Abstract
Nature uses a diversity of glycoside hydrolase (GH) enzymes to convert polysaccharides to sugars. As lignocellulosic biomass deconstruction for biofuel production remains costly, natural GH diversity offers a starting point for developing industrial enzymes, and fungal GH family 7 (GH7) cellobiohydrolases, in particular, provide significant hydrolytic potential in industrial mixtures. Recently, GH7 enzymes have been found in other kingdoms of life besides fungi, including in animals and protists. Here, we describe the in vivo spatial expression distribution, properties, and structure of a unique endogenous GH7 cellulase from an animal, the marine wood borer Limnoria quadripunctata (LqCel7B). RT-quantitative PCR and Western blot studies show that LqCel7B is expressed in the hepatopancreas and secreted into the gut for wood degradation. We produced recombinant LqCel7B, with which we demonstrate that LqCel7B is a cellobiohydrolase and obtained four high-resolution crystal structures. Based on a crystallographic and computational comparison of LqCel7B to the well-characterized Hypocrea jecorina GH7 cellobiohydrolase, LqCel7B exhibits an extended substrate-binding motif at the tunnel entrance, which may aid in substrate acquisition and processivity. Interestingly, LqCel7B exhibits striking surface charges relative to fungal GH7 enzymes, which likely results from evolution in marine environments. We demonstrate that LqCel7B stability and activity remain unchanged, or increase at high salt concentration, and that the L. quadripunctata GH mixture generally contains cellulolytic enzymes with highly acidic surface charge compared with enzymes derived from terrestrial microbes. Overall, this study suggests that marine cellulases offer significant potential for utilization in high-solids industrial biomass conversion processes.
Keywords: gribble, carbohydrate degrading enzymes
Increasing worldwide demand for energy and concerns over greenhouse gas emissions have driven the search for renewable alternatives to transportation fuels from fossil origins, and liquid biofuel production from cellulose, the most abundant biopolymer on Earth, has emerged as a promising option. Cellulose comprises the main structural component of plant biomass and is composed entirely of glucose, but the crystalline nature of this material makes its cost-effective conversion into fermentable sugars challenging (1–3). Cellulose is arranged as crystalline microfibrils cross-linked to hemicellulose and lignin to form a recalcitrant biopolymer matrix (4, 5). Improving methods of sugar release from lignocellulose by manipulation of biomass composition and new chemical treatments are areas of intense research (6); additionally, a key barrier to large-scale conversion of lignocellulose to biofuels is the limited availability of low-cost enzymes to efficiently depolymerize biomass from various sources while maximizing sugar release. Typically, both natural enzyme mixtures and industrial enzyme preparations for biomass deconstruction consist of a mixture of glycoside hydrolases (GHs) that exhibit synergistic properties (7). Filamentous fungi in particular have traditionally been used as a source of GHs, due to their ability to produce high titers of secreted, active cellulolytic enzymes (8, 9). In addition to fungi, bacteria and arthropods, including termites, cockroaches, and beetles, are also recognized for their lignocellulose hydrolysis capabilities (10–12). More recently, the hydrolytic capacity of the wood-boring crustacean Limnoria quadripunctata, which thrives on a diet of wood, was revealed. Unusually, Limnoria do not rely on gut microbes for wood digestion, but instead have a digestive system free of microbial populations, and digest lignocellulose using endogenous enzymes (13). At least three classes of GHs are necessary to deconstruct cellulose (7), and analysis of a cDNA library constructed from the L. quadripunctata digestive gland revealed candidate transcripts that putatively represent all three classes of hydrolases, namely cellobiohydrolases, endoglucanases, and β-glucosidases. Specifically, endoglucanases cleave internal points of cellulose chains in accessible regions to create new, free chain ends; cellobiohydrolases processively hydrolyze cellulose chains from either the reducing or nonreducing end and release cellobiose as the major product; and β-glucosidases hydrolyze cellobiose to glucose. The exposure of chain ends by endoglucanases for cellobiohydrolase attachment and detachment is an important synergism between these two cellulase classes, and the hydrolysis of cellobiose by β-glucosidases mitigates cellobiose inhibition (7, 14–16). The L. quadripunctata transcriptome was dominated by a few GH families, with transcripts representing GH7s being particularly abundant, representing ∼12% of total ESTs sequenced. This was unexpected as GH7s were previously only reported from fungi and symbiont protists from termites, but not from animals (13).
GH family 7 cellobiohydrolases are of particular importance, as they typically represent the most significant hydrolytic potential in many natural and industrial enzyme mixtures (7), and most GH7s studied to date originate from fungi. Along with the discovery of complementary enzymes (17–21), prospecting and engineering GH7s for higher activity represents one of the most important research activities to improve enzyme mixtures, which will in turn significantly aid in the commercialization of biofuel processes based on enzymatic depolymerization of polysaccharides (2, 7, 8). To that end, here, we examine multiple aspects of lignocellulose breakdown by a GH7 cellulase from L. quadripunctata (LqCel7B). We use reverse transcription, quantitative PCR (RT-qPCR) and Western blot analysis to demonstrate that LqCel7B is expressed within the L. quadripunctata hepatopancreas and subsequently secreted into the gut for biomass digestion. To understand the ability of LqCel7B to operate in saline conditions, we use X-ray crystallography to solve four high-resolution LqCel7B structures and then compare the LqCel7B structure to the well-characterized Hypocrea jecorina Cel7A (HjCel7A) structure using molecular dynamics (MD) simulations. The structures overall exhibit several key differences to HjCel7A, including a strikingly acidic surface charge and a tyrosine residue at the tunnel entrance, which likely plays a role in substrate acquisition and processivity. We also demonstrate that LqCel7B is a cellobiohydrolase rather than an endoglucanase due to its structure and activity profile on several model cellulose substrates, and that LqCel7B is active at high salt concentrations, likely due to its high surface charge distribution. Based on the results presented here for LqCel7B, we examine the sequences of known enzymes in the cellulolytic mixture of L. quadripunctata, which suggest that the additional cellulolytic enzymes from this organism all exhibit high surface charge compared with GHs from terrestrial and freshwater organisms. This study suggests that L. quadripunctata and other marine organisms represent interesting targets for prospecting cellulolytic enzymes for industrial biomass conversion and for understanding cellulose conversion in marine environments.
