlus thermocycler (Applied Biosystems, Life Technologies, Carlsbad, CA) and fold changes were calculated using the Comparative CT (ΔΔCT) method and Step One software v2.0.1, Applied Biosystems, Life Technologies, Carlsbad, CA. Primers were designed specifically to exon 8 of both FGFR1 and FGFR2, targeting the iiib isoforms. The primer sequences for FGFR1 are: forward 5′-CCAAGAAGAGCGACTTCCAT-3′; reverse 5′-CATGGATGCACTGGAGTCAG-3′ and for FGFR2 are: forward 5′-CCTGCGGAGACAGGTAACAG-3′; reverse 5′-CGCGTTGTTATCCTCACCA-3′. Prior to analysis within sorted luminal and basal populations from the adult virgin mammary gland, RNA was amplified by the BCM Genomic and RNA Profiling Core using the NuGen WT-Ovation Kit (NuGen, San Carlos, CA).
Adenovirus Transduction and Limiting Dilution Transplantation
Primary MECs isolated from adult female virgin mice of >10 weeks of age as previously described [17] were incubated in suspension culture with adenovirus-CMV-Cre(Ad-Cre) recombinase or adenovirus-CMV-LacZ(Ad-LacZ) viral vector [18] at an multiplicity of infection of 100 in 3 ml of serum-free Dulbecco’s modified Eagle’s medium/F12 for 1 hour at 37°C. Cells were then collected through centrifugation at 300g, washed, and resuspended in appropriate number in 1:1 phosphate buffered saline/Matrigel Basement Membrane Matrix (BD Biosciences, Franklin Lakes, NJ) with individual injection volumes of 10 μl. Cells were then immediately injected into the #4 inguinal mammary gland of 3-week-old C57BL/6 or Scid/Beige female mice (Charles River, Wilmington, NJ) following removal of endogenous gland. All experimental and control groups were transplanted into contralateral cleared fat pads of individual mice. For limiting dilution analysis (LDA), outgrowth was allowed for 8 weeks.
Wholemount Analysis and Quantification
Wholemounts and X-gal staining of β-galactosidase-positive epithelium were carried out as described previously [19]. TEBs and branch points were quantitated by manually counting high magnification wholemount images, tiling the entire gland for each time point as described previously [20]. Ductal penetration was measured as the distance of the leading epithelium from the lymph node [3], and wholemount analysis of mT-mE fluorescent glands was carried out as previously described [19].
Fluorescence-Activated Cell Sorting
Fluorescence-activated cell sorting (FACS) analysis was conducted as previously described [17]. Single cells were counted and lineage-reduced through magnetic bead separation using antibodies listed in Supporting Information Table S1. Single cells were resuspended in Hanks’ balanced saline solution (HBSS) 2% fetal bovine serum (FBS) and incubated with primary antibodies (diluted 1:100). Cells were washed with HBSS 2% FBS and analyzed on a LSRII analyzer (BD Biosciences, Franklin Lakes, NJ). Analysis was conducted using FlowJo8.7.3 (Tree Star). For FACS of luminal and basal mammary epithelial populations, 8–10 weeks virgin mammary glands were isolated as described above and resuspended at a concentration of 1 × 108 cells per milliliter in HBSS supplemented with 10 mM HEPES and 2% FBS. Lineage-positive cells (CD45, Ter119, CD31, and BP-1) were removed using the EasySep Mouse Epithelial Cell Enrichment Kit (Stem Cell Technologies, Vancouver, BC, Canada). MECs were subsequently resuspended at a density of 1 × 107 cells per milliliter and stained with anti-mouse CD24 PE (Stem Cell Technologies, Vancouver, BC, Canada 1:100) and anti-CD49f FITC (Stem Cell Technologies, 1:100), and sorted using a BD FACS Aria Cell Sorter.
Statistical Analysis
Quantification of wholemounts, qPCR, immunohistochemistry, and flow cytometry are presented as mean ± SEM, and genotype groups were compared using Student’s t tests, with p ≤ .05 being deemed statistically significant. t tests and graphical representation of data were performed using GraphPad Prism version 4.0c GraphPad Software, San Diego, CA. LDA was performed using binomial generalized linear modeling with the complementary log-log link [21, 22]. Fat pad data are counts of fat pad exhibiting various degrees of outgrowth filing (i.e., 0%, 10%, 25%, 50%, 75%, and 100%). Cochran-Mantel-Haenszel statistics were used to test for an ordinal difference in fat pad filling between genotypes, after controlling for cell dose. In the analysis, fat pads with 0% filling are omitted, in order to compare filling when growth occurs. CMH ANOVA analysis was carried out using SAS 9.3 (Carry NC). Transplantation experiments were analyzed using the R statistical software [23].
Results
Embryonic Deletion of FGFR1 Delays Normal Mammary Gland Development
To investigate the role of FGFR1, a conditional deletion strategy was used by generating mice expressing a previously characterized FGFR1 floxed allele (FGFR1fl/fl), the Rosa 26 lacZ reporter (R26R) as well as Cre-recombinase under expression of the keratin 14 promoter (K14-Cre) [10–12]. K14 is expressed in the skin as well as the placodes, which originate from the skin, and has been observed in the myoepi-thelial and luminal cells of the developing mammary epithelium. Additionally, it is believed that some MaSCs are K14+ [12, 24, 25]. To investigate the activity and efficiency of the K14-Cre, mammary buds were harvested from K14-Cre; R26R mice at embryonic day 18 (E18). Wholemount X-gal staining demonstrated extensive β-galactosidase activity, indicating Cre-mediated recombination within the skin and early primitive ductal tree (Supporting Information Fig. S1, arrow).
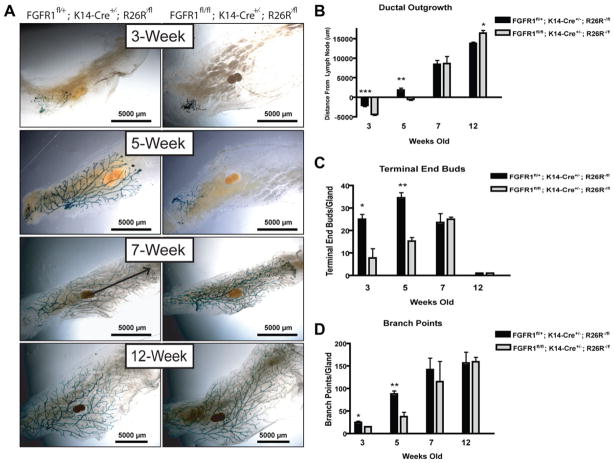
FGFR1+/fl; K14-Cre+/−; R26R−/fl and FGFR1fl/fl; K14-Cre+/−; R26R−/fl mice were generated and mammary gland development was monitored at 3, 5, 7, and 12 weeks of age. FGFR1fl/fl; K14-Cre+/−; R26R−/fl mammary glands showed a significant reduction in ductal outgrowth as compared to heterozygous controls (Fig. 1A, 1B). Additionally, a significant decrease in TEBs and branch points was observed in FGFR1fl/fl; K14-Cre+/−; R26R−/fl mammary glands (Fig. 1C, 1D). This developmental delay was no longer observed at 7 weeks of age and the two groups were indistinguishable from 7 to 12 weeks. K14-Cre-mediated recombination as evidenced by β-galactosidase activity was maintained throughout 12 weeks of development in both groups (Supporting Information Fig. S2).
