Abstract
Sleep is regulated by homeostatic mechanisms, and the low-frequency power in the electroencephalogram (delta power) during non-rapid eye movement sleep reflects homeostatic sleep need. Additionally, sleep is limited by circadian and environmentally influenced arousal. Little is known, however, about the underlying neural substrates for sleep homeostasis and arousal and about the potential link between them. Here, we subjected C57BL/6 mice to 6 h of sleep deprivation using two different methods: gentle handling and continual cage change. Both groups were deprived of sleep to a similar extent (>99%), and, as expected, the delta power increase during recovery sleep was quantitatively similar in both groups. However, in a multiple sleep latency test, the cage change group showed significantly longer sleep latencies than the gentle handling group, indicating that the cage change group had a higher level of arousal despite the similar sleep loss. To investigate the possible biochemical correlates of these behavioral changes, we screened for arousal-related and sleep need-related phosphoprotein markers from the diencephalon. We found that the abundance of highly phosphorylated forms of dynamin 1, a presynaptic neuronal protein, was associated with sleep latency in the multiple sleep latency test. In contrast, the abundance of highly phosphorylated forms of N-myc downstream regulated gene 2, a glial protein, was increased in parallel with delta power. The changes of these protein species disappeared after 2 h of recovery sleep. These results suggest that homeostatic sleep need and arousal can be dissociated behaviorally and biochemically and that phosphorylated N-myc downstream regulated gene 2 and dynamin 1 may serve as markers of homeostatic sleep need and arousal, respectively.
Keywords: phosphoproteomics, two-dimensional difference gel electrophoresis
Sleep–wakefulness is continuously regulated by circadian and homeostatic mechanisms. In addition, multiple factors affect the level of arousal, including emotional, environmental, and physiological influences (1). During non-rapid eye movement (NREM) sleep, the homeostatic sleep need is correlated with the power in the delta wave band (i.e., 1–4 Hz) in the electroencephalogram (EEG). Delta power is augmented in proportion to previous wakefulness time and is dissipated during sleep. These findings imply that homeostatic sleep need is regulated by the durations of prior sleep and wakefulness (2). On the other hand, the level of arousal is inversely related to the likelihood of falling asleep and can be measured in terms of the sleep latency in the multiple sleep latency test (MSLT) (3). Because prolonged waking increases sleep need in addition to lowering the level of arousal, sleep latency often varies inversely with EEG delta power expressed during sleep (3). However, the level of arousal can be affected independently of time spent awake (for example, with stressful or exciting experiences during waking), implying that, under conditions of identical sleep deprivation time, the sleep latency may vary with the experience during sleep deprivation. Sleep need can thus be dissociated from the level of arousal when the duration of waking is kept constant. Depression, anxiety, or traumatic stress can induce a high degree of arousal and cause insomnia (4). Additionally, providing an incentive to sleep shortens sleep latency in normal human subjects (5). In rodent studies, changing the home cage is a routine manipulation that evokes emotional, behavioral, and physiological arousal (6), resulting in wakefulness. These reports indicate that, although the level of arousal can be affected by the duration of a prior waking episode, it can additionally be modulated by behavioral conditions. However, it is not known how the level of arousal is related to sleep need when the duration of lost sleep is the same. No studies have focused on the relation between the degree of arousal and NREM delta power independently of the duration of prior waking (or sleep loss).
In this study, to assess the relationship between the level of arousal and homeostatic sleep need, we hypothesized that the degree of arousal can be different, even under conditions of similar sleep need. To test this hypothesis, we used two different sleep deprivation (SD) treatments, gentle handling (GH) and cage change (CC), and compared the degree of arousal as assessed by the sleep latency and the sleep need as assessed by NREM delta power that occurred in response to the SD. We then examined, through a phosphoproteomic approach, biochemical markers associated with arousal and homeostatic sleep need, respectively, to search for biochemical correlates of these behavioral parameters.
Results
Sleep Latency and NREM Delta Power Can Be Dissociated After Sleep Deprivation.