Results
Distribution of LqCel7B in Digestive System Organs.
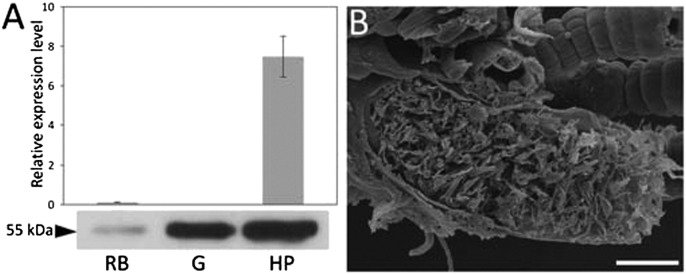
Limnoriids have a bipartite digestive system in which enzymes are produced in the hepatopancreas, and subsequently secreted into the lumen of this gland and then transferred to the hindgut, their presumptive site of action. The distribution and abundance of LqCel7B in the L. quadripunctata digestive system was investigated through RT-qPCR and Western blot analysis (Fig. 1A). RT-qPCR studies demonstrated that LqCel7B transcripts were abundant in the hepatopancreas, but low or absent from the hindgut and rest of the body. In contrast, probing protein extracts with anti-LqCel7B antibodies revealed a unique 55-kDa band in both gut and hepatopancreas extracts (Fig. 1A). SEM of the gut tissue showed the hindgut lumen tightly packed with fine wood particles (Fig. 1B), indicating that LqCel7B functions in a high-solids loading environment. These studies not only provide direct evidence that LqCel7B gene expression is confined to the hepatopancreas, as previously suggested by in situ hybridization (13), but that the protein is subsequently translocated into the hindgut.
Fig. 1.
Abundance and functional environment of the cellobiohydrolase LqCel7B in the L. quadripunctata digestive system. (A) Reverse transcription, quantitative PCR (RT-qPCR) using RNA extracted from rest of the body (RB), hindgut (G), and hepatopancreas (HP). Expression levels presented are normalized to ubiquitin (Upper). Western blot of protein extracts prepared from these organs probed with purified anti-LqCel7B (Lower). (B) Scanning electron micrograph showing an obliquely sectioned hindgut packed with wood particles. (Scale bar, 50 µm.)
Protein Production, Physical Properties, and Enzyme Kinetics.
LqCel7B, a single domain protein lacking a carbohydrate-binding module (CBM) and linker sometimes observed in GH7 enzymes, was expressed in Aspergillus oryzae and Aspergillus niger and subsequently purified using anion exchange and size exclusion chromatography (Fig. S1 A and B). Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) analyses of purified samples confirmed all peptides detected matched the LqCel7B sequence (SI Results, LC-MS/MS Analysis). The protein behaves as a monodisperse monomer with a molecular weight of 45 kDa from size exclusion chromatography multiangle laser light scattering, corresponding closely to the 46.5 kDa predicted from the sequence of the signal-peptide cleaved protein (Fig. S1C). Velocity analytical ultracentrifugation (AUC) confirmed the monomeric nature in solution and yielded a sedimentation coefficient of 4.45 S (Fig. S1D). pH and temperature optima were determined using the chromogenic substrates p-nitrophenyl-β-d-cellotrioside (pNP-G3), and p-nitrophenyl-β-d-cellopentaoside (pNP-G5) (Fig. S2 A and B). LqCel7B exhibits a variable pH optima range (Fig. S2A): pH 4–6.5 for pNP-G3 and pH 4–8 for pNP-G5. High activity was maintained over a wide temperature range (Fig. S2B). Furthermore, the kinetic properties of LqCel7B were determined using pNP-G3 and pNP-G5 (Fig. S2C) due to very low activity on p-nitrophenyl-β-d-cellobioside (pNP-G2) (Fig. S2D). The higher Vmax and kcat together with lower Km and higher catalytic efficiency (kcat/Km) indicate that pNP-G5 is a better substrate. The LqCel7B Km value for pNP-G5 is within the range reported for other GH7 cellobiohydrolases (22–26). The inhibitory effect of cellobiose, which is commonly observed in GH7 enzymes from fungi (15, 16, 26–28), on the hydrolytic activity of LqCel7B was investigated using pNP-G5 (Fig. S2F), and a Ki value of 1.40 mM was measured.
LqCel7B Substrate Specificity.
LqCel7B hydrolytic activity was tested on phosphoric acid swollen cellulose (PASC) and Avicel, respectively, at low [0.1% (wt/vol) PASC or Avicel] and high [1% (wt/vol) PASC or 2% (wt/vol) Avicel] solids loadings (Table 1). The highest rate of substrate conversion was detected in the high-solids loading assays, possibly because of the lack of a CBM, which results in more association events due to reduced diffusion limitations (14, 29). A similar value for PASC hydrolysis ratio was reported for HjCel7A and Thermoascus aurantiacus Cel7A (assayed with CBMs) as well as Melanocarpus albomyces Cel7B (which naturally lacks a CBM), whereas LqCel7B showed lower Avicel conversion ratios compared with the same cellobiohydrolases without CBMs (30, 31). Also, the main product from PASC and Avicel hydrolysis was cellobiose (G2), followed by glucose (G1) and trace cellotriose (G3). G1 and G2 were also the main reaction products detected by high performance anion exchange chromatography (HPAEC) analysis on hydrolysis of cellooligosaccharides (Fig. S2G). As G2 is produced primarily through processive hydrolysis, whereas G1 and G3 are the main products released upon initial hydrolysis events (32), processivity was semiquantitatively estimated as the ratio G2/(G1 + G3) (33, 34). Processivity ratios above 7 were achieved in almost all reactions containing PASC and in extended reactions containing Avicel at a low-solids loading, which are within the range measured for Phanerochaete chrysosporium Cel7D and HjCel7A (33). Additionally, LqCel7B was able to hydrolyze carboxymethyl cellulose (CMC), albeit at much lower conversions than PASC (Fig. S2E), indicating that LqCel7B can conduct endoinitiation, which is a common feature of most GH7 cellobiohydrolases (35, 36). We note that more “open” GH7 cellobiohydrolases also exhibit low activity on CMC (37). These results suggest that LqCel7B is a cellobiohydrolase, which is supported by the structural characteristics described below.
