Abstract
Tumor suppressors known to date impede cancer growth by arresting the cell cycle or promoting apoptosis. Here we show that unphosphorylated human STAT5A functions as a tumor suppressor capable of repressing multiple oncogenes via heterochromatin formation. Unphosphorylated STAT5A binds to heterochromatin protein 1α (HP1α) and stabilizes heterochromatin. Expressing unphosphorylated STAT5A or HP1α inhibits colon cancer growth in mouse xenograft models. Transcriptome profiling shows that expressing an unphosphorylatable STAT5A has similar effects to overexpressing HP1α in global gene expression. Notably, the majority of the genes commonly repressed by unphosphorylated STAT5A and HP1α have been implicated in cancer development. Finally, down-regulation, somatic mutations, and deletions of STAT5 genes are found in certain human cancers. These results suggest that unphosphorylated STAT5A may epigenetically suppress tumor growth by promoting heterochromatin formation.
Keywords: JAK/STAT, Drosophila, fluorescence recovery after photobleaching (FRAP), transcription
Heterochromatin plays a role in chromosomal compaction and transcriptional silencing and is emerging as a mechanism of tumor suppression. Certain tumor suppressors, such as breast cancer type 1 susceptibility protein (BRCA1) and the retinoblastoma protein (RB), have been shown to promote heterochromatin formation at specific loci or maintain the stability of constitutive heterochromatin (1–4). On the other hand, oncogenic JAK disrupts heterochromatin formation in both Drosophila and human cells (5, 6). In addition, a reduction in the levels of heterochromatin protein 1 (HP1), the major component and a determinant of heterochromatin (7), is associated with human cancer progression, including colon and breast cancers and leukemia (8). The target genes normally repressed by heterochromatin, however, have not been systematically investigated.
Previous studies using a Drosophila leukemia model have demonstrated a noncanonical mode of JAK/STAT signaling, in which the unphosphorylated form of STAT is localized in heterochromatin in association with HP1; STAT activation (by phosphorylation) causes its dispersal from heterochromatin, leading to HP1 delocalization and heterochromatin loss (9–11). Moreover, unphosphorylated STAT and heterochromatin are essential for maintaining genomic stability and counteracting aging in Drosophila (12, 13). To investigate whether unphosphorylated STAT in mammals also promotes heterochromatin formation, and whether this constitutes a mechanism of tumor suppression, we examined the interaction between human STAT5A and HP1α (also known as “chromobox protein homolog 5,” or CBX5) in human cells, tested their effects on tumorigenesis in mouse xenograft models, and examined their effects on global gene transcription by means of expression profiling.
Results
STAT5A and HP1α Physically Interact.
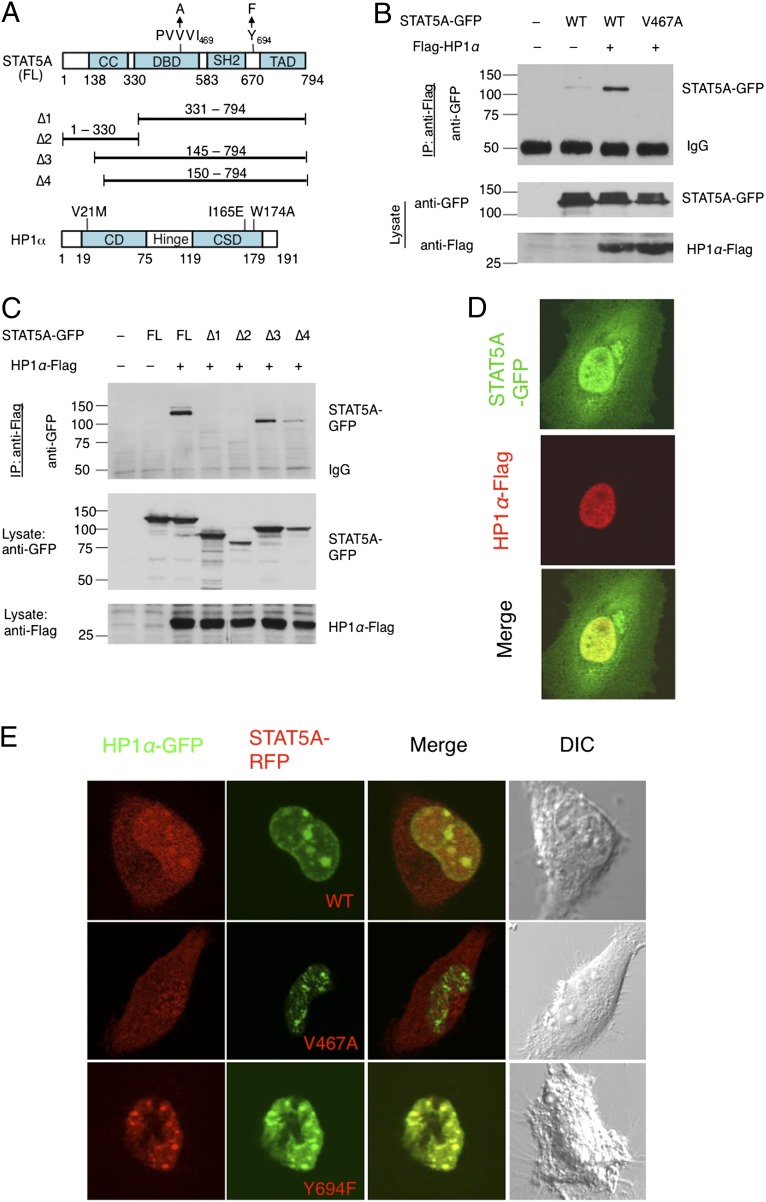
To test whether human STAT5A physically interacts with human HP1α, we used a series of plasmids to express HP1α-Flag and wild-type or mutant GFP-tagged STAT5A (Fig. 1A) in human embryonic kidney 293 (HEK293T) cells, and performed coimmunoprecipitation (co-IP) experiments. STAT5A contains an HP1-binding motif, PxVxI, which is conserved in Drosophila STAT92E (9) (Fig. S1) and is important for binding to HP1 (9, 14). We found that wild-type STAT5A-GFP, but not STAT5AV467A-GFP, which contains a V to A mutation in its HP1-binding motif (Fig. 1A), was coimmunoprecipitated with HP1α-Flag (Fig. 1B). This result suggests that STAT5A might interact directly with HP1α, because their co-IP depended on an intact HP1 binding motif in STAT5A.