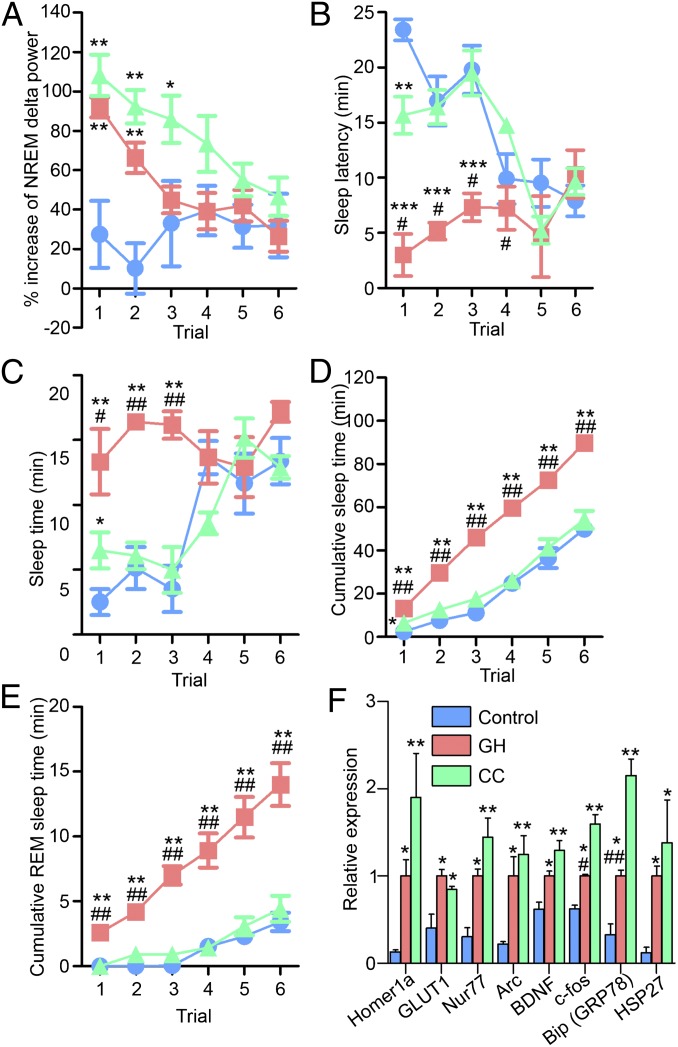
Mice were divided into three groups (n = 4–7): no manipulation (control), and SD by two different kinds of arousing stimuli, GH and CC (see Materials and Methods for details). The resulting behavioral responses were assessed using the MSLT and EEG sleep state monitoring with power analysis of the delta frequency band (1–4 Hz) during NREM sleep. The MSLT is used to determine the degree of arousal as indexed by the sleep latency, i.e., the time to fall asleep (3). Thus, a low (high) level of arousal shortens (lengthens) sleep latency. Here, mice were disturbed for 5 min and then left to sleep freely for 25 min, and this 30-min cycle was repeated six times (3) (Fig. S1). To avoid possible compounding effect of habituation to the GH or CC manipulations used during the SD period, a third method for preventing sleep, i.e., orbital shaker (7), was used during the MSLT. EEG monitoring during the 6-h SD period indicated that both GH and CC groups were similarly deprived of sleep by more than 99% whereas the control group was awake for 25% of this time (Table 1). Consistent with previous reports (3, 7, 8), both SD groups showed significant increases in NREM delta power compared with the control group in MSLT trials 1 and 2 (Fig. 1A). However, despite similar wakefulness times during SD, sleep latency, as an index of the level of arousal, differed markedly between the GH and CC groups (Fig. 1B). As expected, the GH group showed significantly shorter sleep latency times than the control group during MSLT trials 1–3. In contrast, sleep latency times of the CC group were similar to the control group and significantly longer than those of the GH group. In accordance with the sleep latency results, at the end of the MSLT, the GH group had significantly increased total sleep time compared with the control and CC groups (Fig. 1 C and D). Recorded sleep time in the CC group was similar to that in the control group. It is worth noting that the CC group showed a trend of higher delta power compared with the GH group (Fig. 1A). This difference is likely in response to increased wake time observed in the CC group in trials 1–4 compared with the GH group (Fig. 1 C and D). Thus, under these conditions, the change in sleep latency, indicating the level of arousal, does not correlate with the previous duration of wakefulness. In addition, the cumulated REM sleep rebound time of the GH group was significantly longer than those of the control and CC groups (Fig. 1E).
Table 1.
Sleep time during ZT0–6 with or without sleep deprivation (n = 4–7)
| Experimental group | Sleep time |
|
| Min ± SEM | % ± SEM | |
| Control | 297.0 ± 10.3 | 75.0 ± 2.9 |
| GH | 1.2 ± 0.3 | 0.3 ± 0.1 |
| CC | 1.9 ± 1.2 | 0.5 ± 0.3 |
Fig. 1.
Dissociation of delta power and sleep latency following SD by two different methods. NREM delta power (A), sleep latency (B), and total and REM sleep times (C–E) during MSLT following 6-h SD. Mice were deprived of sleep from ZT0–6 by either GH or CC. The control group was allowed to sleep freely during ZT0–6. MSLT, performed from ZT6–9, was composed of six repeats of 30-min trials (i.e., a 5-min period of forced wakefulness followed by a 25-min spontaneous sleep period). EEG/EMG monitoring was performed continuously from ZT0–9. (F) Previously described SD-inducible transcripts were increased in correlation association with delta power as an index of homeostatic sleep need after 6-h SD at ZT6. RNA was extracted from whole brain at ZT6. Values were normalized to Cyclophilin B mRNA levels and expressed relative to the means of the GH group. In Figs. 1–3, data represent means ± SEM, *P < 0.05, **P < 0.01 compared with the control group; #P < 0.05, ##P < 0.01 between the GH and CC groups by ANOVA followed by Tukey’s test (n = 3–7). Arc, activity-regulated cytoskeletal-associated protein; BDNF, brain-derived neurotrophic factor; GLUT1, glucose transporter 1; HSP27, heat shock protein 27.