Figure 1.
Embryonic deletion of FGFR1 transiently attenuates normal mammary gland development. (A): FGFR1+/fl; K14-cre+/−; R26R−/fl and FGFR1fl/fl; K14-cre+/−; R26R−/fl mammary glands were removed at 3, 5, 7, and 12 weeks of age and X-gal stained to demonstrate efficient K14-Cre activity. FGFR1fl/fl; K14-cre+/−; R26R−/fl glands showed a developmental delay from 3 to 5 weeks as compared to FGFR1+/fl; K14-cre+/−; R26R−/fl glands. This developmental delay was no longer observed by 7–12 weeks of age. (B): Ductal outgrowth was measured from the lymph node (LN) (Fig. 1A, arrow). A reduction in ductal outgrowth from the LN was observed at 3 and 5 weeks of development and eventually recovered by 7 weeks (*, p < .04; **, p < .01; ***, p < .001). (C): K14-Cre-mediated deletion of FGFR1 reduces terminal end buds in the developing mammary gland from 3 to 5 weeks of age and is recovered by 7–12 weeks. (*, p < .007; **, p < .001). (D): K14-Cre-mediated deletion of FGFR1 reduces branching in the developing mammary gland from 3 to 5 weeks of age (*, p < .01; **, p < .04). All graphs represent mean ± SEM.
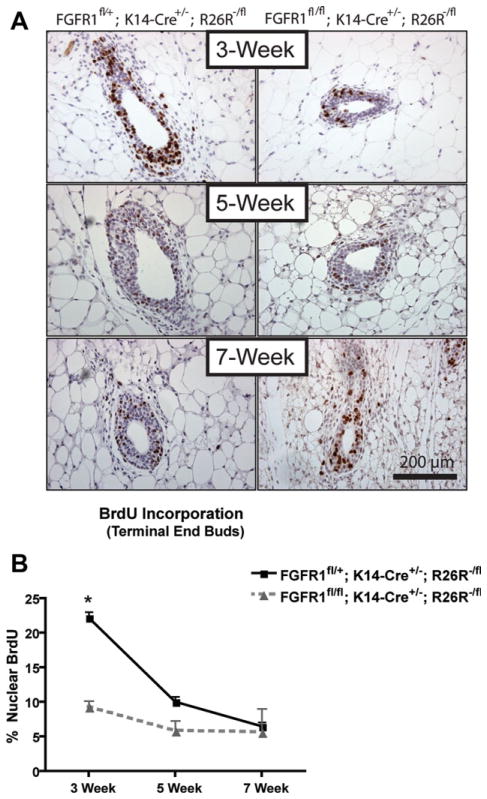
In agreement with the observed early developmental defects, the FGFR1fl/fl; K14-Cre+/−; R26R−/fl mammary glands showed decreased BrdU incorporation in the TEBs at 3 weeks of age as compared to FGFR1+/fl; K14-Cre+/−; R26R−/fl glands (Fig. 2A, 2B). This difference in proliferation was absent by 5–7 weeks and remained unchanged at 12 weeks of age, correlating with the time points where ductal outgrowth, TEB number, and branch points became indistinguishable between the two groups. These findings indicate that FGFR1 plays a significant role in driving the cellular proliferation necessary for normal mammary gland development, but that this requirement was transient.
Figure 2.
Conditional deletion of FGFR1 reduces proliferation in terminal end buds (TEBs) during early mammary gland development. (A): FGFR1+/fl; K14-cre+/−; R26R−/fl and FGFR1fl/fl; K14-cre+/−; R26R−/fl mice were injected with BrdU for 2 hours prior to sacrifice. Immunohistochemistry for BrdU incorporation on histological sections of FGFR1+/fl; K14-cre+/−; R26R−/fl and FGFR1fl/fl; K14-cre+/−; R26R−/fl demonstrates a reduction in cellular proliferation in TEBs at 3 weeks of age. All wholemounts were H&E counterstained. This difference is reduced and lost from 5 to 7 weeks of age. (B): Quantification of BrdU incorporation in developing FGFR1+/fl; K14-cre+/−; R26R−/fl and FGFR1fl/fl; K14-cre+/−; R26R−/fl TEBs (*, p < .004). Graph represents mean ± SEM.
Conditional Deletion of FGFR1 Has a Minimal Effect on Take Rate or Functional Outgrowth Upon LDA
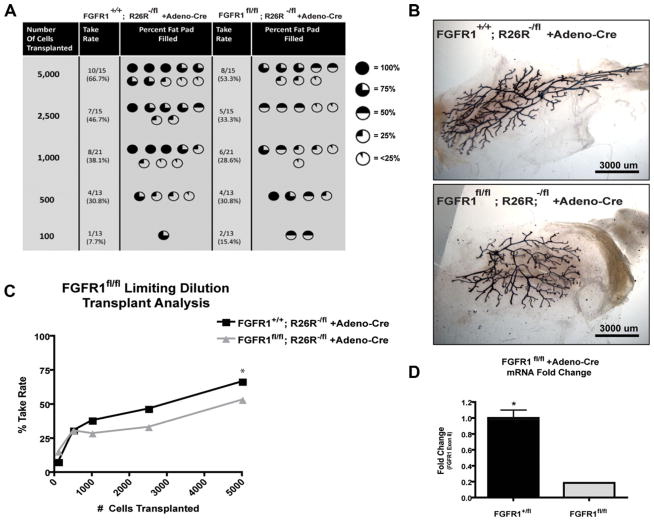
LDA has been extensively used to determine mammary stem cell (MaSC) repopulation activity and normal developmental processes under many experimental conditions [17, 26–28]. In order to investigate the potential role of FGFR1 on stem/progenitor cell activity, LDA was performed upon conditional deletion of FGFR1. Accordingly, FGFR1+/+; R26R−/fl and FGFR1fl/fl; R26R−/fl mice were generated, and primary MECs were obtained as described previously [17, 18]. MECs were transduced with Ad-Cre, cultured in vitro, and mRNA was isolated to verify FGFR1 deletion. Ad-Cre-treated FGFR1fl/fl epithelial cells had a fivefold reduction in FGFR1 mRNA, indicating efficient recombination and deletion of FGFR1 (Fig. 3D). Immediately following transduction, MECs were also transplanted at limiting dilution into contralateral cleared fat pads of 3-week-old recipient mice. Following 8 weeks of growth, the transplanted glands were analyzed for take rate and percent fat pad filled (Fig. 3). A statistically significant difference in outgrowth potential in FGFR1-deleted mammary glands was not observed at the various cell dilutions (Fig. 3A, 3C). Analysis of percentage of fat pad filled revealed a slight but not significant (p = .07) reduction in total mammary gland outgrowth (Fig. 3A). Wholemount analysis of X-gal stained mammary glands demonstrated positive lacZ staining (blue) throughout the reconstituted gland suggesting that there was efficient recombination following Ad-Cre transduction and that recombined cells were still able to fill the fat pad (Fig. 3B).
