Abstract
IgM is the first antibody produced during the humoral immune response. Despite its fundamental role in the immune system, IgM is structurally only poorly described. In this work we used X-ray crystallography and NMR spectroscopy to determine the atomic structures of the constant IgM Fc domains (Cµ2, Cµ3, and Cµ4) and to address their roles in IgM oligomerization. Although the isolated domains share the typical Ig fold, they differ substantially in dimerization properties and quaternary contacts. Unexpectedly, the Cµ4 domain and its C-terminal tail piece are responsible and sufficient for the specific polymerization of Cµ4 dimers into covalently linked hexamers of dimers. Based on small angle X-ray scattering data, we present a model of the ring-shaped Cµ4 structure, which reveals the principles of IgM oligomerization.
Keywords: antibody oligomerization, hybrid approach, dimer interfaces
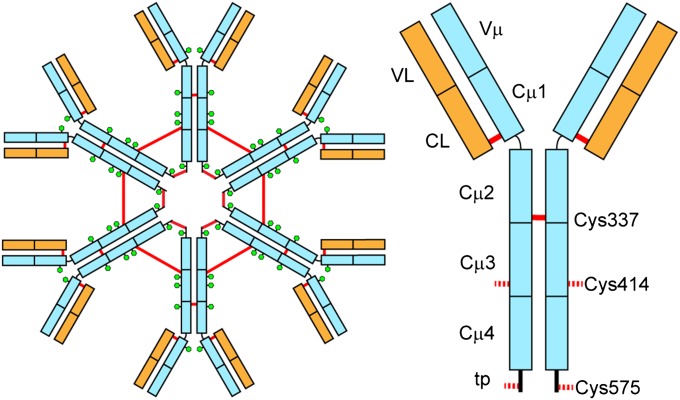
Antibodies, also referred to as Igs, defend against infection by the inactivation of viruses or bacteria and by recruiting downstream effectors such as the complement system or cells specialized to kill invading microorganisms (1, 2). IgM is the first antibody to be produced during the humoral immune response (3). Early IgM antibodies are secreted before B cells have undergone somatic hypermutations and therefore tend to be of low affinity. To compensate for the reduced binding efficiency of the monomers, IgM forms oligomers (Fig. 1) whose multiple antigen-binding sites confer high overall avidity (4). Moreover, the complex structure of IgM makes it especially effective in activating the complement system (5).
Fig. 1.
Schematic view of hexameric IgM (Left) and a single subunit (Right). Heavy chains are depicted in light blue and light chains in orange. Red lines represent intersubunit disulfide bridges and the green hexagons show glycosylation sites. The cysteine residues that form interdomain disulfide bridges between the Cµ2 domains (C337) and covalently link the IgM hexamer in Cµ3 (Cys414) and the C-terminal tp (C575) are depicted.
Whereas the antigen-binding fragment (Fab) of different Ig classes follow a common topology, the constant fragment (Fc) parts differ substantially in domain composition and architecture. In contrast to IgG, the Fc part of IgM is composed of three Ig domains (Cμ2, Cμ3, and Cμ4) and an additional C-terminal tail piece (tp). The IgM polymer is composed of subunits in which two heavy chains (µ) are covalently paired with two light chains (L). IgM is present either as pentamers (µ2L2)5J in the presence of the J chain, or as hexamers (µ2L2)6 in the absence of the J chain, a small protein that is involved in IgM assembly and secretion (6–8). Upon assembly, the heavy chains are covalently linked in the Cμ2, Cμ3, and Cμ4 (tp) domains by interchain disulfide bridges (Fig. 1).
The Fc region of IgM is of outstanding interest because its structure, oligomerization, and effector protein binding clearly differs from other Ig Fc regions. Our current understanding of the IgM structure largely originates from negative-stain EM (9), which identified the pentamer as a planar, star-shaped oligomer with the Fab fragments pointing away from the inner core composed of the Fc regions. Early structural models of complement activation were based on small angle X-ray scattering (SAXS) data and modeling (10) and suggest a planar Fc disk containing a central Cµ4 ring. The Cµ3 and Cµ2 domains are attached in a star-shaped manner. The latest model makes use of the high similarity of IgM and IgE. IgM shares the basic IgE domain architecture with three domains in the Fc part (11). Fluorescence data (12) and the subsequently determined crystal structure (11) of IgE Fc revealed that the IgE Fc is sharply bent. Interestingly, the latest IgM model suggests a similar Fc region with the Cμ4 domains protruding out of the plane defined by Cμ2, Cµ3, and the Fab domains (13).