Levels of SD-Inducible Transcripts Reflect Sleep Need.
The very different levels of arousal between the GH and CC groups led us to search for potential biomarker transcripts as correlates of sleep latency time. We examined specific transcripts that are known to increase in the brains of sleep-deprived rodents, as shown in previous microarray or real-time quantitative PCR (qPCR) studies (8–10). Primer pairs for qPCR are described in Table S1. We found that the mRNAs we examined were all significantly up-regulated in both SD groups compared with the control group. Among these mRNAs, we did not find any candidates that exhibited selective changes with the level of arousal (Fig. 1F); rather, all of the screened mRNAs changed in association with NREM delta power in response to SD. In particular, c-fos and bingding immunoglobulin protein/glucose-regulated protein 78 (BiP/GRP78) mRNA levels were significantly higher in the CC groups compared with GH. This difference may reflect the higher level of NREM delta power in the CC group (Fig. 1A). Alternatively, considering the fact that these mRNAs are stress-induced, their levels may be associated with the apparent levels of stress (see Fig. 4C).
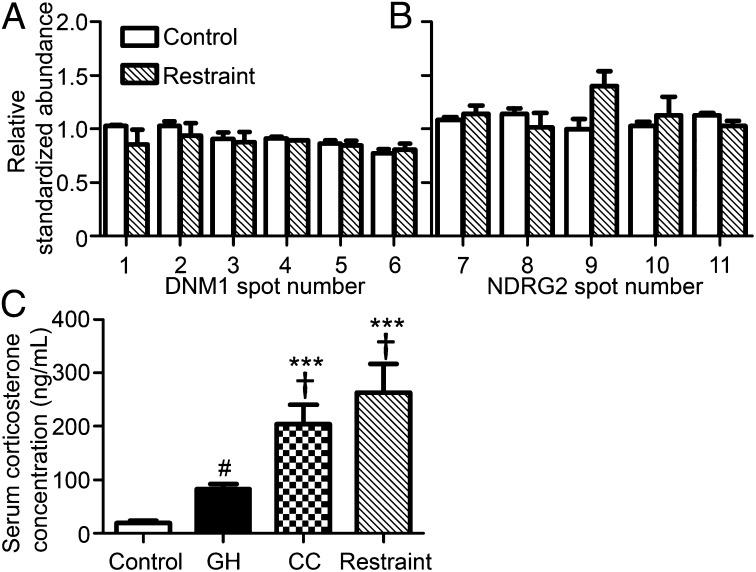
Fig. 4.
Stress stimuli do not affect the abundances of phospho-DNM1 (A) and NDRG2 (B) proteins. Mice in the restraint group were subjected to a 30-min restraint stress from ZT5.5–6.0. Serum corticosterone levels in the GH, CC, and restraint groups showed significant increases compared with the control group (C). All samples were collected at ZT6. Phosphoprotein-enriched samples were purified from the diencephalon. Data represent means + SEM. ***P < 0.001 compared with the control group, and †P < 0.05 compared with the GH group by ANOVA followed by Tukey’s test; #P < 0.05 between the control and GH groups by t test (n = 6–12).
Screening of Phosphoprotein Markers for Level of Arousal and Sleep Need.
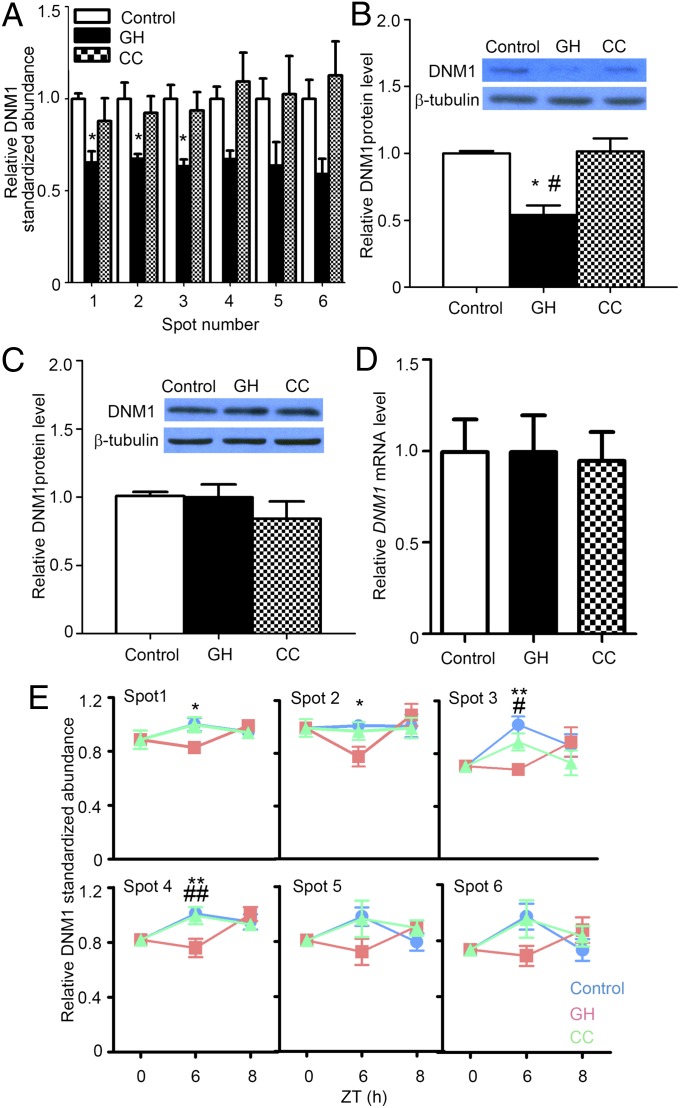
To screen for additional biochemical markers associated with the degree of arousal indexed by sleep latency and homeostatic sleep need indexed by delta power, we turned to proteomic analysis of brain samples using 2D difference gel electrophoresis (2D-DIGE). Because 2D-DIGE–based analysis of total proteins did not yield informative results (despite some previous reports; refs. 11–13), we then tried 2D-DIGE of phosphorylated proteins (Fig. S2). Phosphorylated proteins were enriched from diencephalon extracts from the control, GH, and CC groups using phosphoprotein purification columns (Fig. S3); 2D-DIGE and subsequent protein identification revealed that abundances of six protein spots for dynamin 1 (DNM1) (Table 2) were significantly decreased only in the GH group at Zeitgeber time (ZT) 6 (immediately after SD), associated with sleep latency as an index