Figure 3.
FGFR1 deletion does not inhibit transplantation or mammary outgrowth upon limiting dilution transplantation analysis. (A, C): FGFR1+/+; R26R−/fl and FGFR1fl/fl; R26R−/fl Ad-Cre-transduced mammary epithelial cells were transplanted in limiting dilution from 5,000, 2,500, 1,000, 500, and 100 cells. Take rate (*, p = .338, binomial generalized linear model) and ductal outgrowth (p = .07, CMH ANOVA) were not significantly affected by FGFR1 deletion (C: Whiskers show exact binomial 95% confidence intervals). (B): X-gal stained representative wholemounts of FGFR1+/+; R26R−/fl and FGFR1fl/fl; R26R−/fl outgrowths. (D): FGFR1 fl/+ and FGFR1fl/fl primary epithelial cells transduced with Ad-Cre were allowed to grow in two-dimensional culture for ~ 10 days. RT-qPCR analysis of FGFR1fl/+ and FGFR1fl/fl cell lysates was conducted to determine FGFR1 mRNA loss in FGFR1fl/fl cultures, n = 3; *, p < .002). Graphs represent mean ± SEM. Abbreviation: FGFR, fibroblast growth factor receptor.
Simultaneous Deletion of FGFR1 and FGFR2 Reduces Ductal Outgrowth Upon Limiting Dilution Transplantation Analysis
The similar phenotypes between our studies of prenatal FGFR1 deletion through K14-Cre-mediated recombination, and previously published studies on the postnatal deletion of FGFR2, suggested that these two FGF receptors might perform overlapping roles during mammary gland development. Note that similar studies with FGFR2 floxed mice could not be performed with K14-Cre because of the reported neonatal lethality [29]. To determine whether both FGFR1 and FGFR2 are required for mammary stem cell function, we obtained FGFR1/R2 double floxed mice and conducted limiting dilution transplantation analysis in comparison to deletion of FGFR1 alone [30]. FGFR1fl/fl and FGFR1fl/fl/R2fl/fl mice were generated and primary MECs were isolated and transduced with Ad-Cre. Again the transplantation approach was required because of neonatal lethality observed upon deletion of FGFR2 using K14-Cre. Furthermore, Ad-Cre has been shown previously to transduce MECs including MaSCs. Transduced MECs were transplanted into contralateral cleared fat pads of 3-week-old recipient immunocompromised SCID/Beige mice in limiting dilution and analyzed 8 weeks later. The LDA results are reported as both the take rate, that is, the number of outgrowths observed as well as the extent to which the outgrowths filled the mammary fat pad at each dilution. Interestingly, FGFR1fl/fl/R2fl/fl MECs demonstrated a significant reduction in take rate as compared to FGFR1fl/fl Ad-Cre-transduced MECs with approximately a 2.4-fold decrease (p = .002) in mammary repopulating units (MRUs), suggesting a decrease in MaSC repopulating activity (Fig. 4A, 4C). The fat pad filled percentage was not statistically significant between the two groups (p = .40) with representative outgrowths shown in Figure 4B. As previously observed, qPCR analysis of mRNA isolated from cultured Ad-Cre-transduced MECs verified successful deletion of both FGFR1 as well as FGFR2 (Fig. 4D).
Figure 4.
Deletion of both FGFR1 and FGFR2 inhibits transplantation and mammary outgrowth. (A, C): FGFR1fl/fl and FGFR1fl/fl/R2fl/fl Ad-Cre-transduced mammary epithelial cells were transplanted in limiting dilution from 5,000, 2,500, 1,000, 500, and 100 cells. Take rate (*, p = .002, binomial generalized linear model) was reduced (mammary repopulating unit decreased 2.4-fold) in FGFR1fl/fl/R2fl/fl as compared to FGFR1fl/fl while ductal outgrowth did not differ by genotype among fat pads with any growth (p = .40, CMH ANOVA) (C: Whiskers show exact binomial 95% confidence intervals). (B): Neutral red-stained representative wholemounts of FGFR1fl/fl and FGFR1fl/fl/R2fl/fl-transduced mammary epithelial outgrowths. (D): FGFR1fl/+/R2fl/+ and FGFR1fl/fl/R2fl/fl primary epithelial cells, transduced with Ad-Cre were allowed to grow in two-dimensional culture for ~ 10 days. RT-qPCR analysis of cell lysates was conducted to determine FGFR1 and FGFR2 mRNA loss in FGFR1fl/fl/R2fl/fl cultures. (FGFR1: n = 3; **, p < .0001). (FGFR2: n = 3; *, p < .002). Graphs represent mean ± SEM.
Functional FGFR Signaling Is Required in the Mammary Epithelium During Mammary Gland Development
Deletion of both FGFR1 and FGFR2 simultaneously led to a significant reduction in ductal outgrowth as compared to FGFR1 deletion alone. However, incomplete transduction or inefficient recombination of some FGFR1fl/fl/R2fl/fl cells may allow for unrecombined wild-type “escapers,” to contribute to the outgrowth observed in the FGFR1fl/fl/R2fl/fl LDA studies. This escaper hypothesis has been described previously for FGFR2 deletion alone [3]. If this were the case, the successful outgrowth of transplanted FGFR1/R2-deleted cells previously observed may be due to these wild-type cells exclusively, demonstrating a complete competitive advantage for functional FGFR retention during in vivo mammary gland development. To investigate this possibility, FGFR1fl/fl/R2fl/fl mice were bred to double fluorescent reporter mice, Rosa mT-mG (mT-mG) [14], in which tomato red protein is expressed in the absence of Cre-recombinase. However, upon Cre-mediated recombination, tomato red protein expression is silenced and enhanced green fluorescent protein (eGFP) is expressed (Fig. 5A). The utilization of these mice allows for the identification of both wild-type and recombined cell populations in chimeric outgrowths that have been subjected to Cre-mediated excision [20]. To determine the competitive disadvantage of loss of FGFR1 on mammary gland outgrowth, FGFR1fl/fl; mT-mG fl/+ MECs were transduced with either Ad-Cre or Ad-βgal (control vector) and transplanted at equal numbers (2,500 cells) into the cleared fat pads of recipient mice (Fig. 5B). Interestingly, we observed a similar outgrowth potential between Ad-βgal-transduced FGFR1fl/fl; mT-mG fl/+ (Fig. 5B, red) and Ad-Cre-transduced FGFR1fl/fl; mT-mGfl/+ (Fig. 5B, green) in almost all transplants observed. These results indicate that loss of FGFR1 alone was not sufficient to dramatically reduce outgrowth potential during transplantation.