Fig. 1.
Physical interaction between STAT5A and HP1α. (A) Schematic representations of human STAT5A and HP1α constructs used in the assays are shown. The coiled-coil (CC), DNA-binding domain (DBD), Src-homolog 2 (SH2), and transactivation domain (TAD) motifs of full-length (FL) STAT5A are shown. Single letter representations of amino acid residues are shown for the HP1-binding motif and the tyrosine phosphorylation (Y) site; their substitutions are shown with arrows. Four truncated STAT5A constructs (∆1 to ∆4) are shown as solid lines, with corresponding amino acid numbers. HP1α contains a chromodomain (CD), hinge region, and chromo shadow domain (CSD). Amino acid substitutions are shown. (B and C) HP1α-Flag and STAT5A-GFP or its truncated or mutant variants were cotransfected into 293T cells in the combinations indicated. HP1α-Flag was immunoprecipitated with anti-Flag antibodies and was subjected to SDS/PAGE. The gel was blotted with anti-GFP to detect the presence of STAT5A-GFP in the immunoprecipitates. (D) STAT5A-GFP and HP1α-FLAG were transiently cotransfected into HeLa cells. The cells were fixed and immunostained with anti-Flag (red) and imaged with a confocal microscope. Note that STAT5A-GFP (green) is distributed in both the cytoplasm and nucleus and that the nuclear STAT5A-GFP is colocalized with HP1-Flag. (E) Stable HeLa cell lines expressing STAT5AWT-RFP, STAT5AY694F-RFP, or STAT5AV467A-RFP (red) were generated by lentiviral infection. HP1α-GFP (green) was transfected into these stable cell lines and the cells were imaged using confocal microscopy. Representative images of each stable cell line are shown. Note that strong colocalization of HP1α-GFP with STAT5AY694F-RFP and with STAT5AWT-RFP, but not with STAT5AV467A-RFP, was detected.
Because HP1 is almost exclusively localized in the nucleus (7), whereas STAT proteins shuttle between nuclear and cytoplasmic compartments (15), we tested the importance of STAT5A nuclear translocation for its interaction with HP1α, using a series of truncated STAT5A-GFP variants (∆1–∆4) (15) (Fig. 1A) for co-IP experiments. We found that the STAT5A-GFP variants that were missing the 145–150 nuclear localization sequence (∆1 and ∆4) or the C terminus (∆2) did not coimmunoprecipitate with HP1α-Flag (Fig. 1C, lanes 4, 5, and 7), whereas the full-length and ∆3 versions of STAT5A-GFP, which retained both the HP1-binding and nuclear localization motifs, were coimmunoprecipitated with HP1α-Flag (Fig. 1C, lanes 3 and 6). These data suggest that both an intact HP1-binding motif and nuclear localization of STAT5A are required for its interaction with HP1α.
To understand which regions of HP1α are important for interacting with STAT5A, we carried out co-IP experiments using wild-type STAT5A-GFP and different HP1α-Flag variants with substitutions of amino acid residues V21, I165, or W174, which are important for H3K9me binding, HP1α dimerization, and interaction with the PxVxL motif, respectively (14). We found that the interaction of STAT5A-GFP with wild-type HP1α-Flag was much stronger than with any of the three HP1α mutants (Fig. S2A). This result suggests that STAT5A might have a higher affinity for HP1α when it is a chromatin-bound dimer. Taken together, these results suggest that STAT5A, via its PVVVI motif, may physically associate with HP1α.
Unphosphorylated STAT5A Colocalizes with HP1α in the Nucleus.
For further confirmation that STAT5A and HP1α physically associate, we examined the subcellular localization of the STAT5A and HP1α proteins by immunostaining of HeLa cells cotransfected with STAT5A-GFP and HP1α-Flag. We found that STAT5A-GFP is predominantly localized in the nucleus, consistent with a previous report (15) (Fig. 1D, Top). HP1α-Flag, however, was exclusively detected in the nucleus (Fig. 1D, Middle), also consistent with previous reports (7). Strikingly, we found that most of the nuclear portion of STAT5A-GFP was colocalized with HP1α-Flag (Fig. 1D, Bottom). Colocalization of endogenous STAT5A and HP1α were also found in untransfected HeLa cells (Fig. S2B).
To investigate whether colocalization of STAT5A and HP1α depends on their physical interaction, we examined the subcellular localization of the mutant STAT5AV467A, which does not coimmunoprecipitate with HP1α (Fig. 1B). We found that, compared with the wild-type STAT5A-RFP, which colocalized with HP1α-GFP (Fig. 1E, Top), STAT5AV467A-RFP was evenly distributed in the cytoplasmic and nuclear compartments and was not obviously colocalized with HP1α-GFP (Fig. 1E, Middle). These results confirm the importance of the PVVVI motif in the colocalization of STAT5A and HP1α and, additionally, suggest that association with HP1α is important in the localization of STAT5A to heterochromatic regions.