Owing to its flexibility, crystallization trials of the IgM oligomer have not been successful to date. In this study, we applied X-ray crystallography and NMR spectroscopy to determine the atomic details of all individual mouse IgM Fc domains (Cμ2, Cμ3, and Cμ4). Analysis of their quaternary arrangements in solution revealed unexpectedly that Cμ4 forms defined ring-like hexamers of dimers. SAXS data of the hexameric domain assembly together with modeling suggest a model for the IgM Fc structure.
Results
Characterization of the IgM Fc Domains.
To gain insight into the elusive structure of the IgM Fc segment, we produced the individual Fc domains recombinantly in Escherichia coli and characterized their properties after purification. Investigation of the influence of intersubunit disulfide bonds on oligomerization was addressed by serine mutations of the corresponding cysteine residues in Cμ2 (C337), Cμ3 (C414), and Cμ4tp (C575). During thermal denaturation, all domains unfold cooperatively with transition midpoints ranging from 59–65 °C (Table 1 and Fig. S1). The lack of thermodynamic stability parameters is caused by the irreversibility of the transitions. However, guanidium chloride (GdmCl)-induced equilibrium unfolding (Fig. S1) resulted in reversible transitions of all domains, and thermodynamic parameters are given in Table 1. All domains behave like two-state folders, not significantly populating any equilibrium intermediates.
Table 1.
Oligomeric state and conformational stability of the IgM Fc domains
| Domain | Mass, kDa | Kd, µM | ΔGunfolding, kJ⋅mol−1 | meq, kJ⋅mol−1⋅M−1 | Tmelt, °C |
| Cµ2 | 24.6 | — | 36.2 | 18.6 | 64.5 |
| Cµ2C337S | 23.6 | 2.1 ± 0.1 | 26.0 | 16.6 | 60.0 |
| Cµ3C414S | 11.3 | — | 15.8 | 10.2 | 57.9 |
| Cµ4 | 17.8* | 86 ± 3 | 15.8 | 14.4 | 58.9 |
| Cµ4tpC575S | 15.4* | 224 ± 7 | 21.1 | 17.5 | 60.9 |
Molar masses and Kd values for domain dimerization were calculated from SV and SE aUC runs, respectively.
For Cµ4 and Cµ4tpC575S, monomeric and dimeric species could not be separated due to fast exchange rates. ΔGunfolding and the cooperativity parameter (meq) originate from GdmCl denaturation experiments and melting temperature (Tmelt) from thermal denaturation experiments.
To investigate the quaternary structure of the isolated domains, we first performed size-exclusion chromatography (SEC) experiments (Fig. S2). The Cμ2 domain was purified as a disulfide-linked dimer, as determined by nonreducing SDS/PAGE, which is in agreement with the elution profile of the SEC column (Fig. S2A, dotted line). Moreover, our data demonstrate that dimer formation was independent of the protein concentration. However, when the interchain disulfide bond was removed by mutating Cys337 to Ser, the elution time became concentration-dependent. The single peak is indicative of a fast monomer/dimer equilibrium (Fig. S2A). Both the wild type and the C414S mutant of the Cμ3 domain eluted predominantly as monomers and did not show any peak-shift (Fig. S2 B and C). However, in the case of wild-type Cµ3, small amounts of covalent dimers were observed. The Cμ4 domain behaved similar to the Cμ2C337S mutant. It also exhibits a fast monomer/dimer equilibrium, although with lower affinity as estimated from the smaller shift at higher protein concentrations (Fig. S2D). When the C-terminal tp with the penultimate C575 was attached to the Cμ4 domain, at least one additional prominent species with substantially increased molecular weight appeared (Fig. S2E). This peak could be minimized by mutating Cys575 to Ser (Fig. S2F), indicating that this species seems to correspond to a covalently linked Cµ4tp dimer or higher-order oligomers (discussed below).