Figure 5.
Competitive mammary epithelial outgrowth analysis reveals an absolute requirement for both FGFR1 and FGFR2, but not FGFR1 alone. (A): Schematic diagram demonstrating the functionality of the mT-mG fluorescent reporter construct (adapted from Muzumdar et al.). (B): FGFR1fl/fl; mT-mGfl primary mammary epithelial cells (MECs) were transduced with Ad-Cre or Ad-LacZ control and a 50:50 (2,500 Ad-Cre and 2,500 Ad-LacZ) mix was transplanted and allowed to grow out. Recombined (green) and unrecombined (red) cells contributed equally to ductal outgrowths. (C): FGFR1fl/fl/R2fl/fl; mT-mG fl primary MECs were transduced with Ad-Cre or the Ad-LacZ control and a 50:50 (2,500 Ad-Cre and 2,500 Ad-LacZ) mix was transplanted and evaluated after 8 weeks. Recombined (green) cells did not contribute significantly to the ductal outgrowths and almost all were composed of unrecombined (red) cells exclusively. (D): FGFR1fl/fl; mT-mGfl MECs were transduced with Ad-Cre, transplanted, and evaluated after 8 weeks. All outgrowths were composed of 100% recombined (green) epithelium. (E): FGFR1fl/fl/R2fl/fl; mT-mGfl MECs were transduced with Ad-Cre, transplanted, and evaluated after 8 weeks. All outgrowths were composed of either unrecombined (red) epithelium or a combination of recombined (green) and unrecombined (red) epithelium. Abbreviations: FGFR, fibroblast growth factor receptor; GFP, green fluorescent protein; RFP, tomato red fluorescent protein.
To investigate the necessity of functional retention of both FGFR1 and FGFR2 during postnatal mammary gland development, MECs isolated from FGFR1fl/fl/R2fl/fl; mT-mGfl/+ mice were transduced with Ad-Cre or Ad-βgal (control), and equal numbers of cells (2,500 cells) were transplanted into the cleared fat pads of recipient mice. In contrast to FGFR1 deletion alone, extensive outgrowth was observed in the unrecombined, tomato red+ outgrowths (Fig. 5C, red), while no outgrowth of the Ad-Cre-transduced eGFP+ recombined cells occurred (Fig. 5C, green), indicating the complete loss of mammary outgrowth potential upon deletion of both FGFR1 and FGFR2. These results indicate that FGFR1/R2 attenuated outgrowths are composed exclusively of cells that have failed to efficiently recombine and delete either FGFR1, FGFR2 or both receptors.
To test the hypothesis that only cells that were not efficiently transduced were contributing to ductal outgrowth upon transplantation of Ad-Cre-treated FGFR1fl/fl/R2fl/fl; mT-mGfl/+ MECs, transplantation experiments were performed using a reduced number (1,000 cells) of FGFR1fl/fl; mT-mGfl/+ and FGFR1fl/fl/R2fl/fl; mT-mGfl/+ donor cells following only Ad-Cre transduction. As observed with the R26R reporter construct, FGFR1fl/fl; mT-mGfl/+ epithelium was completely tolerated, and 100% of the outgrowths was eGFP+ and tomato red−, indicating a lack of effect for FGFR1 loss alone (Fig. 5D, green). Interestingly, transplantation of Ad-Cre-transduced FGFR1fl/fl/R2fl/fl; mT-mGfl/+ MECs resulted in no outgrowths with recombined (eGFP+) epithelium, in the absence of a supporting population of nonrecombined (tomato red+) cells (Fig. 5E, green and red). All mammary glands observed within this group were either exclusively tomato red+ or a chimeric population of tomato red+ and eGFP+ cells (Fig. 5E, merge). These results suggest that rescue of recombined FGFR1/2-null cells with wild-type cells may occur potentially by a paracrine mechanism.
FGFR1fl/fl/R2fl/fl Mammary Epithelium Lacks the Lin−CD24+/CD29hi MaSC Population
LDA experiments demonstrated a significant decrease in functional MRUs in vivo upon FGFR1/R2 deletion. However, because of the results observed by analyzing chimeric outgrowths and the potential for escaper cells to contribute to the outgrowths, we speculated that this decrease may have under-represented the effects of FGFR1/2 deletion on MaSCs. Before we investigated this, we analyzed the relative expression levels of FGFR1 and FGFR2 in FACS sorted and validated basal and luminal epithelial cell populations by qPCR (Supporting Information Fig. S3A, S3B). These results demonstrate that FGFR1 and FGFR2 are expressed in both basal and luminal cells, with a higher level of expression of both receptors observed in the luminal subpopulation (Supporting Information Fig. S3C). Because of the lack of specific antibodies, it is not possible, however, to determine whether both receptors are expressed in the same cells by immunohisto-chemical or fluorescent staining.
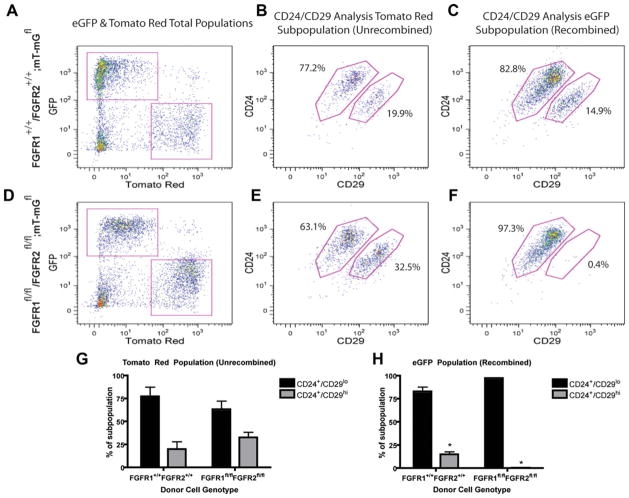
Next, FACS analysis of well-established mammary stem/progenitor cell populations [17] was performed. Five thousand Ad-Cre-transduced FGFR1+/+/R2+/+; mT-mGfl and 5,000 FGFR1fl/fl/R2fl/fl; mT-mGfl MECs were transplanted into recipient mice, and stem/progenitor cell markers, CD24 and CD29, [27] were analyzed in both unrecombined (tomato red+) and recombined (eGFP+) cells (Fig. 6A, 6D). Due to the larger cell number transplanted, almost all mammary glands contained both recombined eGFP+ cells and unrecombined tomato red+ escaper cells. The Lin−CD24+CD29hi subpopulation has been well characterized and is known to be enriched for MRUs, while the Lin−CD24hiCD29lo subpopulation has limited repopulation activity [27]. As expected, the FGFR1+/+/R1+/+; mT-mGfl tomato red+ and eGFP+ populations contained similar levels of Lin−CD24+CD29hi cells, indicating no difference as a result of activation of the mT-mG reporter construct (Fig. 6B, 6C). Interestingly, within the FGFR1fl/fl/R2fl/fl; mT-mGfl cells, a marked reduction in the Lin−CD24+CD29hi sub-population was observed as compared to the FGFR1+/+/R1+/+; mT-mGfl subpopulation (Fig. 6C, 6F, 6H). This analysis revealed a significant −37-fold reduction in the recombined eGFP+, Lin−CD24+CD29hi population in the FGFR1fl/fl/R2fl/fl; mT-mGfl glands as compared to the wild-type FGFR1+/+/R1+/+; mT-mGfl glands (Fig. 6H). Importantly, this loss of MaSCs was not observed in the tomato red+, unrecombined cells between the FGFR1+/+/R1+/+; mT-mGfl and FGFR1fl/fl/R2fl/fl; mT-mGfl populations (Fig. 6B, 6E, 6G).