Table 1.
Substrate conversion ratio and processivity estimated for LqCel7B
| PASC |
Avicel |
||||
| [S] | Time (h) | Conversion (%) | Processivity | Conversion (%) | Processivity |
| Low loading | 1 | 3.69 ± 0.42 | 9.20 ± 0.12 | 0.30 ± 0.02 | 4.74 ± 0.26 |
| 24 | 5.02 ± 0.68 | 10.65 ± 0.71 | 0.43 ± 0.05 | 6.95 ± 0.96 | |
| High loading | 1 | 5.21 ± 0.72 | 6.62 ± 0.07 | 0.22 ± 0.01 | 4.63 ± 0.15 |
| 24 | 10.34 ± 1.14 | 3.88 ± 0.14 | 0.66 ± 0.11 | 3.11 ± 0.04 | |
Low loading: 0.1% (wt/vol) PASC or Avicel; high loading: 1% (wt/vol) PASC; 2% (wt/vol) Avicel. Processivity = G2/(G1 + G3). Conversion rates shown are a product mass percentage relative to substrate mass. [S] denotes substrate concentration. All values were determined by HPAEC and represent mean ± SE.
Structural and Simulation Studies.
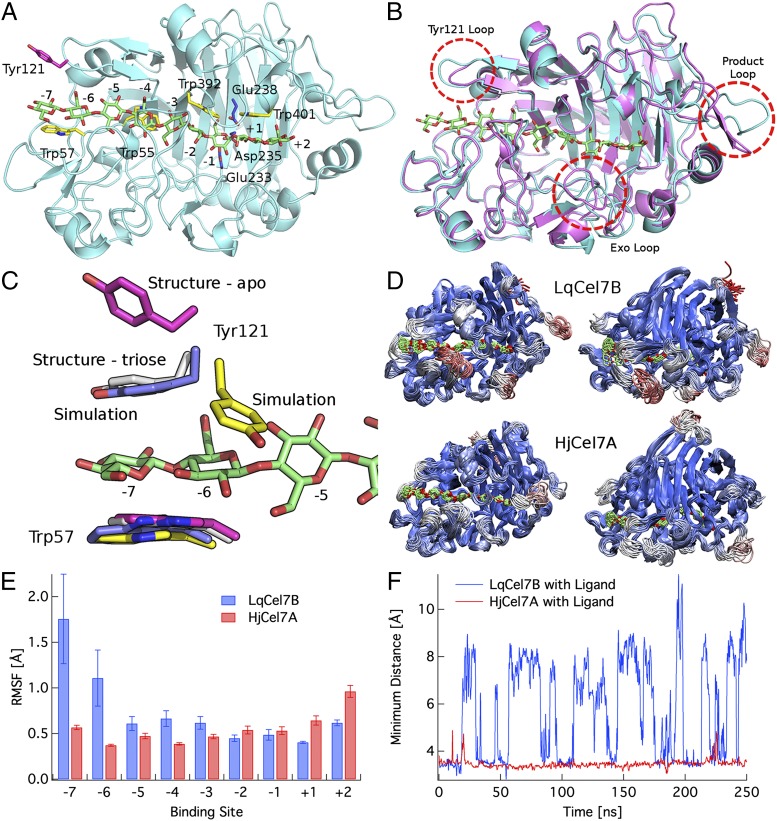
The structural characterization of LqCel7B is based on the solution of four X-ray structures, one apo form at 1.6-Å resolution, and three ligand bound complexes ranging from 1.9 Å to 1.14 Å. The lattermost structure of LqCel7B at 1.14-Å resolution in complex with thiocellobiose reveals close to atomistic detail from the high-quality electron density maps (Fig. S3). Crystallography statistics of the four models are presented in Table S1. The apo structure is shown in Fig. 2A with the cellononaose ligand from the HjCel7A structure (PDB ID code 8CEL) docked into the tunnel. Four highly conserved tryptophan residues in GH7 cellobiohydrolases are in yellow, and the catalytic residues are in blue. The overall fold is homologous to other GH7 cellobiohydrolases, comprising a globular and equatorially elongated protein, but with several interesting features particularly in the surface loop regions (Fig. 2B) (33, 38–41). The active site tunnel, formed by four pairs of curved antiparallel β-sheets packed face to face, extends from the −7 binding site at Trp57 to the +2 binding site near Phe362, representing a distance of ∼48 Å between the tunnel entrance and exit. The conserved triad of catalytic residues (EXDXXE; Fig. S4A), Glu233, Asp235, and Glu238 use a two-step, retaining mechanism for glycosidic bond hydrolysis (11, 33, 38–43).
Fig. 2.