To gain further insight into Cµ domain association, we performed sedimentation equilibrium (SE) and sedimentation velocity (SV) analytical ultracentrifugation (aUC). Data from SE runs performed at different rotor speeds and protein concentrations were fitted with a model for self-association (14) to calculate dissociation constants for the monomer/dimer equilibrium and were confirmed by SV-aUC (Table 1 and Fig. S3). For the Cµ2C337S mutant without the interdomain disulfide bridge, a Kd of 2.1 ± 0.1 µM was assigned. The weaker dimerization of the Cµ4 domain deduced from the SEC experiment was verified by aUC, resulting in a Kd of 86 ± 3 µM. The presence of the C-terminal tp (Cµ4tpC575S) decreased the affinity of the Cµ4 domain dimer to a Kd of 224 ± 7 µM. This finding might be the consequence of a not-fully-unstructured tp that is interacting with the Cµ4 domain. In contrast, the Cµ3C414S domain showed no interaction in the SEC and aUC experiments. Taken together, these experiments conclude that the Cµ2 and Cµ4 domains form dimers in solution, whereas the Cµ3 domain is monomeric. Next, we performed structural analysis of the respective Cµ domains.
Crystal Structure of the Cµ2 Domain.
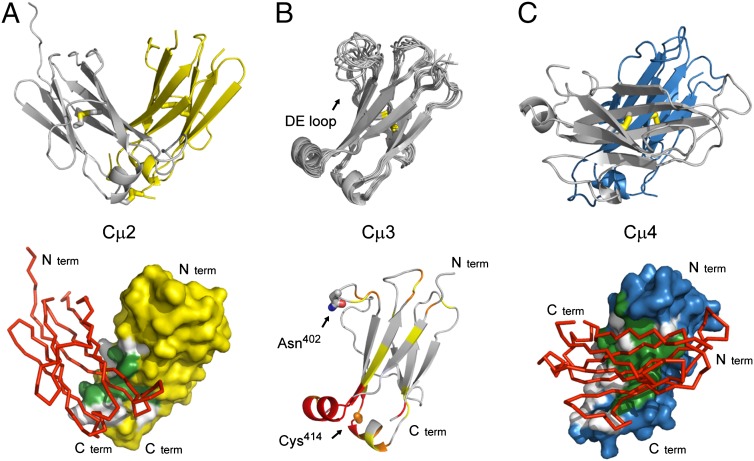
Crystals of the Cµ2 domain were obtained in the space group C2, with the unit cell dimensions a = 92.4 Å, b = 44.8 Å, and c = 54.6 Å, and two molecules in the asymmetric unit accounting for 37% solvent content. The phase problem of the IgM Cµ2 domain was determined by Patterson search calculations using the IgE Cε2 (1O0V) (11) as a starting model and the structure was refined to an Rfree of 18.1% at 1.3 Å resolution (Table S1). Overall the model is well defined in its electron density map, yet some loops (gap between chain-A Glu-293 to Thr-298 and chain-B Phe-248 to Lys-255 and Thr-293 to Gln-300) were structurally disordered and have therefore been omitted. The Cµ2 domain crystallized as a dimer, and the conformation of Cµ2 shows a typical Ig fold with a two-layer sandwich of seven antiparallel β-strands arranged in two β-sheets with a Greek key topology (Fig. 2A). Structural superposition of the Cα-atoms of both chains yields a rms deviation of 0.6 Å. Both chains possess an Ig fold typical internal disulfide bond between Cys-260 and Cys-320. The electron density map shows high occupancy and low Debye–Waller factors in the disulfide bridges, which are oriented perpendicular to the β-sheets. A common feature of many members of the Ig superfamily is the presence of at least one cis proline residue in the native structure. Both domains harbor this cis peptide bond between Thr-266 and Pro-267. However, in chain A there is an additional cis peptide bond involving Pro-253, whereas the corresponding loop in chain B is not resolved.
Fig. 2.
Structures of the individual IgM Fc domains. (A) Cartoon representation of the crystal structure of the Cµ2 domain (Upper, Protein Data Bank ID code 4JVU), the arrangement of the two molecules in the crystallographic asymmetric unit forming a stable dimer. Inter- and intramolecular disulfide bonds are shown in stick representation. The surface of one Cµ2 domain (chain A, yellow), showing the contact area with the second Cµ2 domain (chain B, red), is represented as Cα-trace (Lower). (B) Solution structure of the Cµ3 domain (Protein Data Bank ID code 4BA8). Superposition of the 10 lowest-energy NMR structures are shown as cartoon (Upper). The highly flexible DE loop is labeled. Chemical shift perturbations in the Cµ3 domain, when bound to the Cµ4 domain, are shown (Lower). The color scale is as follows: yellow, 0.025–0.05 ppm; orange, 0.05–0.1 ppm; and red, >0.1 ppm. The positions for the interdomain disulfide bridge (Cys414) and the glycosylation site (Asn402) are labeled and shown in stick representation. (C) Cartoon representation of the crystal structure the Cµ4 domain (Upper, Protein Data Bank ID code 4JVW). Intramolecular disulfide bonds are shown in stick representation. The surface of one Cµ4 domain (chain A, blue), showing the contact area with the second Cµ4 domain (chain B, red), is represented as Cα-trace (Lower). Color coding for the interfaces: gray, hydrophilic contact residues; green, hydrophobic contact residues.