of degree of arousal (Fig. 2A). The levels of these DNM1 spots returned to baseline after 2 h of recovery sleep in the GH group (Fig. 2E). To confirm that these spots represent phosohorylated forms of DNM1, we treated the phosphoprotein-enriched samples with λ-phosphatase and determined target spot positions between the original and dephosphorylated samples in 2D-DIGE (Fig. S4). As expected, the DNM1 spots 1–4 disappeared in the λ-phosphatase-treated sample (Fig. S4B). Western blots of phosphorylated protein samples confirmed that phospho-DNM1 proteins were decreased in the GH group (Fig. 2B) although the level of total DNM1 proteins (both phosphorylated and unphosphorylated forms) did not appreciably change (Fig. 2C). To further confirm the identity of these DNM1 spots, we performed immunoprecipitation analysis using a monoclonal anti-DNM1 antibody that preferentially precipitates highly phosphorylated DNM1 (Fig. S5A). The immunoprecipitates were subjected to Western blots with another DNM1 antibody, confirming the decrease in the GH group compared with the control or CC groups (Fig. S5B). The level of DNM1 mRNA did not change in any of the three groups (Fig. 2D).
Table 2.
Results of protein identificaton by nano-LC/MS/MS
| Spot no. | Protein identification | Accession no. (NCBI database) | Calculated MW/pI | Mascot score |
| 1, 2 | DNM1 | NP_034195.2 | 97.3/6.73 | 2001 |
| 3, 4 | DNM1 | NP_034195.2 | 97.3/6.73 | 1875 |
| 5, 6 | DNM1 | NP_034195.2 | 97.3/6.73 | 370 |
| 7 | NDRG2 isoform1 | NP_038892.1 | 40.8/5.23 | 306 |
| 8 | NDRG2 isoform1 | NP_038892.1 | 40.8/5.23 | 279 |
| 9 | NDRG2 isoform2 | NP_001139431.1 | 39.2/5.94 | 291 |
| 10 | NDRG2 isoform2 | NP_001139431.1 | 39.2/5.94 | 261 |
| 11 | NDRG2 isoform2 | NP_001139431.1 | 39.2/5.94 | 250 |
NCBI, National Center for Biotechnology Information.
Fig. 2.
Abundance of phospho-DNM1 in diencephalon negatively associates with the degree of arousal indexed by sleep latency. (A) Abundances of phospho-DNM1 species at ZT6, expressed relative to the average of control group. (B and C) Verification of 2D-DIGE results by Western blots. Western blots confirmed decreased DNM1 in the phosphoprotein sample from the GH group (B) but no changes in total DNM1 protein in the three groups (C). Values were normalized to β-tubulin and expressed relative to the control. (D) DNM1 mRNA levels at ZT6. The DNM1 mRNA level was measured by qPCR and normalized to Cyclophilin B mRNA level. (E) Effect of recovery sleep on phospho-DNM1 abundance as determined by 2D-DIGE. Following SD, either by GH or CC from ZT0–6, all mice were allowed to sleep freely for 2 h from ZT6–8. After 2 h of recovery sleep, phospho-DNM1 amounts in the GH group returned to baseline by ZT8. In the control and CC groups, there was not significant change in phospho-DNM1 amounts during the time course of experiments.