Figure 6.
Quantitative analysis of stem cell populations in FGFR1+/+/R2+/+; mT-mGfl and FGFR1fl/fl/R2fl/fl; mT-mGfl mammary outgrowths using flow cytometry. (A): Magnetic bead, lineage-reduced wild-type FGFR1+/+/R2+/+; mT-mGfl outgrowths were separated into mT-mG-recombined eGFP+ and mT-mG-unrecombined tomato red+ populations to be analyzed separately. (B, C): In wild-type FGFR1+/+/R2+/+; mT-mGfl mammary gland outgrowths, the mT-mG recombined, eGFP+ population and unrecombined mT-mG tomato red+ population demonstrated similar levels of MRU-containing, Lin−CD24+CD29hi cells. Percentages of each subpopulation represent the average between all runs. (D): Magnetic bead, lineage-reduced floxed FGFR1fl/fl/R2fl/fl; mT-mGfl outgrowths were separated into mT-mG-recombined eGFP+ and mT-mG-unrecombined tomato red+ populations to be analyzed separately. (E, F): In floxed FGFR1fl/fl/R2fl/fl; mT-mGfl mammary gland outgrowths, the mT-mG recombined, eGFP+ population contained dramatically reduced levels of Lin−CD24+CD29hi, MRU-containing cells as compared to the mT-mG unrecombined tomato red+ population. Percentages of each subpopulation represent the average between all runs. (G): mT-mG unrecombined, tomato red+ cells from both the wild-type FGFR1+/+/R2+/+ and floxed FGFR1fl/fl/R2fl/fl; mT-mGfl mammary glands did not show significantly different levels of MRU-containing Lin−CD24+CD29hi population. (H): mT-mG recombined, eGFP+ cells from the floxed FGFR1fl/fl/R2fl/fl; mT-mGfl mammary glands show a significant reduction in MRU-containing Lin−CD24+CD29hi cells as compared to mT-mG recombined eGFP+ wild-type FGFR1+/+/R2+/+; mT-mGfl (*, p < .04, n = 2). Graphs represent mean ± SEM. Abbreviations: FGFR, fibroblast growth factor receptor; GFP, enhanced green fluorescent protein.
FGFR1fl/fl/R2fl/fl MECs Are Not Tolerated in the Basal/Myoepithelial Compartment
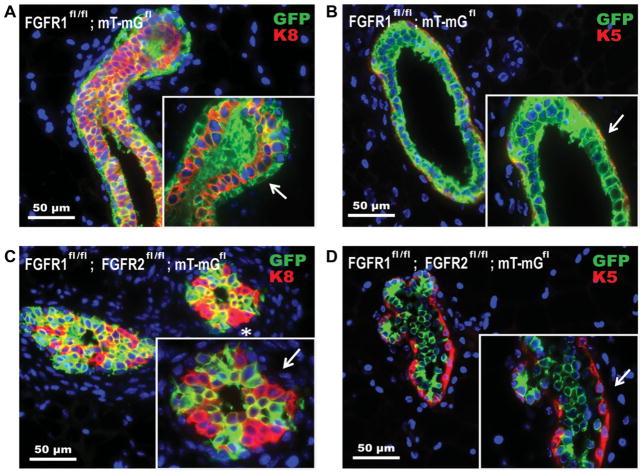
These results demonstrate the complete loss of the MRU-containing Lin−CD24+CD29hi subpopulation in FGFR1/R2-deleted mammary epithelium and support a requirement for FGFR signaling in MaSCs. Furthermore, there was a requirement for a population of nonrecombined escaper cells within the Cre-transduced FGFR1fl/fl/R2fl/fl; mT-mGfl glands in order to facilitate any outgrowth of the FGFR1/2-null cells. Importantly, these escaper cells are the only population that contains the Lin−CD24+CD29hi cells, as all eGFP+ cells in the FGFR1fl/fl/R2fl/fl; mT-mGfl glands were severely deficient for the MRU-containing population. Based on our previous results, FGFR1 deletion alone did not lead to a significant reduction in MaSC function (Fig. 3) or a reduction in outgrowth potential as evidenced by the lack of a competitive advantage between cells with intact FGFR1 and cells without intact FGFR1 (Fig. 5B, 5D). In addition, we observed that FGFR1fl/fl/R2fl/fl; mT-mGfl outgrowths continually contained a population of both nonrecombined tomato red+ and recombined eGFP-positive cells, most often occupying the same mammary ducts and TEBs (Fig. 5E). With these results in mind, we decided to compare our phenotypically normal FGFR1fl/fl; mT-mGfl outgrowths with our FGFR1fl/fl/R2fl/fl; mT-mGfl outgrowths at the cellular level. MaSCs defined by CD24 and CD29 expression have been shown to be primarily basal cells, characterized by expression of keratin 14 (K14), K5, and a lack of estrogen receptor. In contrast, luminal cells are primarily Lin−CD24hiCD29lo [27]. We, therefore, used double immunofluorescent staining for eGFP and K5 (basal) or eGFP and K8 (luminal) in both the FGFR1fl/fl; mT-mGfl and FGFR1fl/fl/R2fl/fl; mT-mGfl outgrowths (Fig. 7). As expected, within the FGFR1fl/fl; mT-mGfl glands, the eGFP+ recombined cells contributed equally to both the luminal (K8+) and the basal (K5+) compartments (Fig. 7A, 7B, arrows). Importantly, eGFP+ recombined FGFR1fl/fl; mT-mGfl cells colocalized with K5+ cells, indicating that FGFR1-null cells are present within the basal compartment (Fig. 7B, arrow). Interestingly, eGFP+ recombined FGFR1fl/fl/R2fl/fl; mT-mGfl cells only colocalized with the luminal K8+ cells (Fig. 7C) and were completely absent from the basal K5+ compartment (Fig. 7C, 7D, arrows). These results indicate a requirement for FGFR signaling within the basal cell compartment in agreement with the absence of the Lin−CD24+CD29hi stem cell population within the eGFP+ cells from FGFR1fl/fl/R2fl/fl; mT-mGfl reconstituted glands. Interestingly, within the luminal cell compartment of the FGFR1fl/fl/R2fl/fl; mT-mGfl glands, not all the cells were recombined as evidenced by a lack of some eGFP+ costaining (Fig. 7C, asterisk). These data suggest that there also is some requirement for FGFR1/R2 signaling perhaps within luminal progenitor cells.