To analyze the biological assembly state of the structure, the PISA server (15) was used and confirmed that Cµ2 forms a physiological dimer comprising a buried area of ∼1,660 Å2. The Cµ2 dimer shares the mode of domain pairing seen in IgE Cε2 with its less extensive interaction surface and interchain disulfide bonds (11). However, unlike IgE Cε2, the interface is dominated by a central hydrophobic core (Fig. 2A) as typically seen in other Ig domain structures. The dimer is further stabilized by six hydrogen bonds, one salt bridge (between chain A Glu-261 OE2 and chain B Lys-257 NZ, 2.77 Å) as well as an additional disulfide bridge between the C-terminal Cys-337 residues of chain A and B (Fig. 2A). The hydrophobic contact interface also accounts for the observed dimer formation in the absence of the C-terminal disulfide bridge.
Solution Structure of the Cµ3 Domain.
Because crystallization trials of the Cµ3C414S domain failed, this structure was elucidated by heteronuclear NMR spectroscopy (Table S2). We determined the overall tumbling correlation time of Cµ3C414S to be 6.0 ± 0.4 ns from 15N NMR relaxation data (Fig. S4). Comparison with an expected value of 7.5 ns for the monomer and 14.4 ns for the dimer confirms that Cµ3C414S is monomeric, in agreement with the sedimentation data described above. The solution structure of Cµ3C414S consists of the typical Ig fold with a β-sheet sandwich that includes an intramolecular disulfide bridge between Cys367 and Cys426 (Fig. 2B). NMR signals of residues in the loop between strands D and E connecting the two β-sheets are line-broadened owing to chemical exchange, further confirmed by comparison of the transverse R2 15N relaxation rates to offset-corrected R1ρ rates (Fig. S4). The increased dynamics of this loop had been already observed in the corresponding IgG Cγ2 domain in the absence of glycosylation in the DE loop (16), suggesting that the DE loop in Cµ3C414S may be stabilized by glycosylation of Asn402. Chemical shift perturbations (CSP) of Cµ3C414S and Cµ3C414S–Cµ4 domain constructs (Fig. S4) have been obtained by comparing the chemical shifts of their 1HN, 15N resonance from the backbone assignments. The CSPs have been calculated according to Eq. S2. In the spectra of the tandem construct the region between Phe354–Phe358 has been broadened out completely owing to the interaction in the interface, and therefore no exact CSP could be assigned. Residues Thr374–Leu378 and Val396–Ser399 have been excluded from the analysis, because their 1HN, 15N resonances could not be assigned owing to line broadening in the single domain construct. This allowed mapping of the binding interface of the Cµ3 domain with Cµ4 (Fig. 2B). Interestingly, this interface on Cµ3 comprises predominantly the two helices at the C-terminal side of Cµ3 and is very similar to the interactions seen between the corresponding domains in other Ig classes. The mapped interface on Cµ3 was then used for modeling experiments, eventually yielding the overall domain arrangements below.
Crystal Structure of the Cµ4 Domain.
The Cµ4 domain crystallized in the space group C2 with the cell dimensions a = 169.2 Å, b = 41.2 Å, and c = 67.1 Å. Four molecules are located in the asymmetric unit leading to 40% solvent content. Using the structure of the Cµ2 domain as a starting model, the IgM Cµ4 could be determined by molecular replacement methods and refined to a Rfree of 23.4% at 2.0 Å resolution (Table S1). The conformations of all four IgM Cµ4 monomers show a typical Ig topology that is stabilized by a disulfide bond between Cys474 and Cys536 (Fig. 2C) and a cis peptide bond involving Pro481 is observed.