To investigate whether the STAT5A that was colocalized with HP1α was phosphorylated (active) or unphosphorylated (transcriptionally inactive), we generated stable HeLa cell lines expressing HP1α-GFP and the unphosphorylatable mutant STAT5AY694F-RFP. We found that, strikingly, STAT5AY694F-RFP was almost exclusively localized to the nucleus and strongly colocalized with HP1α-GFP (Fig. 1E, Bottom). These results are consistent with previous reports of nuclear localization of unphosphorylatable STAT mutants (15) and their association with HP1 and heterochromatin (9) and suggest that, similarly to its Drosophila counterpart, unphosphorylated human STAT5A has a propensity for physically associating with HP1α in the nucleus.
Unphosphorylated STAT5A Promotes Heterochromatin Formation and Stabilizing Heterochromatic HP1α.
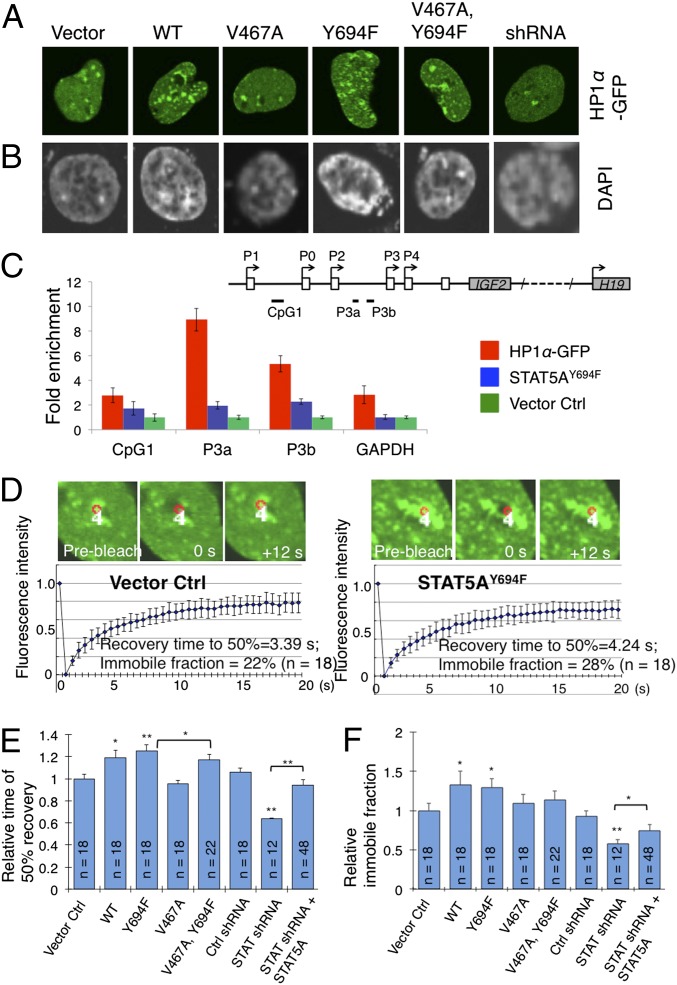
We next investigated the biological significance of the physical association of unphosphorylated STAT5A with HP1α. We first examined heterochromatin levels by observing the number of HP1 or heterochromatic foci in the nucleus of HeLa cells stably expressing STAT5A variants and HP1α-GFP (Fig. 2A). Compared with the vector control, HeLa cells expressing wild-type STAT5A exhibited an increase in the number of HP1α-GFP–positive heterochromatic foci (Fig. 2A and Fig. S3). An even larger increase in the number of HP1α-GFP foci was observed in cells expressing the unphosphorylatable STAT5AY694F mutant (Fig. 2A and Fig. S3). However, expressing either the STAT5AV467A mutant or the STAT5AV467A,Y694F double mutant, which does not associate with HP1α (Fig. 1B), did not result in significant changes in the number of heterochromatic foci compared with the vector control (Fig. 2 A and B and Fig. S3). We obtained similar results upon staining of the same series of stable cell lines with the DNA dye DAPI, another measure of heterochromatin (Fig. 2B and Fig. S3). These data are consistent with the idea that unphosphorylated STAT5A promotes heterochromatin formation by binding to HP1α.
Fig. 2.
Unphosphorylated STAT5A promotes heterochromatin formation. (A and B) Stable HeLa cell lines overexpressing the vector control, RFP-tagged STAT5AWT, STAT5AV467A, STAT5AY694F, or STAT5AV467A,Y694F were generated by lentiviral infection. The samples were analyzed using confocal microscopy. (A) HP1α-GFP or (B) DAPI staining was used to visualize heterochromatin foci. Representative images of each stable cell line are shown. Quantifications are shown in Fig. S4. (C) DLD-1 cells overexpressing the vector control (vector), HP1α, or STAT5AY694F were subjected to ChIP analysis in triplicate with anti-H3K9me3 antibodies. The fold enrichment method was used for the calculation, with normalization to the vector control. Error bars represent the SEM. (Inset, Top) Schematic representation of the organization of the human IGF2/H19 genomic region showing the positions of the IGF2 promoters (P0–P4), CpG islands (black lines), exons (open boxes), and ORF (gray boxes). Solid lines below the genomics region indicate PCR primers used in ChIP assays. (D) HeLa cells stably expressing a vector control or the indicated STAT5A variants were transfected with HP1α-GFP and were analyzed for FRAP using a confocal microscope. The red circle delineates the area in heterochromatin (HP1α-GFP focus) that was bleached with a laser beam. White number 4 indicates sample number. The recovery of fluorescence in the bleached area was imaged over time. The indicated number of FRAP experiments was averaged and the mean FRAP curves are shown. Error bars indicated SDs. Two FRAP experiments are shown. Additional experiments are shown in Fig. S5. (E and F) Quantifications of FRAP experiments, showing the mean recovery times of HP1α-GFP to 50% of before-bleached fluorescence intensity in different cell lines, and the mean immobile fractions of HP1α-GFP for different cell lines. n represents the number of cells of the indicated cell line on which FRAP was done. Error bars are SDs. *P < 0.05 and **P < 0.01 (Student t test), respectively, compared with the vector control or with the indicated cell type.