Structural and computational analysis of LqCel7B. (A) Overall fold of the apo LqCel7B structure with the catalytic residues highlighted in blue and a selection of the planar Trp residues that line the ligand binding tunnel in yellow. Tyr121 is shown in pink. The cellononaose ligand is docked into the LqCel7B structure from the HjCel7A structure and is shown in green (38). (B) Structural comparison of the apo LqCel7B structure and HjCel7A, with the ligand from 8CEL (38). (C) View of the −7 to −5 binding sites in two crystallographic and two simulation conformations, including the ligand from 8CEL (38) in green. Tyr121 binds directly to the ligand in the −7 glucose binding site. The color coding for Tyr121 and Trp57 is as follows: pink, apo structure conformations; gray, cellotriose LqCel7B structure; blue, MD simulation snapshot of LqCel7B with Tyr121 stacking on the −7 glucose unit; and yellow, MD simulation snapshot of LqCel7B with Tyr121 binding to the −6 glucose unit. Fig. S5E shows the cluster view of Tyr121 over the 250-ns MD simulation. (D) Cluster representations of LqCel7B and HjCel7A over a 250-ns MD simulation. HjCel7A data are taken from ref. 44, and the enzymes are colored by root mean square fluctuations (RMSFs), wherein red represents fluctuations up to 4 Å and blue represents the minimum fluctuations. The Root square mean deviation (RSMD) and RMSF of both enzymes are shown in Fig. S5 A–C. (E) RMSF of LqCel7B and HjCel7A (44) ligands. Error bars represent one SD from the average value measured by block averaging. (F) Time series of the exo-loop opening and closing events in LqCel7B and HjCel7A MD simulations over 250 ns with a cellononaose ligand present (33, 38). The exo-loop motions are the minimum distance between the exo loop and the corresponding loops on the other side of the tunnel: for LqCel7B, the minimum distance from the exo loop (residues 265–274) to residues 394–399 and for HjCel7A, the minimum distance from the exo loop (residues 244–253) to residues 369–373. A histogram of this time series is shown in Fig. S5D.
Three holo crystal structures not only provide reference sites for docking of the full cellononaose ligand, but a detailed picture of the hydrogen bonding networks at both the product and entrance sites. Cocrystallization of LqCel7B with cellopentaose resulted in a structure with cellobiose bound in the +1 and +2 product sites, anchored through a combination of hydrogen bonds between the hydroxyl groups of cellobiose and the glycosyl binding residues Arg271, His249, Arg417 and the catalytic residues Asp235 and Glu238 as well as hydrophobic stacking between Trp401 and the pyranose rings of cellobiose (Fig. S4B). The absence of cellopentaose and presence of cellobiose indicates that activity remained intact during crystallization. Additionally, thiocellobiose was cocrystallized with LqCel7B crystals in an alternative P2 21 21 space group by seeding trials (Fig. S3). Thiocellobiose is located in the same position as the cellobiose in the previous structure, potentially contributing a stabilizing role in the crystal and aiding data collection at high resolution. Further detailed features were apparent in these electron density maps, including dual conformation of the proton donor Glu238 in the active site, an observation that correlated directly with MD simulations (Fig. S5F). Additionally, soaking native crystals of the apo form of LqCel7B with cellotriose, revealed both cellobiose in the product site as before, with the addition of cellotriose in the active site entrance from the −7 to −5 sites, stabilized via hydrogen bonding occurring between Asp124 and Lys202 and hydrophobic stacking between Trp55 and Trp57 and the pyranose rings (Fig. S4C).
Fig. 2B shows the structural alignment of LqCel7B and the HjCel7A 8CEL crystal structure (38) and Fig. S6 B and C show the surface representation of the same structures. The LqCel7B active site tunnel is mostly enclosed, as opposed to the more exposed cleft of the endoglucanase HjCel7B (PDB ID code 1EG1; Fig. S6A), and it is, therefore, similar to other known structures of GH7 cellobiohydrolases (38–41), suggesting that LqCel7B is a cellobiohydrolase as indicated by the activity measurements described above (35). Structurally, the most significant differences in the active site tunnels between LqCel7B and HjCel7A are observed at the −7 subsite, the exo loop (33), and the product sites, which are discussed below in turn.
As shown in Fig. 2C, the −7 subsite in LqCel7B exhibits a potential, additional aromatic-carbohydrate interaction site due to Tyr121. In the apo structure of LqCel7B, Tyr121 is significantly displaced from the −7 subsite (pink) when the ligand is not present. However, in the LqCel7B structure with the cellotriose ligand bound in the −7 to −5 sites, Tyr121 is bound directly to the glucose residue in the −7 site (gray). Additionally, rapid binding of Tyr121 to the −7 glucose unit is observed (Fig. 2C, in blue) in MD simulations constructed from the LqCel7B apo structure with the 8CEL ligand docked into the active site tunnel. As shown in Fig. 2C, excellent agreement is observed between MD simulations and the structural data with Tyr121-glucosyl binding at the −7 site. Further quantitative characterization of these results is provided in Fig. S5 A–D. Lastly, the Tyr121 residue is observed in the MD simulations to undergo a dihedral rotation about the χ1 dihedral such that the aromatic ring of Tyr121 binds to the sugar in the −6 site (Fig. S5E and Movie S1). This dihedral rotation in turn induces larger fluctuations in the ligand, as shown in Fig. 2 D and E. This additional aromatic binding platform at the entrance of a GH7 tunnel has not been observed previously in contact with a ligand, although it has been suggested to play a role in guiding a cellulose chain into the tunnel in previous structural studies of the apo structure of M. albomyces (40) and in a recent computational study of the Heterobasidion irregulare cellobiohydrolase Cel7A (44). These results suggest that an additional aromatic residue at the tunnel entrance may play a role in substrate acquisition and processivity.
Additionally, Fig. 2D shows that the exo loop (33) in LqCel7B is able to undergo significant conformational changes relative to the exo loop in HjCel7A. Fig. 2F shows the distance between the exo loops and the corresponding loop across the enzyme tunnel as a function of time. As shown, the HjCel7A loop does not open during the 250-ns MD simulation (44), whereas the LqCel7B exo loop undergoes a conformational change between a closed and open state quite readily on the nanosecond time scale. This difference in exo-loop behavior is likely to increase the propensity of LqCel7B to engage in endotype initiation on the cellulose surface relative to HjCel7A (35), as suggested in previous studies of P. chrysosporium Cel7D and HjCel7A (35, 44). Lastly, we note that there are significant protein fluctuations at the product site of LqCel7B not observed in HjCel7A. This, however, does not affect ligand fluctuations (Fig. 2E), but may impact product inhibition, which will be examined in future work. An illustration of the LqCel7B bound to cellulose is shown in Movie S2.