The analysis of the biological assembly using the Protein Interfaces, Surfaces and Assemblies (PISA) server predicts that the monomers A and D as well as B and C form stable dimers. The conformation of both dimers in the asymmetric unit match to each other with an rmsd on the Cα-atoms of 0.3 Å and 0.4 Å, respectively. Approximately 1,900 Å2, which corresponds to 17% of the surface area, are buried in the interface that comprises hydrophobic protein interactions resulting in a stable dimer of Cµ4 (Fig. 2C). Interestingly, compared with the dimerization behavior of other Ig folds, the two molecules are arranged in a parallel manner (Fig. 2C). Particularly, the N termini are located on the same side of the dimer, and the C termini on the opposite end, which is important for the oligomer formation. A summary of the different homo–dimer association modes of Ig Fc domains is shown in Fig. S5.
Cμ4 Oligomerization.
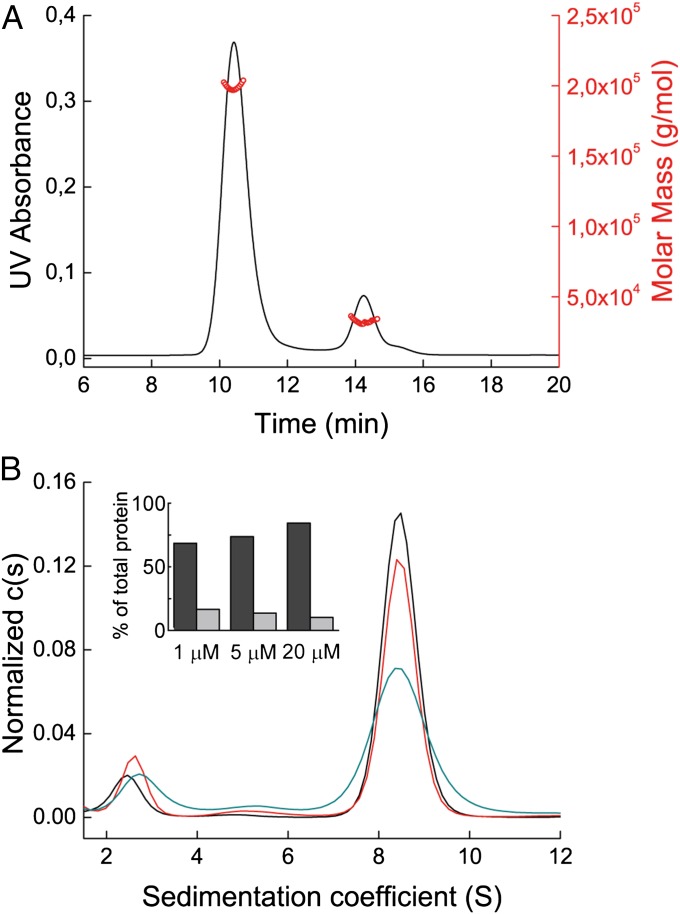
As shown above, the Cµ4tp domain forms weak noncovalent dimers in the absence of Cys575 in the tp. In the presence of Cys575, we also observed noncovalently linked dimers in dilute solutions. At higher concentrations, however, those dimers form disulfide-linked oligomers. The analysis of the native Cµ4tp by SEC revealed a concentration-dependent equilibrium between dimers and oligomers with masses of 31 kDa and 198 kDa applying multiangle light scattering (MALS) (Fig. 3A). Interestingly, the latter species corresponds well to a hexamer of Cμ4 dimers, as observed for native IgM in the absence of the J chain (6, 17, 18). Thus, the isolated Cµ4tp domain is sufficient for the assembly of defined hexamers of dimers.
Fig. 3.
Characterization of the Cμ4tp oligomer. (A) SEC MALS profile of the Cµ4tp oligomer. UV absorbance is shown in black, and calculated masses in red. (B) Cµ4tp oligomer characterized by aUC SV runs at 20 μM (black), 5 μM (red), and 1 μM (cyan). The histogram shows the percentages of hexamer of dimers (dark gray) and dimers (light gray) of the total protein in solution.
Further support of oligomer formation was received by SV aUC runs at different protein concentrations (1–20 μM) (Fig. 3B), revealing a major species that sediments with 8.8 S and a minor one with 2.6 S. Both the sedimentation coefficients and the fitted frictional coefficient f/f0 of 1.34 calculated the masses for the two species to be 190 kDa and 30 kDa, respectively. These values validate the dimer/hexamer of dimer equilibrium of Cµ4tp dimers. Furthermore, also the relative ratios could be shifted toward the hexamer of dimers in a concentration-dependent manner (Fig. 3B, Inset).
Model for the Cμ4 Hexamer Structure.