Next, we examined the effect of unphosphorylated STAT5A on heterochromatin formation at the promoter level. Using HeLa cells and chromatin immunoprecipitation (ChIP) assays, we analyzed how unphosphorylated STAT5A affects heterochromatin formation in the promoter region of the well-characterized human insulin-like growth factor 2 (IGF2) gene. In cultured human cells, IGF2 expression is governed mainly by promoter 3 (P3) (16). HeLa cells stably overexpressing HP1α, STAT5AY694F, or the vector control were subjected to ChIP assays using antibodies against the heterochromatin marker H3K9me3. HP1α overexpression resulted in a ninefold enrichment of H3K9me3 in the DNA sequences around the two C-G dinucleotide (CpG) islands located upstream of the IGF2 P3 promoter (Fig. 2C). Overexpression of unphosphorylated STAT5AY694F also resulted in significant enrichment of H3K9me3 in the same regions (Fig. 2C), albeit at lower levels than HP1α overexpression. These results support the idea that unphosphorylated STAT5A promotes heterochromatin formation.
To gain insight into how unphosphorylated STAT5A affects heterochromatin dynamics, we turned to fluorescence recovery after photobleaching (FRAP) (17, 18) (Fig. S4 A and B) to quantitatively determine heterochromatin stability by measuring the diffusion kinetics of HP1α-GFP molecules localized in heterochromatin. In control cells, fluorescence recovery in heterochromatic regions was rapid and reached 50% of the prebleach intensity in 3.4 s (Fig. 2D), consistent with previous reports (17, 18). The immobile fraction, defined as the portion of fluorescence that was not recovered after photobleaching, and which reflects the portion of HP1α-GFP stably associated with heterochromatin, comprised about 20% of total HP1α-GFP (Fig. 2D).
Expressing STAT5AWT resulted in a 20% slower recovery of HP1α-GFP flourescence in photobleached heterochromatin, whereas expressing the unphosphorylatable mutant STAT5AY694F slowed recovery even more, with the 50% recovery time slowed by ∼30% (Fig. 2 D and E). A slower fluorescence recovery rate indicates reduced mobility of HP1α-GFP, or an increase in the stability of its localization in heterochromatin. Expressing the STAT5AV467A mutant did not significantly affect recovery time after photobleaching (Fig. 2E and Fig. S4C), consistent with the finding that STAT5AV467A does not interact with HP1α (Fig. 1). The STAT5AV467A mutation, however, moderately suppressed the effects of STAT5AY694F on HP1α-GFP mobility, such that the double mutant STAT5AV467A,Y694F was not as effective as STAT5AY694F in slowing down the recovery of HP1α-GFP fluorescence (Fig. 2E), consistent with the fluorescent microscopic studies (Fig. 1). Corresponding effects were found for the immobile fraction (Fig. 2F). These results suggest that unphosphorylated STAT5A inhibits HP1α mobility, thereby stabilizing its localization in heterochromatin.
In HeLa cells stably expressing shRNA targeting STAT5A, HP1α-GFP fluorescence in heterochromatin was much reduced, and FRAP analysis indicated an overall increase in mobility of HP1α-GFP compared with its mobility in control cells receiving a scrambled shRNA (Fig. 2E and Fig. S4C). Moreover, shRNA knockdown of STAT5A decreased the immobile fraction, indicating that less HP1α-GFP was stably associated with heterochromatin (Fig. 2F and Fig. S4C).
Taken together, the observed dependence of HP1α-GFP mobility on STAT5A levels, as shown in the FRAP experiments, substantiates a fundamental role for STAT5A in stabilizing HP1α localization in heterochromatin and supports the idea that unphosphorylated STAT5A plays an essential role in promoting heterochromatin formation.
Unphosphorylated STAT5A Suppresses Growth of Mouse and Human Colon Cancer Cells in Vivo.
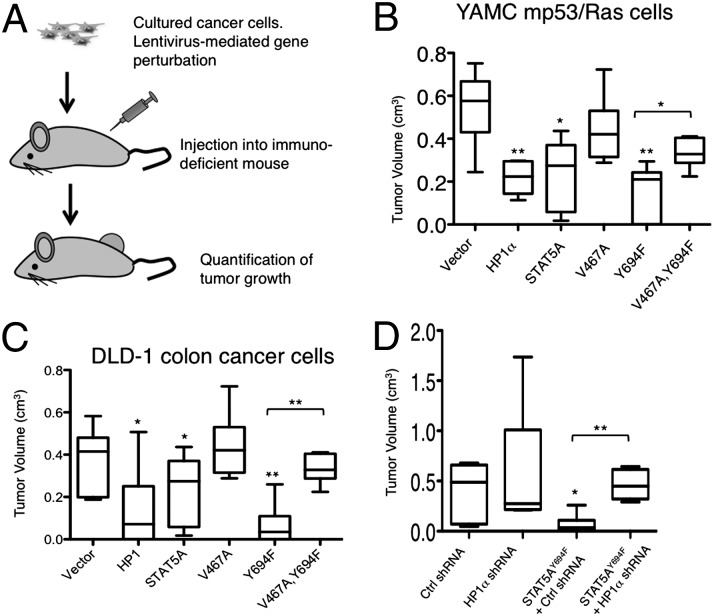
To investigate the effects of unphosphorylated human STAT5A on the growth of mouse and human cancer cells, we used mouse xenograft models. We first investigated the effects of unphosphorylated STAT5A on in vivo tumor growth of young adult murine colon (YAMC) cells simultaneously expressing mutant p53175H and oncogenic H-RasV12 (referred to as mp53/Ras), two cooperating oncogenic mutations that drive cellular transformation (19, 20). By means of lentiviral infection, we generated YAMCmp53/Ras cell lines stably overexpressing human HP1α, STAT5AWT, STAT5AV467A, STAT5AY694F, or STAT5AV467A,Y694F. We then assessed the ability of these cells to grow tumors after s.c. injection into immunodeficient (nude) mice (Fig. 3A). Using these xenograft models, we found that overexpression of HP1α or STAT5AY694F significantly reduced the ability of YAMCmp53/Ras cells to grow tumors, whereas overexpressing STAT5AWT partially reduced tumor growth (Fig. 3B and Fig. S5). The effects of STAT5A molecules on tumor growth closely correlate with their effects on heterochromatin levels. Expressing STAT5AV467A had only a small effect on tumor growth (Fig. 3B and Fig. S5), consistent with its inability to bind to HP1α and its minimal effects on heterochromatin levels (Figs. 1 and 2). When the V467A mutation was combined with the Y694F mutation in the double mutant STAT5AV467A,Y694F, however, the V467A mutation neutralized the effects of the Y694F mutation on tumor growth; that is to say, the effect of the double mutant STAT5AV467A,Y694F on tumor growth resembled that of STAT5AV467A more than that of STAT5AY694F (Fig. 3B and Fig. S5). Similar results were found using human colon cancer DLD-1 cells (Fig. 3C and Fig. S5). These results are consistent with the idea that binding to HP1 and stabilizing heterochromatin are essential for the tumor suppressive effect of unphosphorylated STAT5A.