LqCel7B Salt Tolerance.
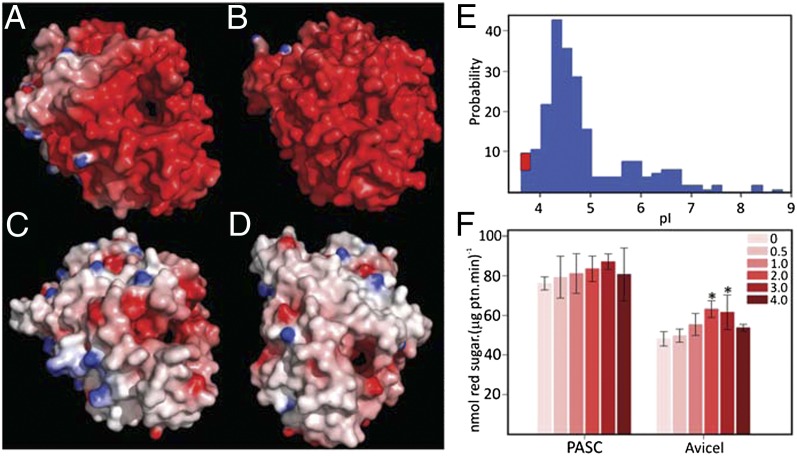
Analysis of the surface properties of LqCel7B reveals a striking predominance of negatively charged residues as shown in Fig. 3A. Homology modeling and surface charge analysis suggest that the GH7 enzyme from the marine crustacean Chelura terebrans exhibits negatively charged residues that create an almost continuous acidic surface, as shown in Fig. 3B. Conversely, the homology model constructed for the GH7 ortholog from the related freshwater crustacean Daphnia magna exhibits a more neutral composition with both acidic and basic residues less numerous and more evenly distributed in common with the fungal HjCel7A (Fig. 3 C and D). The high number of acidic surface residues also endows the enzyme with a low isoelectric point (pI), far below that calculated for the vast majority of other identified GH7s as shown through a comparison of pI values across GH7 family members (Fig. 3E). The acidic nature of Limnoria and Chelura GH7s may be a consequence of their marine environment and the physiological need of the enzymes to remain active in the gut of these crustaceans, which likely contain seawater. This hypothesis was further corroborated by the construction of homology models of the entire cellulolytic mixture of L. quadripunctata, which suggests that the additional known GH7, GH5, and GH9 enzymes from this organism all have high negative surface charges compared with terrestrial microbial equivalents (Fig. S6 F–H). Highly negative electrostatic potential surface is one of the hallmarks of halophilic proteins, which not only require salts for optimum activity, but also remain active at high ionic strength values (45). We also examined the hydrolytic activity of LqCel7B in high salt conditions using PASC and Avicel as substrates (Fig. 3F). Starting at 0 M NaCl, then 0.5 M to reflect the ionic strength of seawater, then progressively increasing concentrations up to 4 M, NaCl does not inhibit enzyme activity, and activity increases significantly at some salt concentrations.
Fig. 3.
LqCel7B salt tolerance. Electrostatic potential distribution on the solvent accessible surface of Limnoria quadripunctata LqCel7B (A), Chelura terebrans GH7 (B), Daphnia magna GH7 (C), and Hypocrea jecorina Cel7A (D). Electrostatic potential between −7 kT/e and 7 kT/e was shown as a colored gradient from red (acidic) to blue (basic). (E) pI clusters showing GH7s from three GH7 L. quadripunctata enzymes grouped together with the C. terebrans ortholog (red square) as a unique cluster at low pI value (3.8 < pI < 3.9). (F) Hydrolytic activity of LqCel7B as a function of sodium chloride concentration expressed as reducing sugar released from PASC and Avicel. Asterisks represent statistically significant values following one-way ANOVA DSF (*P < 0.01).
Discussion and Conclusions
LqCel7B is a uniquely and extensively characterized nonfungal GH7 cellulase, and it exhibits a number of features that differ from well-characterized fungal examples of this important cellulase family. From a protein sequence standpoint, previous work showed that the Limnoriid GH7s formed a well-separated branch from fungal and protist equivalents in phylogenetic analysis of protein sequences (13). The lack of a CBM is intriguing and together with the high abundance of LqCel7B in the gut lumen (Fig. 1) could reflect an evolutionary adaptation to the gut environment, which is densely packed with wood particles (13). CBMs are known to increase the concentration of the enzymes in the vicinity of the substrate, but because of the high substrate loading in the gut (Fig. 1B), the need for a CBM to bind the enzyme to the cellulose surface has presumably been made redundant (Fig. S6 D and E) (14, 29). The absence of a CBM may also permit the retention of the enzyme after wood digestion by the animal rather than being lost with the excreted, processed biomass. Despite not possessing a CBM, the kinetic properties of the LqCel7B and its extent of processivity are broadly similar to other GH7 cellobiohydrolases.
An intriguing feature of LqCel7B is its ability to retain activity at high salt concentrations, a property that could lend itself to saccharification following or during biomass pretreatment with ionic liquids (ILs), which are strong enzyme-denaturing agents (46). For this reason, IL pretreated biomass requires rinsing in water before enzymatic hydrolysis. Alternatively, direct saccharification of IL pretreated biomass without separate cellulose recovery and wash has also been attempted but enzyme activity was dependent on IL dilution (47). It has been shown that a GH5 enzyme from a halophilic archeon, with a synonymous pattern of acidic surface charges to LqCel7A, demonstrated significant IL compatibility (48). In either approach, LqCel7B and other marine cellulases could represent promising candidate enzymes. In summary, LqCel7B is a unique functionally characterized animal family GH7 enzyme, and the data presented in this report demonstrate unusual features that likely reflect the environment within which the enzyme operates.