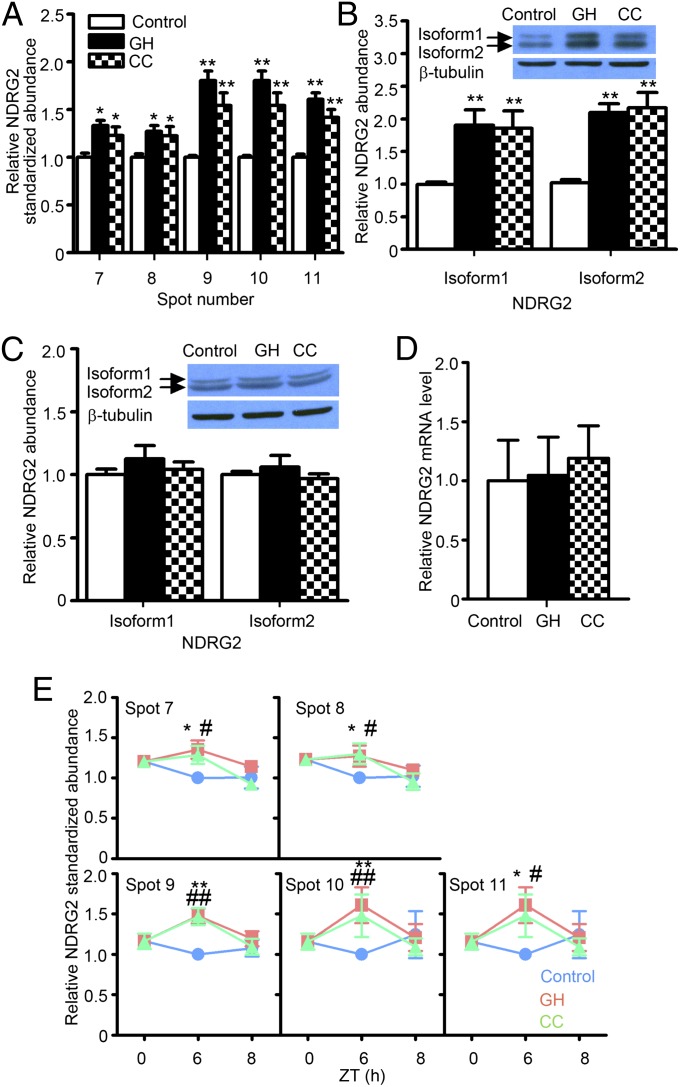
In the same set of diencephalic phosphoprotein samples, the abundance of each of the five protein spots representing N-myc downstream regulated gene 2 (NDRG2) was found to be similarly increased in both the GH and CC samples at ZT6 (Fig. 3A and Table 2). The intensities of these NDRG2 spots returned to baseline levels after 2 h of recovery sleep in both GH and CC groups (Fig. 3E). The 2D-DIGE comparison of λ-phosphatase-treated and untreated samples confirmed that all of the NDRG2 spots that were increased in the SD groups were highly phosphorylated forms (Fig. S4C). Western blots of phosphoprotein samples confirmed that phospho-NDRG2 proteins were increased in both the GH and CC groups (Fig. 3B) although the level of total NDRG2 proteins did not change (Fig. 3C). Furthermore, the level of NDRG2 mRNA did not change in either GH or CC groups (Fig. 3D). Taken together, these results indicate that the abundances of highly phosphorylated DNM1 and NDRG2 forms reflect biochemical changes in the brain that are associated with the level of arousal and the homeostatic sleep need, respectively.
Fig. 3.
Prolonged wakefulness accompanies increase of NDRG2 hyperphosphorylation in the diencephalon. Same sets of protein samples were used in Figs. 2 and 3. (A) Five phospho-NDRG2 spots were significantly increased after 6 h of SD in GH and CC groups, as determined by 2D-DIGE and protein identification (Table 2). This increase in NDRG2 in the SD groups returned to control levels after 2 h of recovery sleep at ZT8 (E). NDRG2 appeared in two distinct bands of isoform 1 (spots 7 and 8) and isoform 2 (spots 9–11) (B and C). Western blots indicated that the phosphorylated forms, but not the total amount of either isoform of NDRG2, were elevated in both SD groups (B and C). The NDRG2 mRNA was unchanged by SD (D).
Effects of Stress on Phosphorylated DNM1 and NDRG2 Abundances.
SD inevitably causes stress (14–18). Several stress response genes and corticosterone concentrations as a stress marker have been shown to be increased in the brains and sera, respectively, of sleep-deprived rodents (8–10, 14–18). We tested the effect of physical restraint stress on the levels of our phospho-DNM1 and phospho-NDRG2 spots on 2D-DIGE. A significant increase of serum corticosterone levels was observed in the mice subjected to the restraint stress, as well as in the CC group and, to a lesser degree, in the GH group (Fig. 4C). However, our target DNM1 and NDRG2 spots failed to show any changes after the restraint stress (Fig. 4 A and B). Thus, although mice in the CC and GH groups were “stressed” during SD as measured by corticosterone levels, stress per se did not influence the phosphorylation levels of DNM1 and NDRG2 proteins. Therefore, stress and stress-induced release of corticosterone is unlikely to be a major cause for the DNM1 and NDRG2 phosphorylation changes observed in sleep-deprived brains.