Fig. 3.
Unphosphorylated STAT5A and HP1α inhibit tumor growth in vivo. (A) Schematic illustration of the tumor cell xenograft assay. (B–D) Box plots of tumor volumes four weeks (Right) after s.c. injection of the indicated derivatives of YAMC mp53/Ras cells (B) or human colon cancer DLD-1 cells (C and D) expressing the vector control (vector), HP1α, STAT5AWT, STAT5AV467A, STAT5AY694F, or STAT5AV467A&Y694F (B and C) or expressing shRNA-HP1α, scramble shRNA, or shRNA-HP1α along with STAT5AY694F (D) are shown. Each box shows the range of the second and third quartiles of tumor volumes. The line in the box indicates the median tumor volume. The bars (“whiskers”) represent the largest and smallest tumors. Six injections (n = 6) were done for each of the indicated transgene and cell line combination. n represents the number of injections for the indicated cell line. *P < 0.05 and **P < 0.01 (Student t test), respectively, compared with the vector control or with the indicated cell type.
Interestingly, the in vitro proliferation rates of YAMCmp53/Ras and DLD-1 cells were unaffected by expression of the human HP1α, STAT5AWT, STAT5AV467A, STAT5AY694F, or STAT5AV467A,Y694F genes (Fig. S6), suggesting that these transgenes did not cause changes in cell proliferation or survival in vitro. Thus, greater selective pressure may be exerted by the environment in vivo than in vitro, perhaps due to a limited availability of nutrients in vivo than in culture media. Measurements of mRNA levels indicated that expression levels of the different transgenes were similar in tumor cells before injection and at harvest (Fig. S6), suggesting that the difference in tumor size was not due to differences in transgene expression.
To test whether HP1α mediates the tumor suppressive function of unphosphorylated STAT5A, we used shRNA to reduce the levels of HP1α in DLD-1 cells that stably express STAT5AY694F. Analysis of tumor growth in the s.c. xenograft model revealed that expressing STAT5AY694F no longer suppressed tumor growth when HP1α expression was knocked down (Fig. 3D and Fig. S5D). Taken together, these data are consistent with the idea that unphosphorylatable STAT5A suppresses colon cancer development via binding to HP1α and promoting heterochromatin formation.
Unphosphorylated STAT5A and HP1α Down-Regulate Genes Implicated in Human Cancers.
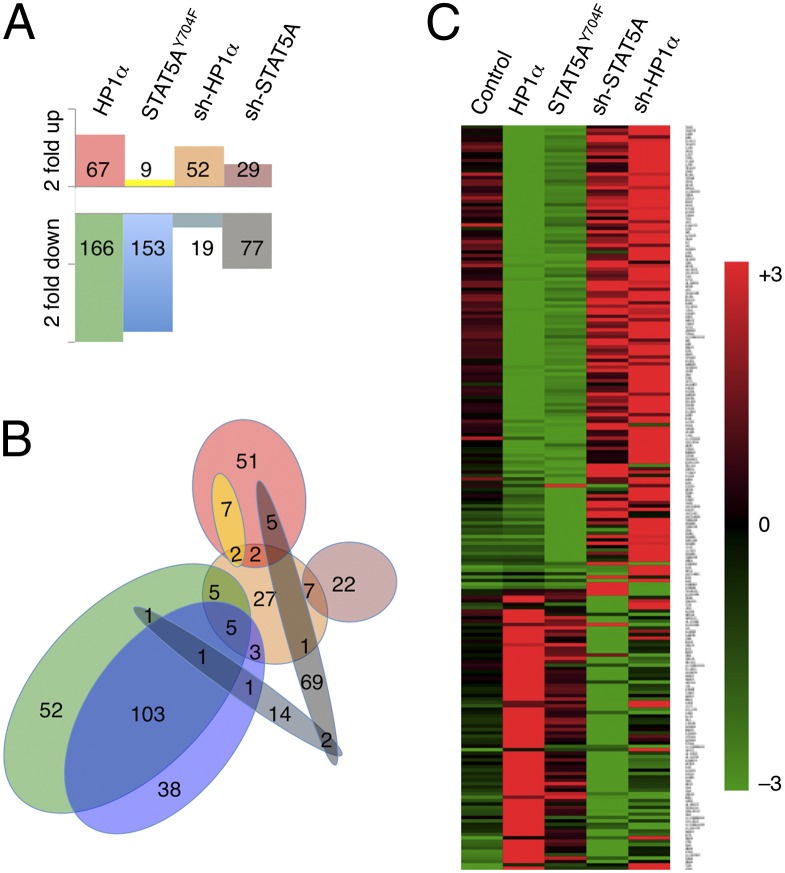
To determine to what extent overexpression of STAT5AY694F promotes heterochromatin formation as opposed to interfering with the canonical transcriptional activity of the JAK/STAT pathway, we used transcriptome profiling to investigate global gene expression under conditions of STAT5AY694F, HP1α, sh-STAT5A, or sh-HP1α transgene expression. To this end, we performed microarray studies on human colon cancer (DLD-1) cell lines stably expressing STAT5AY694F, HP1α, shRNA-STAT5A, shRNA-HP1α, or control constructs (empty vector or shRNA-GFP). We found that overexpressing STAT5AY694F or HP1α, or knocking down endogenous STAT5A or HP1α, caused only a small percentage of genes (<1%) to change their expression levels by more than twofold, whereas expression levels of the vast majority of genes (>95%) remained unchanged (within a ±1.5 fold range) (Fig. 4A and Table S1). When STAT5AY694F was overexpressed, for instance, 153 genes were down-regulated by at least twofold and 9 genes up-regulated by at least twofold, constituting 0.3% and 0.02% of the 47,324 transcripts on the Illumina Human gene chip, respectively (Fig. 4A and Table S1).