Methods
Methods on protein extraction from the Limnoria digestive system and Western blot analysis, RNA extraction and reverse transcription quantitative PCR, biophysical analysis, enzyme kinetics, and HPAEC are discussed in SI Methods.
Expression and Purification.
LqCel7B was expressed in A. oryzae (kindly provided by Novozymes, Bagsvaerd, Denmark) and in A. niger culture supernantants. One hundred milliliters of the A. oryzae culture supernatant was sterile filtered and concentrated to 10 mL using a polyethersulfone Ultraconcentrator MWCO 10 kDa (Novagen) by centrifugation at 3,450 × g rpm at 4 °C. The concentrated fraction was then desalted and buffer exchanged into 20 mM Tris pH 8.0 containing 50 mM NaCl using a Zeba Desalt Spin column (Pierce Biotechnology). The desalted supernatant was purified through a 5-mL anion exchange Q-Trap column (GE Healthcare) using an Äkta 100 (GE Healthcare), equilibrated in 20 mM Tris pH 8.0, 50 mM NaCl. The enzyme was eluted with a 0.05–1 M NaCl gradient in the same buffer at a flow rate of 1 mL/min and the elution was monitored through absorbance at 280 nm. Eluted fractions containing absorbance peaks were analyzed by SDS/PAGE to confirm the presence of the recombinant protein. A. niger protein production protocol is described in SI Methods.
Enzyme Assays.
The standard concentration of the polymeric substrates in the reactions was 0.1% and 1% (wt/vol) PASC or CMC and 0.1% and 2% (wt/vol) Avicel. All reactions containing oligosaccharides were carried out at 30 °C for 4 h or, alternatively, for 1 h or 24 h for polysaccharides. All reactions were conducted in 100 mM citrate buffer pH 4.5 (for pNP- and cellooligomer substrates) or 5.0 (for polysaccharide substrates), 500 mM NaCl, and 1 µg (pNP- and cellooligomer reactions) or 10 µg (polysaccharide reactions) of purified LqCel7B. Enzyme activity was measured according to release of pNP ions at 405 nm or release of reducing sugars at 620 nm (49).
Crystallography.
Purified LqCel7B was concentrated to 15 mg/mL and crystallized by vapor diffusion in 0.2 M MgCl2, 0.1 M Bis⋅Tris pH 5.0 and 25% (wt/vol) PEG 3350 at 289 K. The crystals were cryoprotected in a solution of 25% (vol/vol) glycerol and cryocooled in liquid nitrogen before data collection. Cocrystallization was performed by incubating LqCel7B in a 1:1.5 molar ratio with cellopentaose. The crystals were grown by vapor diffusion in 0.1 M CH3CO2Na pH 4.5, 15% (wt/vol) PEG 6000 and 0.5 M CaCl2 at 289 K. The crystals were transferred to a cryoprotectant containing 6.25% (vol/vol) diethylene glycol, 6.25% (wt/vol) 2-Methyl-2,4-pentanediol (MPD), 18.75% (vol/vol) 1,2-propanediol and 6.25% (vol/vol) DMSO, and cryocooled in liquid nitrogen. Free protein crystals were soaked with 1:10 molar ratio of cellotriose for 90 min before transfer to the same cryoprotectant for cryocooling. Cocrystallization was performed in buffer containing up to a 20-fold molar excess of thiocellobiose with LqCel7B at a concentration of 15 mg/mL. Several rounds of micro- and macroseeding were used to generate larger crystals. The cryoprotectant was 6.25% (vol/vol) diethylene glycol, 6.25% (vol/vol) MPD, 6.25% (vol/vol) 1,2-propanediol, 12.5% (vol/vol) DMSO, 6.25% (vol/vol) glycerol and 6 mM 3-(1-pyridinio)-1-propanesulfonate and contained 5 mM thiocellobiose. Data were processed using either XDS and XSCALE (50) or MosFlm (51) and SCALA (52) and molecular replacement was carried out with Phaser (53) using the Talaromyces emersonii cellobiohydrolase IB (CBH IB) structure (PDB accession code: 1Q9H). Reiterative cycles of model building and refinement were carried out using COOT (54) and REFMAC (55) or Phenix (56), respectively. Figures were produced with the program PyMol with electrostatic surfaces calculated using DELPHI (57).
MD Simulations.
The 1.6-Å resolution apo LqCel7B structure was used to build the LqCel7B simulation input, and the HjCel7A 8CEL ligand was threaded into the active site via protein backbone alignment (38). The simulation setup followed the protocols outlined by Bu et al. (58) and Momeni et al. (44). MD simulations were conducted as described in detail in SI Methods. Briefly, the TIP3P potential was used for water (59) and the CHARMM force fields were used for the protein and carbohydrates (60–62). Periodic boundary conditions were applied and the particle mesh Ewald method was used to treat long-range electrostatics (63). MD simulations were conducted for 250 ns with a 2-fs time step.
Supplementary Material
Acknowledgments
We thank the staff at the Diamond Light Source for their assistance, Jo Diamond for SEM work, Clare Steele-King for critical reading of the manuscript, and Jerry Ståhlberg and Mats Sandgren for helpful discussions. This work was funded by Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/G016178/1. Visits between the research teams were supported by BBSRC–US Partnering Award BB/H531543/1. G.T.B., M.E.H., and C.M.P. acknowledge the US Department of Energy’s BioEnergy Technologies Office for funding. Computer time for this research was provided by the National Institute for Computational Sciences Kraken cluster under the National Science Foundation XSEDE Grant MCB090159 and by the National Renewable Energy Laboratory Computational Sciences Center supported by the Department of Energy, Energy Efficiency and Renewable Energy under Contract DE-AC36-08GO28308.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4GWA, 4HAP, 4HAQ, and 4IPM) and the sequence reported in this paper has been deposited in the GenBank database (accession no. KC776193).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301502110/-/DCSupplemental.