Discussion
The present study has shown that the amplitude of the sleep-homeostatic response of increased delta power following SD is independent of the means of SD (either CC or GH) when the amount of lost sleep remains similar. This observation is consistent with previous studies showing a direct correlation between waking duration and delta power expression during the ensuing NREM sleep episode, even when different methods of SD were used (3, 7, 8). However, we found that the means for inducing SD had a significant effect on the level of subsequent arousal. After SD by CC, sleep latency remained almost at the level of non–sleep-deprived mice, but delta power was increased to a similar degree to that of mice sleep-deprived by GH. These results indicate a differential effect of CC on arousal and homeostatic sleep need. Arousal, as assessed by the sleep latency, can be manipulated independently of the homeostatic sleep response. We also determined biochemical correlates of these behavioral changes. We found that the abundances of hyperphosphorylated DNM1 proteins were selectively related to the level of arousal; i.e., reduced levels were noted only in the GH group expressing shorter sleep latencies. In contrast, the levels of hyperphosphorylated NDRG2 spots were increased in both the GH and CC groups, and thus more closely associated with homeostatic sleep need.
Although GH is widely used for SD studies, the details vary according to the protocol adopted (3, 4, 8, 9). In this study, we specifically avoided the use of novel objects or other methods that might elicit emotional stimuli, locomotion, or voluntary arousal in the GH group (19). During GH SD, we continuously observed the behavior of each mouse and touched or tapped the cage gently to disturb them if they adopted a presleep posture such as starting to recline or lowering their heads to the floor of the cage. For the voluntary awake model, we performed cage changes once an hour to keep the mice awake by using their instinctive, spontaneous exploration of novel environments (4). Although a single cage change has been shown to keep mice awake for about 1.5–2.0 h (4), we also visually monitored the CC group throughout the 6-h SD period.
Although the underlying mechanisms remain unknown, we have shown here that two methods of SD produce differences in subsequent levels of arousal, sleep rebound, and phospho-DNM1 amounts. It has been reported that SD combined with fasting or stress can also lead to distinct patterns of sleep architecture in flies and rats (20, 21). Moreover, flies mutated with cycle exhibited disrupted sleep homeostasis as well as attenuated learning when deprived of sleep by starvation (20). A1 adenosine receptor knockout mice show a reduced delta-power rebound and deficient working memory after SD (22). These findings indicate that different waking experiences, or different qualities of waking (even with similar amounts of lost sleep), can produce appreciable differences in subsequent sleep and cognition.
The apparent dissociation between the level of arousal and homeostatic sleep need, together with the associated biochemical changes in the brain, led us to speculate that nonhomeostatic regulatory factors can induce changes in the level of arousal. Consistent with this concept, during insomnia induced by the stress of olfactory cues, when arousal and waking time are increased, both arousal-related and sleep-promoting neurons are activated (4). This result indicates that, under some circumstances at least, arousal can overcome homeostatic and circadian sleep drives. Indeed, as noted above, the CC group showed high levels of arousal and did not fall asleep despite high homeostatic sleep need. Recent pharmacological and gene-targeting studies suggest possible mechanisms involved. Dopamine antagonists inhibit the sleep latency increase caused by cage changes (23). Similarly, dopamine D2 receptor knockout mice do not exhibit longer sleep latencies after exposure to new environments (23, 24). Furthermore, mice with histamine deficiency also fail to exhibit prolonged sleep latencies during CC manipulation (25). This latter result indicates an orexinergic influence of arousal under these circumstances because the histamine system subserves much of the orexin-induced arousal (26). Further studies are required to determine the role of these neuromodulators in supporting arousal.
DNM1 phosphorylation is decreased in association with the level of arousal rather than homeostatic sleep need. DNM1 is a phosphorylated neural protein and a GTPase enzyme present in most presynaptic terminals (27). Phosphorylation of DNM1 generally inhibits neuronal endocytosis (28). Our findings thus suggest a connection between the DNM1-mediated regulation of presynaptic endocytosis and the level of arousal. Furthermore, we have shown that the level of hyperphosphorylated forms of NDRG2 protein is increased in the diencephalic samples from mice with increased homeostatic sleep need. NDRG2 is an astrocytic protein (29) of which the cellular function is poorly characterized. NDRG2 may be a substrate for a number of protein kinases, and our preliminary data indicate that the hyper-phosphorylated NDRG2 spots are recognized by antibodies against Akt substrates as well as by those against PKC substrates (Fig. S6). Although detailed mechanisms remain unknown, our findings support the emerging concept of glial involvement in sleep regulation (30).