Figure 7.
FGFR1fl/fl/R2fl/fl mammary epithelial cells are not observed in the basal/myoepithelial compartment. (A): Double immunofluorescence staining of FGFR1fl/fl; mT-mGfl glands with anti-GFP (green) and anti-K8 (red) (arrow demonstrating nonoverlapping staining). (B): Double immunofluorescence staining of FGFR1fl/fl; mT-mGfl glands with anti-GFP (green) and anti-K5 (green) (arrow demonstrating overlapping staining). (C): Double immunofluorescence staining of FGFR1fl/fl/R2fl/fl; mT-mGfl glands with anti-GFP (green) and anti-K8 (red) (arrow demonstrating a lack of GFP staining in the myoepithelial, K8- compartment). (D): Double immunofluorescence staining of FGFR1fl/fl/R2fl/fl; mT-mGfl glands with anti-GFP (green) and anti-K5 (red) (arrow demonstrating the lack of GFP staining in the myoepithelial, K5+ compartment).
Discussion
Prior to this study, the role of FGF signaling in mammary stem and progenitor cells had not been defined. A conditional strategy using K14-driven Cre-recombinase-mediated deletion allowed us to delete FGFR1 embryonically in the placode, primitive tree, and prior to placode specification in the skin, resulting in attenuation of FGFR1 throughout all stages of mammary gland development. Through this strategy, we have uncovered a potential role for FGFR1 in driving cellular proliferation in the TEBs during early pubertal development from 3 to 5 weeks of age, resulting in reduced ductal outgrowth, TEB number, and branching of the mammary epithelium. Interestingly, this developmental delay was transient and lost by 7 weeks of age, and the mature virgin mammary gland showed no obvious defects resulting from FGFR1 deletion by 10 weeks of age. Importantly, a similar phenotype was observed upon conditional deletion of FGFR2 through MMTV-driven Cre expression, highlighting comparable roles for each of the two receptors expressed in the mammary epithelium [7]. This raises the question of whether FGFR2 may potentially be able to compensate for the loss of FGFR1 at the later time points in virgin development. The correlation of the developmental phenotype observed in this study, with the onset of puberty, may suggest an interplay or requirement for FGFR1 during the early stages of pubertal development at around 3 weeks of age [31]. It will be important in future studies to investigate the relationship between FGFR1 signaling and embryonic growth factors such as PTHrP, Wnts, and bone morphogenic protein (BMPs) as well as pubertal ovarian hormones such as estrogen and progesterone during the early stages of development [6, 32]. In particular, estrogen is known to regulate the expression of the epidermal growth factor receptor (EGFR) ligand amphiregulin during this stage of development in the mammary gland [33]. It may be possible that multiple cell signaling pathways, including any remaining intact FGFR signaling, can compensate for the loss of one or more FGF receptors. Similarly, the potential compensation by FGFR2 raises the question of how other RTKs such as EGFR may compensate for FGFR1 loss.
In order to investigate the requirement of FGFR1 signaling for MaSC function, we conducted LDA using FGFR1fl/fl MECs but were unable to demonstrate any significant loss in mammary outgrowth potential. These results suggest that FGFR1 alone is not essential in normal stem cell function and branching morphogenesis. We also did not observe a competitive loss of unrecombined cells indicated by reduced competitive outgrowth or loss of β-galactosidase activity as previously observed with deletion of FGFR2 [7]. This along with the developmental and cellular proliferation effects seen following K14-Cre-mediated deletion indicates that FGFR1 signaling alone appears to play a limited role in normal gland development and is not essential for normal mammary stem cell function, perhaps due to compensation from FGFR2.
Based on these findings, we decided to investigate the developmental effects of total elimination of FGF signaling by conducting limiting dilution transplantation analysis on conditionally deleted FGFR1/R2 MECs. Interestingly, we observed a significant loss in take rate upon deletion of both FGFR1 and FGFR2 as compared to deletion of FGFR1 alone. Using the mT-mG double fluorescent reporter construct for Cre-mediated recombination, we consistently observed the contribution of unrecombined escaper cells that were present within all structures of the FGFR1/R2 floxed glands. These results indicate that functional FGFR signaling is required for normal mammary gland outgrowth and stem cell function.
In previous studies from our laboratory, using inducible FGFR1 and FGFR2 signaling in three-dimensional cultures of primary MECs and HC 11 cells, we demonstrated marked differences in the duration of signaling and downstream phenotypes regulated by these closely related RTKs [34]. As demonstrated by the analysis of chimeras where knockout cells were partially rescued by wild-type cells, paracrine mechanisms may be of importance in facilitating the outgrowth of FGFR1-deleted cells in the presence of FGFR2 and vice versa. However, because of the lack of appropriate immunological reagents, it has not been possible to determine the expression of FGFR1 and FGFR2 at the cellular level to ascertain whether these receptors are expressed in the same epithelial cells. Additional studies will be required, therefore, to determine the specific basal and/or luminal cells in which these two receptors are expressed and how they might respond to the numerous FGF ligands expressed in the mammary gland presumably in both a paracrine and autocrine manner. These will be required to make definitive conclusions about whether these two FGF receptors might perform overlapping roles during mammary gland development and the precise compensatory mechanisms between FGFR1 and FGFR2.
Based on LDA, we wanted to investigate the effects of total FGFR deletion on a well-characterized cell population, enriched for MRUs [17, 27]. We conducted quantitative flow cytometry analysis for CD24 and CD29 on both FGFR1-deleted and FGFR1/R2-deleted glands. Using the mT-mG fluorescent reporter, we were able to separate recombined eGFP+ populations from unrecombined tomato red+ populations within the reconstituted mammary glands to observe differences in CD24 and CD29 levels as a direct result of conditional deletion of FGFR1/R2. Interestingly, there was a complete loss of the MRU containing Lin−CD24+CD29hi population within the eGFP+ recombined FGFR1/R2-deleted glands. These results indicate, that any FGFR1/R2-deleted mammary outgrowths, the escaper cell population is required and contains the total population of Lin−CD24+CD29hi stem cells.