References
- 1.Somerville C. Cellulose synthesis in higher plants. Annu Rev Cell Dev Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- 2.Himmel ME, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 3.Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol. 2009;60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 4.Burton RA, Gidley MJ, Fincher GB. Heterogeneity in the chemistry, structure and function of plant cell walls. Nat Chem Biol. 2010;6(10):724–732. doi: 10.1038/nchembio.439. [DOI] [PubMed] [Google Scholar]
- 5.Chundawat SPS, Beckham GT, Himmel ME, Dale BE. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng. 2011;2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 6.Geddes CC, Nieves IU, Ingram LO. Advances in ethanol production. Curr Opin Biotechnol. 2011;22(3):312–319. doi: 10.1016/j.copbio.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Percival Zhang YH, Himmel ME, Mielenz JR. Outlook for cellulase improvement: Screening and selection strategies. Biotechnol Adv. 2006;24(5):452–481. doi: 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DB. Microbial diversity of cellulose hydrolysis. Curr Opin Microbiol. 2011;14(3):259–263. doi: 10.1016/j.mib.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Baldrian P, Valásková V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev. 2008;32(3):501–521. doi: 10.1111/j.1574-6976.2008.00106.x. [DOI] [PubMed] [Google Scholar]
- 10.Geib SM, et al. Lignin degradation in wood-feeding insects. Proc Natl Acad Sci USA. 2008;105(35):12932–12937. doi: 10.1073/pnas.0805257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe H, Tokuda G. Cellulolytic systems in insects. Annu Rev Entomol. 2010;55:609–632. doi: 10.1146/annurev-ento-112408-085319. [DOI] [PubMed] [Google Scholar]
- 12.Bayer EA, Lamed R, White BA, Flint HJ. From cellulosomes to cellulosomics. Chem Rec. 2008;8(6):364–377. doi: 10.1002/tcr.20160. [DOI] [PubMed] [Google Scholar]
- 13.King AJ, et al. Molecular insight into lignocellulose digestion by a marine isopod in the absence of gut microbes. Proc Natl Acad Sci USA. 2010;107(12):5345–5350. doi: 10.1073/pnas.0914228107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalak J, Kurašin M, Teugjas H, Väljamäe P. Endo-exo synergism in cellulose hydrolysis revisited. J Biol Chem. 2012;287(34):28802–28815. doi: 10.1074/jbc.M112.381624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruno M, Väljamäe P, Pettersson G, Johansson G. Inhibition of the Trichoderma reesei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnol Bioeng. 2004;86(5):503–511. doi: 10.1002/bit.10838. [DOI] [PubMed] [Google Scholar]
- 16.Schülein M. Enzymatic properties of cellulases from Humicola insolens. J Biotechnol. 1997;57(1-3):71–81. doi: 10.1016/s0168-1656(97)00090-4. [DOI] [PubMed] [Google Scholar]
- 17.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330(6001):219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 18.Quinlan RJ, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108(37):15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6(12):1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 20.Westereng B, et al. The putative endoglucanase PcGH61D from Phanerochaete chrysosporium is a metal-dependent oxidative enzyme that cleaves cellulose. PLoS ONE. 2011;6(11):e27807. doi: 10.1371/journal.pone.0027807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selig MJ, Knoshaug EP, Adney WS, Himmel ME, Decker SR. Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresour Technol. 2008;99(11):4997–5005. doi: 10.1016/j.biortech.2007.09.064. [DOI] [PubMed] [Google Scholar]
- 22.Igarashi K, Samejima M, Eriksson KEL. Cellobiose dehydrogenase enhances Phanerochaete chrysosporium cellobiohydrolase I activity by relieving product inhibition. Eur J Biochem. 1998;253(1):101–106. doi: 10.1046/j.1432-1327.1998.2530101.x. [DOI] [PubMed] [Google Scholar]
- 23.Lahjouji K, et al. Biochemical and molecular characterization of a cellobiohydrolase from Trametes versicolor. Appl Microbiol Biotechnol. 2007;75(2):337–346. doi: 10.1007/s00253-006-0824-5. [DOI] [PubMed] [Google Scholar]
- 24.Lee K-M, et al. Production and characterization of cellobiohydrolase from a novel strain of Penicillium purpurogenum KJS506. Appl Biochem Biotechnol. 2011;163(1):25–39. doi: 10.1007/s12010-010-9013-1. [DOI] [PubMed] [Google Scholar]
- 25.Ståhlberg J, et al. Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1996;264(2):337–349. doi: 10.1006/jmbi.1996.0644. [DOI] [PubMed] [Google Scholar]
- 26.Tuohy MG, et al. Kinetic parameters and mode of action of the cellobiohydrolases produced by Talaromyces emersonii. Biochim Biophys Acta. 2002;1596(2):366–380. doi: 10.1016/s0167-4838(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 27.Lee K-M, et al. Characterization of cellobiohydrolase from a newly isolated strain of Agaricus arvencis. J Microbiol Biotechnol. 2011;21(7):711–718. doi: 10.4014/jmb.1102.02001. [DOI] [PubMed] [Google Scholar]
- 28.Shin K, Kim YH, Jeya M, Lee J-K, Kim Y-S. Purification and characterization of a thermostable cellobiohydrolase from Fomitopsis pinicola. J Microbiol Biotechnol. 2010;20(12):1681–1688. [PubMed] [Google Scholar]
- 29.Zhang J, Moilanen U, Tang M, Viikari L. The carbohydrate-binding module of xylanase from Nonomuraea flexuosa decreases its non-productive adsorption on lignin. Biotechnol Biofuels. 2013;6(1):18. doi: 10.1186/1754-6834-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voutilainen SP, et al. Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol Bioeng. 2008;101(3):515–528. doi: 10.1002/bit.21940. [DOI] [PubMed] [Google Scholar]