Fig. 4.
Gene expression profiles of DLD-1 cells with altered HP1α and STAT5A levels. Total RNA isolated from DLD-1 cells stably expressing the vector control, HP1α, STAT5AY694F, shRNA-GFP, shRNA-STAT5A, or shRNA-HP1α were subjected to microarray analysis using an Illumina HumanHT-12 v4 Expression BeadChip, which provides coverage of 47,324 transcripts in the human transcriptome. (A) The number of transcripts whose expression levels were up- or down-regulated by at least twofold relative to the control (out of a total 47,324 transcripts) is shown. (B) Venn diagram showing the relationship between sets of transcripts delineated in A. Note that the sets of transcripts up- or down-regulated due to expression of HP1α largely overlap with the corresponding sets resulting from expression of STAT5AY694F, but are different from those resulting from shRNA knockdown of STAT5A. (C) Heat map representation of transcripts whose expression levels differ by at least threefold when cells expressing different transgenes are compared. Note that when cells expressing HP1α are compared with cells expressing STAT5AY694F, many genes show concordant changes, whereas in cells where STAT5A or HP1α was knocked down by sh-RNA expression, many of these same genes show changes in the opposite direction.
To identify genes that were regulated in common by expression of the different transgenes, we analyzed those genes whose expression levels were either altered by at least twofold, relative to control as a result of transgene expression (method 1; Fig. 4 A and B), or whose expression levels differed by at least threefold when cells expressing different transgenes were compared (method 2; Fig. 4C). The two methods yielded qualitatively similar results, with method 1 identifying different numbers of genes for each cell type (Fig. 4A) and method 2 selecting a total of 220 genes for all cell types (Fig. 4C). Strikingly, the set of genes whose expression levels changed by at least twofold as a result of STAT5AY694F expression overlapped, to a large extent, with the corresponding set resulting from HP1α expression (Fig. 4B). Concordant changes were also seen in genes whose expression differed by threefold or more across different cell types, as analyzed by the second method (Fig. 4C). These results suggest that STAT5AY694F and HP1α regulate genes in common, and that knockdown of STAT5A had similar effects on global gene transcription to knockdown of HP1α.
In contrast, when comparing knockdown of endogenous STAT5A with expression of the STAT5AY694F transgene, the sets of genes whose expression levels changed by at least twofold did not, for the most part, overlap (Fig. 4 B and C). In cases where the same gene was found in both sets, the changes in transcript levels were in opposite directions (Fig. 4C, columns 3 and 4). These data demonstrate that expression of STAT5AY694F and knocking down of endogenous STAT5A had opposite effects on transcription, ruling out the possibility that STAT5AY694F interferes with the transcriptional activity of endogenous STAT5A.
We next analyzed the genes regulated by STAT5A and HP1α in common. Strikingly, we found that 70% (86/123) of the genes commonly down-regulated by STAT5A and HP1α have been previously shown to be overexpressed in or are drivers of human cancer development (Table S2). About half of these genes (41/86) have been implicated in colorectal cancers (Fig. S7A and Table S2), including lymphoid enhancer-binding factor 1 (LEF1), a prognostic biomarker for colorectal liver metastases (21). This group includes additional known oncogenes or drivers of cancer development, such as C-X-C motif chemokine 5 (CXCL5) (22), forkhead box Q1 (FOXQ1) (23), matrix metallopeptidase 7 (MMP7) (24), transforming growth factor beta-1 (TGFB1) (25), and tyrosine-protein kinase KIT (26). This group also includes the imprinted gene H19, which is normally repressed by heterochromatin formation and DNA methylation, and whose derepression has been implicated in cancer development, including colorectal cancers (27). In contrast, expression of genes involved in cell cycle progress or apoptosis was not significantly changed in these cells (Fig. S7 A and B), suggesting that STAT5A and HP1α inhibit tumor growth by repressing cancer-promoting genes and not by affecting cell cycle or apoptotic genes.
Loss of STAT5 and HP1α in Human Cancers.