Studies using the double fluorescent reporter indicated that while FGFR1 deletion alone failed to affect gland development, there was an absolute requirement for unrecombined escaper cells within FGFR1/R2-deleted gland transplants. Double immunofluorescence staining for the luminal marker K8 and the basal/myoepithelial marker K5 within both FGFR1-deleted and FGFR1/R2-deleted glands revealed a complete dependence on FGFR1/R2 expression within the basal/myoepithelial compartment, as indicated by a lack of eGFP co staining with K5. The FGFR1/R2-deleted cell population exclusively occupied the luminal compartment. Importantly, these results may explain the loss of the Lin−CD24+CD29hi stem cell population within the eGFP+ cells from the FGFR1/R2-deleted glands, as the basal/myoepithelial compartment is known to contain this population [27]. The luminal compartment within the FGFR1/R2-deleted glands also maintained a significant population of unrecombined eGFP− cells as well, which may indicate a necessity for FGFR signaling potentially to sustain the FGFR1/R2-deleted luminal cells through paracrine signaling mechanisms. Recent studies using an inducible iFGFR1 transgenic model have revealed a rapid induction of both amphiregulin and epiregulin following FGFR activation [35]. Thus, it is conceivable that these EGFR ligands may provide one potential mechanism involved in the rescue of FGFR1/2 null MECs. Future studies will be required to investigate the mechanisms by which FGFR signaling acts within the luminal compartment in this model. Furthermore, it is not totally surprising that FGFR signaling would be required to a greater extent in the basal/myoepithelial cells than the luminal cells, as FGFRs within the mammary epithelium are known to be activated by FGF ligands, such as FGF7 and FGF10 secreted by the stroma [36]. The basal/myoepithelial cells, unlike the luminal cells, are in direct cell-to-cell contact with the mammary stroma and may require paracrine FGF signaling from the stroma for proper maintenance and development.
The importance of paracrine interactions not only between stromal and epithelial cells but also between different epithelial cell compartments in mammary gland development has been elegantly demonstrated by the analysis of chimeras containing a mixture of wild-type and null cells [37]. For example, paracrine interactions have been suggested to be important for the observed effects of a conditionally activated smoothened in MECs on the proliferation of nearby cells [20]. Similar interactions between c-Kit in luminal progenitors and its ligand SCF have been suggested [38]. The effects of steroid hormones, estrogen, and progesterone on the proliferation of both adjacent MECs as well as the regulation of mammary stem cells are mediated by local growths factors, such as Wnt 4 and Rankl [39]. Similar mechanisms may also be present in breast cancer. For example, estrogen has been reported to expand breast cancer stem-like cells through paracrine FGF/TBX3 signaling [40]. Interestingly, in studies performed in MCF-7 cells, FGFR3 knockdown was able to abolish estrogen-induced expansion of the breast cancer stem-like populations. Therefore, signaling through FGFR3 and FGFR4 [41] as well as FGFR1 and FGFR2 also appears to be important in breast cancer.
Wnt proteins are self-renewal factors for mammary stem cells and the combined use of Wnt, EGF, and Matrigel can promote MaSC expansion in vitro [42]. In addition, the Fgf and Wnt pathways have been shown to cooperate in mouse mammary tumor virus-induced tumors [43] and in genetically engineered mouse models of human breast cancer [44]. Based upon the current studies, it is apparent that FGF signaling through FGFR1 and FGFR2 also appears to play a critical role in the maintenance of mouse MaSCs. The apparent paracrine rescue of FGFR1 and FGFR2 null cells by adjacent wild-type cells suggests that other ligands, perhaps amphiregulin as hypothesized above, may compensate for the loss of Fgf signaling perhaps by activation of EGFR. These results support observations obtained using mammosphere assays in serum-free medium in which FGF, EGF, or insulin-like growth factor (IGF) are all able to promote sphere formation [45]. Thus, in addition to canonical Wnt signaling to promote self-renewal, activation of RTK pathways may be required for expansion of MaSCs. Under physiological conditions in normal mouse mammary gland development, FGFR1 and FGFR2 appear to sustain this function.
Conclusions
In summary, these results highlight the requirement of functional FGFR signaling for proper mammary stem cell function and development. FGF signaling is increasingly implicated in human cancers, including breast cancer [46–49]. In addition, the role of cancer stem cells in the initiation, relapse and recurrence, and therapeutic resistance of breast cancer makes the discovery of genes important for stem cell function critical to future treatments. The importance of compensatory mechanisms of drug resistance is a critical component in therapeutic failure. As a viable drug target, it will be important, therefore, to understand possible compensatory mechanisms within the FGF pathway as well as potential interactions with other developmental pathways.
Supplementary Material
Acknowledgments
This study was supported by Grants R37-CA16303 and P30-CA125123 from the National Cancer Institute. We would like to thank our colleagues Dr. Heather LaMarca Machado and Jason Herschkowitz for their helpful comments as well as Drs. Feng Yang and David Rowley for supplying FGFR floxed mice.
Footnotes
Disclosure of Potential Conflicts of Interest
None.
Author contributions: A.C.P.: conception and design, collection and assembly of data, and manuscript writing; T.B., X.B., and K.R.: collection and assembly of data and manuscript editing; S.H.: data analysis and interpretation; J.M.R.: financial support, conception and design, manuscript writing, and final approval of manuscript.