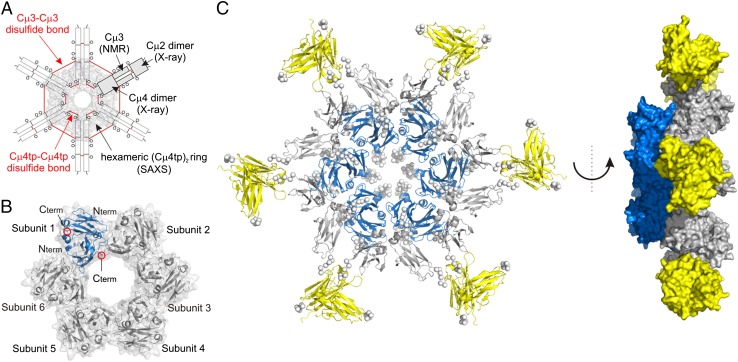
SAXS experiments were used to further characterize the hexameric structure of Cµ4tp dimers (Materials and Methods gives details). The molecular mass of Cμ4tp of 162 ± 4 kDa, determined based on the I(0), fits well to the expected molecular mass of 176 kDa for a hexamer. The radius of gyration (Rg) of 41.6 ± 0.5 and a maximum dimension of 114 Å also support hexamer of dimer formation (Fig. S6A). The SAXS data together with the crystal structure of the Cµ4 domain allowed us to model the hexameric structure of Cµ4tp dimers. Note that the additional Cµ4–Cµ4 interface observed in the crystal structure cannot be used to construct the hexamer (Fig. S6E). Therefore, no additional restraints have been used in the SAXS modeling. All structures share a characteristic toroidal assembly (Fig. 4B). The generated structures fit well to the experimental data (χ = 1.98–3.34) (Fig. S6G). Interestingly, in the oligomer the Cμ4tp dimers contact each other involving the surface of the β-sheets and several loop regions with a surface area of ∼1,000 Å2 (Dataset S1). The N termini of Cμ4tp are pointing outward of the hexameric ring, whereas the C termini, which are structurally constrained by the disulfide bond, are facing its interior (Fig. 4B).
Fig. 4.
SAXS analysis of Cµ4tp and hexameric IgM Fc modeling. (A) Scheme of the IgM Fc hexamer with the information used for model building is indicated. (B) Structural model of the Cµ4tp hexamer of dimers (Protein Data Bank ID code 4BLE). The hexameric subunits and N/C termini of the Cµ4 dimer structure are annotated. One single Cµ4tp dimer is shown in blue. (C) Cartoon representation of the IgM Fc hexamer model (Left). A side view of the Fc hexamer is given (Right), showing that the inner core (Cµ4) is protruding out of the plane defined by the Cµ2 and Cµ3 domains. Cµ2 dimers are shown in yellow, Cµ3 in gray, and the Cµ4 core ring in blue. The termini and linker residues are represented as Cα dummy atoms by the program CORAL and are shown as spheres.
In conclusion, the various structural and functional data described above allowed us to generate a model of the entire Cµ2–Cµ3–Cµ4tp hexamer (Fig. 4A) using (i) the structures of the individual subunits, (ii) the SAXS-derived structure of the hexameric Cµ4tp dimers, (iii) the distances for the inter-Cµ3 disulfide bonds, and (iv) NMR chemical shift mapping of the Cµ3–Cµ4 binding interface (Fig. 2C and Fig. S4). We conclude that the Cµ2–Cµ3 subunits in IgM point apart from the hexameric ring formed by Cµ4tp dimers (Fig. 4C). The domain interactions, including the short linkers between the Cµ2, Cµ3, and Cµ4 domains, sterically restrict the accessible conformational space (Fig. 4C). These findings demonstrate that the Cµ4 domain orchestrates the assembly of the oligomeric IgM, whereas the covalent linkages in the tail piece and the Cµ3 domain stabilize this complex.
Discussion
Whereas the structure and assembly of IgG is well-studied (19), for IgM we largely lack detailed structural information. Here we present a hybrid approach using X-ray crystallography, NMR, and SAXS to solve the structures of the individual domains of the mouse IgM Fc (Cµ2, Cµ3, and Cµ4) and to reconstruct the Fc oligomer. Aside from the expected domain topologies with their characteristic β-sheet sandwich and two short α-helices (20), we observed unexpected domain associations that were not considered in previous models. Based on electron microscopy, X-ray scattering, and mutagenesis studies, we know that the IgM polymer is assembled through interactions of identical domains, that is, Cμ2–Cμ2, Cμ3–Cμ3, and Cμ4–Cμ4, and that the corresponding cysteine residues (C337, C414, and C575) form the interchain disulfide bonds (21–23). A detailed mutagenesis study showed that the cysteine at position 575 is essential for efficient assembly, whereas C337 and C414 are not needed for polymer formation (24). The latest IgM model is based on the IgE structure (13).