To understand the magnitude of STAT5A and HP1α alterations in human cancers, we used the cBio Cancer Genomics Portal (28) to interrogate cancer genomic data for alterations of STAT5 and HP1α in large numbers of tumor samples from cancer studies. Because STAT5A and STAT5B are highly similar in sequence and are closely linked in the human genome, we analyzed both STAT5A and STAT5B for possible coregulation. We thus searched in the tumor samples for information on three genes, STAT5A, STAT5B, and HP1α (also known as CBX5), with regard to mutations, putative copy-number alterations, and changes in mRNA expression levels.
We searched cancer databases in the cBio Cancer Genomics Portal (http://cbioportal.org) and found that in a collection of 216 prostate cancer samples generated by Memorial Sloan-Kettering Cancer Center, nearly 40% had one or all of the three genes down-regulated (Fig. S7C), suggesting that down-regulation of STAT5 and/or HP1α might contribute to prostate cancer initiation or progression. Patient disease-free survival data indicate that patients with altered expression of STAT5A, STAT5B, or HP1α had worse prognosis (Fig. S7D). In a collection of 563 serous ovarian cancer samples from The Cancer Genome Atlas, cBio Cancer Genomics Portal (http://cbioportal.org), about 18% had altered expression of STAT5A, STAT5B, or HP1α, with four cases of putative homozygous deletions of both STAT5 genes and four cases of somatic mutations (Fig. S7C). These data are consistent with the notion that STAT5 and HP1α are tumor suppressors, whose loss contributes to cancer development.
Discussion
We have investigated the functional significance of the interaction between human STAT5A and HP1α in human cells, mouse models, and patient tumor samples. Our biochemical studies have recapitulated the studies done previously in Drosophila (5, 9, 12) and demonstrated that unphosphorylated human STAT5A physically interacts and colocalizes with HP1α (Fig. 1). Using FRAP analysis of HeLa cells, we have demonstrated that unphosphorylated STAT5A plays an important role in HP1α dynamics, stabilizing its localization in heterochromatin and promoting heterochromatin formation (Fig. 2). Moreover, we have shown that unphosphorylated STAT5A inhibits the growth of murine and human colon cancer cells when transplanted into immunodeficient mice (Fig. 3). Our gene expression profiling studies have indicated that unphosphorylated STAT5A and HP1α share common targets for transcriptional regulation, many of which have been implicated in cancer development (Fig. 4, Table S2, and Fig. S7). Finally, bioinformatics studies of human cancer samples indicate that STAT5 and HP1α are down-regulated in certain types of cancers (Fig. S7). Based on these studies, we propose that unphosphorylated STAT5A stabilizes heterochromatin and suppresses tumor growth. These effects are likely to be epigenetic, mediated by the physical interaction of STAT5A and HP1α.
As a transcription factor, activated or phosphorylated STAT has been implicated in many cancers (29). On the other hand, dephosphorylation of STAT has been shown to have a tumor suppressive effect, which has previously been attributed to lack of STAT transcriptional activity or to a dominant-negative effect on STAT transcription (29). In this study, we have provided substantial evidence that unphosphorylated STAT5A suppresses the development of colon cancer via epigenetic gene regulation, mediated through the physical interaction of STAT5A and HP1α. Two key results support this conclusion. One is the finding that many of the effects of unphosphorylated STAT5A were mitigated by knockdown of HP1α (Fig. 3E). The other is the finding that expression of an unphosphorylatable mutant STAT5A affected a set of genes that largely overlapped with those affected by HP1α expression, but not with those affected by knockdown of endogenous STAT5A (Fig. 4). This is consistent with our previous results in Drosophila showing that expressing a nonphosphorylatable STAT mutant has an effect opposite to that of STAT knockdown with regard to the sensitivity of tissues to low-dose irradiation (12). Additional evidence supporting the idea that unphosphorylated STAT5A is involved in epigenetic regulation has come from the finding that unphosphorylated STAT5A enhances heterochromatinization of the imprinted IGF2 allele in HeLa cells. Loss of IGF2 and H19 imprinting has been reported to be a biomarker for increased risk of colorectal cancer (27).
Moreover, our studies suggest that not only the exogenous STAT5AY694F, but also endogenous STAT5A, likely in its unphosphorylated state, functions to promote heterochromatin formation and suppress tumor growth. Knockdown of endogenous STAT5A had the opposite effect on genes regulated by HP1α expression (Fig. 4C, columns 2 and 4), consistent with the idea that endogenous STAT5A and HP1α control many target genes in common. In addition, the observation that expression of STAT5AWT, which remains unphosphorylated in the absence of activating signals, affects tumor growth in a way similar to STAT5AY694F or HP1α (Fig. 3), is consistent with a role for endogenous, unphosphorylated STAT5A in suppression of tumor growth. Previous reports have shown that STAT5A functions as a tumor suppressor in T-cell lymphomas (30) and that STAT5 deficiency causes cancer development in mice (31). These data may be informed by our finding that STAT5A plays a role in heterochromatin formation.
Interestingly, it has been reported that unphosphorylated STAT1 and STAT3 regulate expression of target genes that are distinct from those controlled by their canonical pathways (32). It remains to be determined whether unphosphorylated STAT1 and STAT3 also share transcriptional targets with HP1α and whether they promote heterochromatin formation. In any case, our results support the idea that unphosphorylated STAT5A promotes heterochromatin formation in general.
Strikingly, we found that the majority of the common target genes repressed by unphosphorylated STAT5A and HP1α are known to be overexpressed in various human cancers, some of which are “drivers” of cancer development (Fig. S7A and Table S2). In contrast, genes involved in cell cycle progression or apoptosis were not significantly changed when levels of STAT5A and HP1α were altered in DLD-1 cells (Fig. S7 A and B), suggesting that STAT5A and HP1α as tumor suppressors are distinct from other known tumor suppressors, such as p53 and Rb, whose major functions include inhibiting cell cycle progression and/or promoting cell death. Thus, unphosphorylated STAT5A and HP1α may represent a unique class of tumor suppressors that are capable of, in addition to protecting genome stability (12), suppressing transcription of multiple cancer-promoting genes. This finding should lead to unique therapeutic options for treating human cancers.