- 31.Szijártó N, et al. Hydrolysis of amorphous and crystalline cellulose by heterologously produced cellulases of Melanocarpus albomyces. J Biotechnol. 2008;136(3-4):140–147. doi: 10.1016/j.jbiotec.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Fox JM, Levine SE, Clark DS, Blanch HW. Initial- and processive-cut products reveal cellobiohydrolase rate limitations and the role of companion enzymes. Biochemistry. 2012;51(1):442–452. doi: 10.1021/bi2011543. [DOI] [PubMed] [Google Scholar]
- 33.von Ossowski I, et al. Engineering the exo-loop of Trichoderma reesei cellobiohydrolase, Cel7A. A comparison with Phanerochaete chrysosporium Cel7D. J Mol Biol. 2003;333(4):817–829. doi: 10.1016/s0022-2836(03)00881-7. [DOI] [PubMed] [Google Scholar]
- 34.Horn SJ, et al. Costs and benefits of processivity in enzymatic degradation of recalcitrant polysaccharides. Proc Natl Acad Sci USA. 2006;103(48):18089–18094. doi: 10.1073/pnas.0608909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurašin M, Väljamäe P. Processivity of cellobiohydrolases is limited by the substrate. J Biol Chem. 2011;286(1):169–177. doi: 10.1074/jbc.M110.161059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ståhlberg J, Johansson G, Pettersson G. Trichoderma reesei has no true exo-cellulase: All intact and truncated cellulases produce new reducing end groups on cellulose. Biochim Biophys Acta. 1993;1157(1):107–113. doi: 10.1016/0304-4165(93)90085-m. [DOI] [PubMed] [Google Scholar]
- 37.Uzcategui E, Ruiz A, Montesino R, Johansson G, Pettersson G. The 1,4-beta-D-glucan cellobiohydrolases from Phanerochaete chrysosporium. I. A system of synergistically acting enzymes homologous to Trichoderma reesei. J Biotechnol. 1991;19(2-3):271–285. doi: 10.1016/0168-1656(91)90064-3. [DOI] [PubMed] [Google Scholar]
- 38.Divne C, Ståhlberg J, Teeri TT, Jones TA. High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1998;275(2):309–325. doi: 10.1006/jmbi.1997.1437. [DOI] [PubMed] [Google Scholar]
- 39.Grassick A, et al. Three-dimensional structure of a thermostable native cellobiohydrolase, CBH IB, and molecular characterization of the cel7 gene from the filamentous fungus, Talaromyces emersonii. Eur J Biochem. 2004;271(22):4495–4506. doi: 10.1111/j.1432-1033.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- 40.Parkkinen T, Koivula A, Vehmaanperä J, Rouvinen J. Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding. Protein Sci. 2008;17(8):1383–1394. doi: 10.1110/ps.034488.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ubhayasekera W, Muñoz IG, Vasella A, Ståhlberg J, Mowbray SL. Structures of Phanerochaete chrysosporium Cel7D in complex with product and inhibitors. FEBS J. 2005;272(8):1952–1964. doi: 10.1111/j.1742-4658.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 42.Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3(9):853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 43.White A, Rose DR. Mechanism of catalysis by retaining beta-glycosyl hydrolases. Curr Opin Struct Biol. 1997;7(5):645–651. doi: 10.1016/s0959-440x(97)80073-5. [DOI] [PubMed] [Google Scholar]
- 44.Momeni MH, et al. Structural, biochemical, and computational characterization of the glycoside hydrolase family 7 cellobiohydrolase of the tree-killing fungus Heterobasidion irregulare. J Biol Chem. 2013;288(8):5861–5872. doi: 10.1074/jbc.M112.440891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delgado-García M, Valdivia-Urdiales B, Aguilar-González CN, Contreras-Esquivel JC, Rodríguez-Herrera R. Halophilic hydrolases as a new tool for the biotechnological industries. J Sci Food Agric. 2012;92(13):2575–2580. doi: 10.1002/jsfa.5860. [DOI] [PubMed] [Google Scholar]
- 46.Vancov T, Alston AS, Brown T, McIntosh S. Use of ionic liquids in converting lignocellulosic material to biofuels. Renew Energy. 2012;45:1–6. [Google Scholar]
- 47.Kamiya N, et al. Enzymatic in situ saccharification of cellulose in aqueous-ionic liquid media. Biotechnol Lett. 2008;30(6):1037–1040. doi: 10.1007/s10529-008-9638-0. [DOI] [PubMed] [Google Scholar]
- 48.Zhang T, et al. Identification of a haloalkaliphilic and thermostable cellulase with improved ionic liquid tolerance. Green Chem. 2011;13(8):2083–2090. [Google Scholar]
- 49.Jue CK, Lipke PN. Determination of reducing sugars in the nanomole range with tetrazolium blue. J Biochem Biophys Methods. 1985;11(2-3):109–115. doi: 10.1016/0165-022x(85)90046-6. [DOI] [PubMed] [Google Scholar]
- 50.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leslie AGW. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, Vol 26.
- 52.Bailey S. Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rocchia W, Alexov E, Honig B. Extending the applicability of the nonlinear Poisson-Boltzmann equation: Multiple dielectric constants and multivalent ions. J Phys Chem B. 2001;105(28):6507–6514. [Google Scholar]
- 58.Bu L, et al. Probing carbohydrate product expulsion from a processive cellulase with multiple absolute binding free energy methods. J Biol Chem. 2011;286(20):18161–18169. doi: 10.1074/jbc.M110.212076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 60.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 61.Guvench O, et al. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J Chem Theory Comput. 2011;7(10):3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guvench O, et al. Additive empirical force field for hexopyranose monosaccharides. J Comput Chem. 2008;29(15):2543–2564. doi: 10.1002/jcc.21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Essmann U, et al. A smooth particle mesh ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.