To obtain a better understanding of the elusive structure of the IgM Fc, we first characterized the isolated Fc domains. The Cμ2 domain, the most N-terminal domain of the IgM Fc, replaces the hinge region found in IgG (25). It forms a disulfide-linked dimer with a unique interface dominated by hydrophobic interactions. In the absence of the disulfide bridge, the Kd is around 2 μM. This weak interaction needs to be further stabilized by the covalent linkage (Cys337). The IgE Cε2 domain is functionally equivalent to Cμ2 and it also bears some structural resemblance. Both have a unique association mode with a small interface area, compared with other Ig constant domains, and a distinct rotation angle (110° for Cµ2 and 105° for Cε2) between the domains (Fig. S5A). However, there are important differences. These include the arrangement of the disulfide bridges in Cε2 (11) and the interface that is polar and less pronounced in Cε2, leading to a monomeric protein in the absence of the disulfide bonds (26).
The Cµ3 domain does not make any stable dimer contacts. This is similar to the corresponding IgG Cγ2 and IgE Cε3 domains, which do not have any direct contacts either. In contrast to IgG and IgE, IgM forms larger oligomers. During assembly, the Cµ3 domains come into close proximity and the weak Cµ3–Cµ3 interactions are stabilized via disulfide bridges (Cys414) only in the complete IgM oligomer. The position of the Cys414 at the edge of the Cµ3 domain and its mainly polar environment supports such a weak contact that must be stabilized by a disulfide bridge.
The mode of dimerization elucidated here for Cµ4 has so far not been observed among Igs (Fig. 2 and Fig. S5A): Whereas all other C-terminal antibody domain dimers such as IgG Cγ3, IgA Cα3, and IgE Cε4 are arranged in an antiparallel fashion, with the N and C termini on opposing sides, the IgM Cµ4 topology results in the location of the N termini and C termini on the same side. This is explained by the rotation angles between the domains. Whereas IgG Cγ3, IgA Cα3, and IgE Cε4 have rotation angles of 132°, 138°, and 142°, respectively, the Cµ4 of IgM harbors a rotation angle of 70°, which is important for the self-assembly of the dimers into pentamers or hexamers of dimers. Interestingly, IgA Cα3 and IgE Cε4 share up to 50% sequence identity with Cµ4. Whereas the dimer interfaces of IgM Cµ4, IgA Cα3, and IgE Cε4 share a conserved, mostly hydrophobic, core, most residues lying at the outer part of the Cµ4 interface show little identity or homology to IgA or IgE (Fig. S5B). These differences must lead to the different dimer association of Cµ4. Hence, their different oligomeric states are caused by the unique amino acid composition at the interface that defines the configuration of the dimer.
A hallmark of the current work is the identification of the minimum requirements for IgM oligomerization. Attaching the C-terminal tp to the Cµ4 domain is already sufficient for oligomer formation, including the intermolecular disulfide bridge involving Cys575, the penultimate amino acid in the tp. Our results therefore show that the Cµ4 domain possesses two dimerization interfaces consisting of the four β-strands as well as a weak interaction site distinct from the first one that is stabilized by the interdimer disulfide bridge. Thus, our data reveal the existence of a noncovalently associated Cµ4 dimer that is covalently surrounded by two adjacent Cµ4 dimers. The noncovalent Cµ4 oligomer does not seem especially stable (Fig. 3) at protein concentrations usually found in plasma (ca. 1 µM). However, the oligomer is further stabilized by the Cµ2 dimer interface and the inter-HC disulfide bridge at the C terminus of the Cµ2 domain (23).
Based on the ring-like Cµ4 oligomer and the data from the single domains, this study describes a model for the hexameric IgM Fc, composed of the Cµ4 inner core and the Cµ3 and Cµ2 domains that build a flexible star-shaped arrangement around the inner core. This is similar to previous models based on electron microscopy (9) and SAXS (10) data. However, the model presented here is not planar and the central region (Cµ4) is projecting out of the plane defined by the Cµ2 and Cµ3 domains (Fig. 4C). The dimensions of this structure are in good agreement with previous estimations based on cryo-atomic force microscopy (AFM) data (13). In addition to the nonplanar shape, also the size of the central circular region, composed of the Cµ3 and Cµ4 domains, with a diameter of ∼180 Å, fits well to the core region dimension of 190 ± 20 Å seen in the cryo-AFM experiments.