Materials and Methods
Cell Culture, Transfection, and Plasmid DNA.
YAMC cells expressing both p53175H and HRasV12 (referred to as mp53/Ras) were cultured as previously described (19, 20). HeLa cells, HEK-293T cells, and DLD-1 human colon cancer cells were maintained at 37 °C and with 5% (vol/vol) CO2 in water-jacketed, humidified incubators. Genetically perturbed cells were derived by means of lentiviral infection with virus containing the appropriate cDNA or shRNA expression constructs. Plasmid DNA and additional details are described in SI Materials and Methods.
Immunostaining, Immunoprecipitation, and Western Blotting.
Protocols and antibodies used for immunofluorescence and Western blotting and primers used for ChIP are described in SI Materials and Methods.
FRAP.
See SI Materials and Methods and Fig. S5 for FRAP procedures.
Xenograft Assays, Cell Proliferation Measurements, and Flow Cytometry.
Details of tumor and cell growth measurements are described in SI Materials and Methods.
Quantitative Real-Time PCR and Microarray Studies.
Primers and protocols for quantitative PCR and microarray studies are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Laurel Newman for assistance with mouse xenograft studies; Dr. Nancy Reich (Stony Brook University) for STAT5A plasmids; Dr. Mark S. Lechner (Simon Fraser University) and Lori Wallrath (University of Iowa) for HP1α plasmids; and Drs. Lynn Marquat, Jiyong Zhao, Chris Proschel, Lei Xu, and Shawn Murphy (University of Rochester) for various cell lines and DNA plasmids. This work is supported by National Institutes of Health Grants R01CA131326 (to W.X.L.) and RO1CA138249 (to H.L.), and a Leukemia & Lymphoma Society Research Scholar grant (to W.X.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221243110/-/DCSupplemental.
References
- 1.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412(6846):561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalo S, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7(4):420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 3.Isaac CE, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26(9):3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Q, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477(7363):179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi S, et al. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38(9):1071–1076. doi: 10.1038/ng1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson MA, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461(7265):819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grewal SI, Elgin SC. Heterochromatin: New possibilities for the inheritance of structure. Curr Opin Genet Dev. 2002;12(2):178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 8.Dialynas GK, Vitalini MW, Wallrath LL. Linking Heterochromatin Protein 1 (HP1) to cancer progression. Mutat Res. 2008;647(1-2):13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi S, et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10(4):489–496. doi: 10.1038/ncb1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18(11):545–551. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown S, Zeidler MP. Unphosphorylated STATs go nuclear. Curr Opin Genet Dev. 2008;18(5):455–460. doi: 10.1016/j.gde.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 12. Yan SJ, Lim SJ, Shi S, Dutta P, Li WX (2011) Unphosphorylated STAT and heterochromatin protect genome stability. FASEB J 25(1):232–241. [DOI] [PMC free article] [PubMed]
- 13.Larson K, et al. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8(1):e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiru A, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23(3):489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer J, Reich NC. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. FASEB J. 2008;22(2):391–400. doi: 10.1096/fj.07-8965com. [DOI] [PubMed] [Google Scholar]
- 16.Begley LA, et al. CXCL5 promotes prostate cancer progression. Neoplasia. 2008;10(3):244–254. doi: 10.1593/neo.07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299(5607):721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 18.Festenstein R, et al. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science. 2003;299(5607):719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- 19.Xia M, Land H. Tumor suppressor p53 restricts Ras stimulation of RhoA and cancer cell motility. Nat Struct Mol Biol. 2007;14(3):215–223. doi: 10.1038/nsmb1208. [DOI] [PubMed] [Google Scholar]
- 20.McMurray HR, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453(7198):1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozano J, et al. Deficiency of kinase suppressor of Ras1 prevents oncogenic ras signaling in mice. Cancer Res. 2003;63(14):4232–4238. [PubMed] [Google Scholar]
- 22.Brose MS, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- 23.Bos JL. ras oncogenes in human cancer: A review. Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 24.Sternberg DW, Gilliland DG. The role of signal transducer and activator of transcription factors in leukemogenesis. J Clin Oncol. 2004;22(2):361–371. doi: 10.1200/JCO.2004.10.124. [DOI] [PubMed] [Google Scholar]
- 25.Kirschmann DA, et al. Down-regulation of HP1Hsalpha expression is associated with the metastatic phenotype in breast cancer. Cancer Res. 2000;60(13):3359–3363. [PubMed] [Google Scholar]
- 26.Tian Q, Frierson HF, Jr, Krystal GW, Moskaluk CA. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol. 1999;154(6):1643–1647. doi: 10.1016/S0002-9440(10)65419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneda A, Feinberg AP. Loss of imprinting of IGF2: A common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65(24):11236–11240. doi: 10.1158/0008-5472.CAN-05-2959. [DOI] [PubMed] [Google Scholar]
- 28. Cerami E, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404. [DOI] [PMC free article] [PubMed]
- 29.Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol. 2012;30(9):1005–1014. doi: 10.1200/JCO.2010.31.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Wang HY, Liu X, Wasik MA. STAT5A is epigenetically silenced by the tyrosine kinase NPM1-ALK and acts as a tumor suppressor by reciprocally inhibiting NPM1-ALK expression. Nat Med. 2007;13(11):1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- 31.Hosui A, et al. Loss of STAT5 causes liver fibrosis and cancer development through increased TGF-beta and STAT3 activation. J Exp Med. 2009;206(4):819–831. doi: 10.1084/jem.20080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18(4):443–451. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.