SD is inevitably associated with the stress response mediating activation of the hypothalamic–pituitary–adrenal axis and the systemic release of glucocorticoids (14–18). Previous studies report a link between stress exposure, EEG delta power modulation, and the expression of sleep-related marker transcripts (31, 32). Indeed, corticosterone levels indicated that the GH and CC conditions were both stressful compared with controls. However, we found that restraint stress itself had no appreciable effect on the level of hyperphosphorylated DNM or NDRG2 proteins. Furthermore, there was no appreciable association between the stress levels as measured by serum corticosterone and the levels of the hyperphosphorylated marker proteins in the control, GH, and CC groups (Figs. 2, 3, and 4C).
In summary, the waking experience during SD can affect subsequent sleep in association with either the change in level of arousal or sleep need. Further studies, for example examining cognitive, metabolic, and immunological sequelae, are now required to understand the consequences of this effect in more detail.
Materials and Methods
Further details are described in SI Materials and Methods. All procedures were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center (UTSW) and were carried out in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Male C57BL/6 mice older than 10 wk were individually housed and used for all experiments. EEG/electromyography (EMG) implantation, monitoring of behavioral states (awake, NREM, and REM), and calculation of delta power were performed as described (7). SD was performed during ZT0–6 by GH or CC methods. Subsequently, MSLT and recovery sleep were performed from ZT6–9 and ZT6–8, respectively. Mice were subjected to restraint stress from ZT5.5–6 for 30 min. Phosphoprotein-enriched samples were extracted using a PhosphoProtein Purification kit (Qiagen) from the diencephalon. Protein samples were labeled with CyDyes as described in Table S2. Protein differential display was carried out using a 2D-DIGE system (GE Healthcare), and target protein spots were identified by nano-liquid chromatography-tandem mass spectrometry (nano-LC/MS/MS) operated by the UTSW Protein Chemistry Technology Center. Confirmation of 2D-DIGE results was performed by 1D Western blot using phosphorylated protein and total protein samples against anti-DNM1 (Abcam) and anti-NDRG2 antibodies (29). Values were then expressed relative to the β-tubulin. qPCR was performed with the ABI 7000 Sequence Detection System (Applied Biosystems). mRNA level was normalized to Cyclophilin B expression level. Serum corticosterone concentration was measured with Corticosterone Double Antibody kit (MP Biomedical).
Supplementary Material
Acknowledgments
We thank Dr. Toshiyuki Miyata for the anti-NDRG2 antibody; S. Dixon, R. Floyd, A. Skach, and M. Thornton for technical support; and T. Motoike, Y. Ikeda, H. Kumagai, A. Chang, and A. Tsai for critical discussions. This research is supported by the Japan Society for the Promotion of Science through the Funding Program for World-Leading Innovative R&D on Science and Technology (M.Y.), the Perot Family Foundation (M.Y.), NoA 1R01NS075545 (to R.W.G.), and the Department of Veterans Affairs (R.W.G.). M.Y. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308295110/-/DCSupplemental.
References
- 1.Horne J, editor. Why We Sleep: The Functions of Sleep in Humans and Other Mammals. New York: Oxford Univ Press; 1988. pp. 142–217. [Google Scholar]
- 2.Achermann P. The two-process model of sleep regulation revisited. Aviat Space Environ Med. 2004;75(3) Suppl:A37–A43. [PubMed] [Google Scholar]
- 3.McKenna JT, et al. Sleep fragmentation elevates behavioral, electrographic and neurochemical measures of sleepiness. Neuroscience. 2007;146(4):1462–1473. doi: 10.1016/j.neuroscience.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawlyk AC, Morrison AR, Ross RJ, Brennan FX. Stress-induced changes in sleep in rodents: Models and mechanisms. Neurosci Biobehav Rev. 2008;32(1):99–117. doi: 10.1016/j.neubiorev.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cano G, Mochizuki T, Saper CB. Neural circuitry of stress-induced insomnia in rats. J Neurosci. 2008;28(40):10167–10184. doi: 10.1523/JNEUROSCI.1809-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison Y, Bright V, Horne JA. Can normal subjects be motivated to fall asleep faster? Physiol Behav. 1996;60(2):681–684. doi: 10.1016/s0031-9384(96)80048-5. [DOI] [PubMed] [Google Scholar]
- 7.Sinton CM, Kovakkattu D, Friese RS. Validation of a novel method to interrupt sleep in the mouse. J Neurosci Methods. 2009;184(1):71–78. doi: 10.1016/j.jneumeth.2009.07.026. [DOI] [PubMed] [Google Scholar]
- 8.Terao A, et al. Gene expression in the rat brain during sleep deprivation and recovery sleep: An Affymetrix GeneChip study. Neuroscience. 2006;137(2):593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885(2):303–321. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- 10.Maret S, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc Natl Acad Sci USA. 2007;104(50):20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basheer R, Brown R, Ramesh V, Begum S, McCarley RW. Sleep deprivation-induced protein changes in basal forebrain: implications for synaptic plasticity. J Neurosci Res. 2005;82(5):650–658. doi: 10.1002/jnr.20675. [DOI] [PubMed] [Google Scholar]