References
- 1.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Chodosh LA, Gardner HP, Rajan JV, et al. Protein kinase expression during murine mammary development. Dev Biol. 2000;219:259–276. doi: 10.1006/dbio.2000.9614. [DOI] [PubMed] [Google Scholar]
- 3.Lu P, Ewald AJ, Martin GR, et al. Genetic mosaic analysis reveals FGF receptor 2 function in terminal end buds during mammary gland branching morphogenesis. Dev Biol. 2008;321:77–87. doi: 10.1016/j.ydbio.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mailleux AA, Spencer-Dene B, Dillon C, et al. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129:53–60. doi: 10.1242/dev.129.1.53. [DOI] [PubMed] [Google Scholar]
- 5.Dillon C, Spencer-Dene B, Dickson C. A crucial role for fibroblast growth factor signaling in embryonic mammary gland development. J Mammary Gland Biol Neoplasia. 2004;9:207–215. doi: 10.1023/B:JOMG.0000037163.56461.1e. [DOI] [PubMed] [Google Scholar]
- 6.Eblaghie MC, Song SJ, Kim JY, et al. Interactions between FGF and Wnt signals and Tbx3 gene expression in mammary gland initiation in mouse embryos. J Anat. 2004;205:1–13. doi: 10.1111/j.0021-8782.2004.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsa S, Ramasamy SK, De Langhe S, et al. Terminal end bud maintenance in mammary gland is dependent upon FGFR2b signaling. Dev Biol. 2008;317:121–131. doi: 10.1016/j.ydbio.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Jackson D, Bresnick J, Rosewell I, et al. Fibroblast growth factor receptor signalling has a role in lobuloalveolar development of the mammary gland. J Cell Sci. 1997;110(Pt 11):1261–1268. doi: 10.1242/jcs.110.11.1261. [DOI] [PubMed] [Google Scholar]
- 9.Pirvola U, Ylikoski J, Trokovic R, et al. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 10.Partanen J, Schwartz L, Rossant J. Opposite phenotypes of hypomorphic and Y766 phosphorylation site mutations reveal a function for Fgfr1 in anteroposterior patterning of mouse embryos. Genes Dev. 1998;12:2332–2344. doi: 10.1101/gad.12.15.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 12.Jonkers J, Meuwissen R, van der Gulden H, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 13.Yu K, Xu J, Liu Z, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 14.Muzumdar MD, Tasic B, Miyamichi K, et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 15.Grimm SL, Contreras A, Barcellos-Hoff MH, et al. Cell cycle defects contribute to a block in hormone-induced mammary gland proliferation in CCAAT/enhancer-binding protein (C/EBPbeta)-null mice. J Biol Chem. 2005;280:36301–36309. doi: 10.1074/jbc.M508167200. [DOI] [PubMed] [Google Scholar]
- 16.Heckman BM, Chakravarty G, Vargo-Gogola T, et al. Crosstalk between the p190-B RhoGAP and IGF signaling pathways is required for embryonic mammary bud development. Dev Biol. 2007;309:137–149. doi: 10.1016/j.ydbio.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaMarca HL, Visbal AP, Creighton CJ, et al. CCAAT/enhancer binding protein beta regulates stem cell activity and specifies luminal cell fate in the mammary gland. Stem Cells. 2010;28:535–544. doi: 10.1002/stem.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rijnkels M, Rosen JM. Adenovirus-Cre-mediated recombination in mammary epithelial early progenitor cells. J Cell Sci. 2001;114:3147–3153. doi: 10.1242/jcs.114.17.3147. [DOI] [PubMed] [Google Scholar]
- 19.Landua JD, Visbal AP, Lewis MT. Methods for preparing fluorescent and neutral red-stained whole mounts of mouse mammary glands. J Mammary Gland Biol Neoplasia. 2009;14:411–415. doi: 10.1007/s10911-009-9155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visbal AP, LaMarca HL, Villanueva H, et al. Altered differentiation and paracrine stimulation of mammary epithelial cell proliferation by conditionally activated Smoothened. Dev Biol. 2011;352:116–127. doi: 10.1016/j.ydbio.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnefoix T, Bonnefoix P, Verdiel P, et al. Fitting limiting dilution experiments with generalized linear models results in a test of the single-hit Poisson assumption. J Immunol Methods. 1996;194:113–119. doi: 10.1016/0022-1759(96)00077-4. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 23.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. http://www.R-project.org/ [Google Scholar]
- 24.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 25.Asselin-Labat ML, Sutherland KD, Barker H, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 26.Medina D. The mammary gland: A unique organ for the study of development and tumorigenesis. J Mammary Gland Biol Neoplasia. 1996;1:5–19. doi: 10.1007/BF02096299. [DOI] [PubMed] [Google Scholar]
- 27.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 28.Eirew P, Stingl J, Raouf A, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14:1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 29.Hosokawa R, Deng X, Takamori K, et al. Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. J Exp Zool B Mol Dev Evol. 2009;312B:343–350. doi: 10.1002/jez.b.21274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Meyer M, Muller AK, et al. Fibroblast growth factor receptors 1 and 2 in keratinocytes control the epidermal barrier and cutaneous homeostasis. J Cell Biol. 2010;188:935–952. doi: 10.1083/jcb.200910126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colby DR, Vandenberg JG. Regulatory effects of urinary pheromones on puberty in the mouse. Biol Reprod. 1974;11:268–279. doi: 10.1095/biolreprod11.3.268. [DOI] [PubMed] [Google Scholar]
- 32.Hens JR, Wysolmerski JJ. Key stages of mammary gland development: Molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res. 2005;7:220–224. doi: 10.1186/bcr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciarloni L, Mallepell S, Brisken C. Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc Natl Acad Sci USA. 2007;104:5455–5460. doi: 10.1073/pnas.0611647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xian W, Schwertfeger KL, Rosen JM. Distinct roles of fibroblast growth factor receptor 1 and 2 in regulating cell survival and epithelial-mesenchymal transition. Mol Endocrinol. 2007;21:987–1000. doi: 10.1210/me.2006-0518. [DOI] [PubMed] [Google Scholar]
- 35.Bade LK, Goldberg JE, Dehut HA, et al. Mammary tumorigenesis induced by fibroblast growth factor receptor 1 requires activation of the epidermal growth factor receptor. J Cell Sci. 2011;124:3106–3117. doi: 10.1242/jcs.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dailey L, Ambrosetti D, Mansukhani A, et al. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Brisken C, O’Malley B. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2010;2:a003178. doi: 10.1101/cshperspect.a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regan JL, Kendrick H, Magnay FA, et al. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene. 2012;31:869–883. doi: 10.1038/onc.2011.289. [DOI] [PubMed] [Google Scholar]
- 39.Joshi PA, Jackson HW, Beristain AG, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465:803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 40.Fillmore CM, Gupta PB, Rudnick JA, et al. Estrogen expands breast cancer stem-like cells through paracrine FGF/Tbx3 signaling. Proc Natl Acad Sci USA. 2010;107:21737–21742. doi: 10.1073/pnas.1007863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seitzer N, Mayr T, Streit S, et al. A single nucleotide change in the mouse genome accelerates breast cancer progression. Cancer Res. 2010;70:802–812. doi: 10.1158/0008-5472.CAN-09-3239. [DOI] [PubMed] [Google Scholar]
- 42.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–577. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theodorou V, Kimm MA, Boer M, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–769. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 44.Pond AC, Herschkowitz JI, Schwertfeger KL, et al. Fibroblast growth factor receptor signaling dramatically accelerates tumorigenesis and enhances oncoprotein translation in the mouse mammary tumor virus-Wnt-1 mouse model of breast cancer. Cancer Res. 2010;70:4868–4879. doi: 10.1158/0008-5472.CAN-09-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 48.Reis-Filho JS, Simpson PT, Turner NC, et al. FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas. Clin Cancer Res. 2006;12:6652–6662. doi: 10.1158/1078-0432.CCR-06-1164. [DOI] [PubMed] [Google Scholar]
- 49.Meyer KB, Maia AT, O’Reilly M, et al. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biol. 2008;6:e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.