- 12.Pawlyk AC, Ferber M, Shah A, Pack AI, Naidoo N. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103(6):2301–2313. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- 13.Poirrier JE, et al. Proteomic changes in rat hippocampus and adrenals following short-term sleep deprivation. Proteome Sci. 2008;6 doi: 10.1186/1477-5956-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen ML, Martins PJ, D’Almeida V, Bignotto M, Tufik S. Endocrinological and catecholaminergic alterations during sleep deprivation and recovery in male rats. J Sleep Res. 2005;14(1):83–90. doi: 10.1111/j.1365-2869.2004.00428.x. [DOI] [PubMed] [Google Scholar]
- 15.Peñalva RG, et al. Effect of sleep and sleep deprivation on serotonergic neurotransmission in the hippocampus: A combined in vivo microdialysis/EEG study in rats. Eur J Neurosci. 2003;17(9):1896–1906. doi: 10.1046/j.1460-9568.2003.02612.x. [DOI] [PubMed] [Google Scholar]
- 16.Tartar JL, et al. Experimental sleep fragmentation and sleep deprivation in rats increases exploration in an open field test of anxiety while increasing plasma corticosterone levels. Behav Brain Res. 2009;197(2):450–453. doi: 10.1016/j.bbr.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobler I, Murison R, Ursin R, Ursin H, Borbély AA. The effect of sleep deprivation and recovery sleep on plasma corticosterone in the rat. Neurosci Lett. 1983;35(3):297–300. doi: 10.1016/0304-3940(83)90333-6. [DOI] [PubMed] [Google Scholar]
- 18.Mongrain V, et al. Separating the contribution of glucocorticoids and wakefulness to the molecular and electrophysiological correlates of sleep homeostasis. Sleep. 2010;33(9):1147–1157. doi: 10.1093/sleep/33.9.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo A, Carlin MM, Powell JW, Dinges DF. Chronic restriction of sleep to 4 hours per night for 14 nights changes performance linearly but not subjective sleepiness. Sleep. 1998;21(Suppl):241. [Google Scholar]
- 20.Thimgan MS, Suzuki Y, Seugnet L, Gottschalk L, Shaw PJ. The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLoS Biol. 2010;8(8):e1000466. doi: 10.1371/journal.pbio.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-García F, Beltrán-Parrazal L, Jiménez-Anguiano A, Vega-González A, Drucker-Colín R. Manipulations during forced wakefulness have differential impact on sleep architecture, EEG power spectrum, and Fos induction. Brain Res Bull. 1998;47(4):317–324. doi: 10.1016/s0361-9230(98)00071-9. [DOI] [PubMed] [Google Scholar]
- 22.Bjorness TE, Kelly CL, Gao T, Poffenberger V, Greene RW. Control and function of the homeostatic sleep response by adenosine A1 receptors. J Neurosci. 2009;29(5):1267–1276. doi: 10.1523/JNEUROSCI.2942-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu W-M, et al. Essential role of dopamine D2 receptor in the maintenance of wakefulness, but not in homeostatic regulation of sleep, in mice. J Neurosci. 2010;30(12):4382–4389. doi: 10.1523/JNEUROSCI.4936-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell HA, et al. Behavioral responses of dopamine β-hydroxylase knockout mice to modafinil suggest a dual noradrenergic-dopaminergic mechanism of action. Pharmacol Biochem Behav. 2008;91(2):217–222. doi: 10.1016/j.pbb.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parmentier R, et al. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: Evidence for the role of brain histamine in behavioral and sleep-wake control. J Neurosci. 2002;22(17):7695–7711. doi: 10.1523/JNEUROSCI.22-17-07695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anaclet C, et al. Orexin/hypocretin and histamine: Distinct roles in the control of wakefulness demonstrated using knock-out mouse models. J Neurosci. 2009;29(46):14423–14438. doi: 10.1523/JNEUROSCI.2604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clayton EL, Evans GJO, Cousin MA. Activity-dependent control of bulk endocytosis by protein dephosphorylation in central nerve terminals. J Physiol. 2007;585(Pt 3):687–691. doi: 10.1113/jphysiol.2007.137539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferguson SM, et al. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316(5824):570–574. doi: 10.1126/science.1140621. [DOI] [PubMed] [Google Scholar]
- 29.Okuda T, Kokame K, Miyata T. Differential expression patterns of NDRG family proteins in the central nervous system. J Histochem Cytochem. 2008;56(2):175–182. doi: 10.1369/jhc.7A7323.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halassa MM, et al. Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron. 2009;61(2):213–219. doi: 10.1016/j.neuron.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meerlo P, Pragt BJ, Daan S. Social stress induces high intensity sleep in rats. Neurosci Lett. 1997;225(1):41–44. doi: 10.1016/s0304-3940(97)00180-8. [DOI] [PubMed] [Google Scholar]
- 32.Meerlo P, Turek FW. Effects of social stimuli on sleep in mice: Non-rapid-eye-movement (NREM) sleep is promoted by aggressive interaction but not by sexual interaction. Brain Res. 2001;907(1-2):84–92. doi: 10.1016/s0006-8993(01)02603-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.