In conclusion, the Cµ4tp domain is responsible and sufficient for the specific IgM polymerization. In vivo the situation seems more complicated. In the crowded environment of an antibody-producing cell, a sophisticated quality control system is present (27). In the case of IgM, the formation of pentamer/hexamer complexes is stringently controlled by the ERp44/ERGIC53 assembly platform including a thiol retention mechanism via Cys575 (28, 29). In addition, it was shown that carbohydrates have an influence on oligomerization (30), particularly the glycan linked to Asn563 in the tp of Cµ4 that interacts with ERp44. In its absence, pentamers cannot be formed in vivo and polymers of six or more subunits are secreted (30, 31). Furthermore, the J chain plays an important role in the selective assembly and it is expected that its major function is a place holder for one IgM subunit in the pentamer compared with the hexamer (32). This is supported by the hexameric ERp44/ERGIC53 assembly platform that is involved in the formation of pentameric and hexameric IgM assemblies. Therefore, the hexameric Cµ4 ring presented here might be identical to a pentameric Cµ4/J-chain ring with one Cµ4 dimer subunit exchanged by one J-chain protein. Notably, hexameric IgM that lacks the J chain activates the complement system 15- to 20-fold more efficiently than pentameric IgM (33, 34).
In summary, our study provides a comprehensive structural analysis of the IgM Fc domains as well as of the oligomerization of IgM subparticles. Despite the high structural similarity of individual Ig domains from various classes, interactions between the domains are different and adapted for the desired function. Notably, it is the Cµ4 domain of IgM together with its C-terminal extension that, apart from any antibody domain characterized to date, confers the intrinsic ability to oligomerize into defined hexamers of dimer. Because the sequences of mouse and human IgM Fc are conserved (68% identity), we assume that the model presented here is also true for human IgM Fc.
Materials and Methods
Protein Production and Purification.
Proteins were expressed as inclusion bodies in E. coli, oxidatively refolded, and purified as described in SI Materials and Methods.
Analytical Gel Filtration.
Analytical SEC measurements were performed in PBS as outlined in SI Materials and Methods. For the Cµ4tp domain detection and mass calculations of the observed species a Dawn Heleos MALS detector was used.
Analytical Ultracentrifugation.
Domain association and determination of dissociation constants was assessed with analytical ultracentrifugation as described in SI Materials and Methods. For dimerizing domains, data were fitted to a self-association model (14).
Optical Spectroscopy.
Fluorescence and CD measurements were carried out in PBS as described in SI Materials and Methods. To obtain thermodynamic parameters from the GdmCl-induced unfolding and refolding transitions, a two-state model was applied (35).
Crystallization and Structure Determination.
Crystals of Cµ2 and Cµ4 were grown at 20 °C using the sitting drop vapor diffusion method. Protein solution (10 mg/mL) in 10 mM Tris⋅HCl, pH 7.5, was mixed with an equal volume of buffer C2 [1.0 M lithium chloride, 0.1 M MES (pH 5.4), and 23% PEG 6000] for the Cµ2 domain and buffer C4 [15% PEG 8000 and 0.1M Hepes (pH 7.2)] for the Cµ4 domain, respectively. Diffraction datasets were collected using synchrotron radiation at the X06SA beamline at the Swiss Lightsource (Cµ2) or on a Bruker Microstar/X8 Proteum (Bruker AXS Inc.) with a Cu rotating anode (λ = 1.54 Å; Cµ4 domain). For structure determination, molecular replacement was performed in Phaser (36) using the coordinates of the Cε2 domain of human IgE (1O0V) for Cµ2 and the coordinates of the here determined structure of the Cµ2 domain for Cµ4, respectively. Details are provided in SI Materials and Methods.
NMR Spectroscopy.
NMR data were acquired at 25 °C on a Bruker Avance III 600 MHz spectrometer with a 300-µM uniformly 15N, 13C-labeled Cµ3C414S sample except for the stereospecific assignment of valine and leucine side chains, which was based on a 1H-13C heteronuclear multiple quantum correlation acquired on a 10% 13C-labeled sample as previously described (37, 38). Details are provided in SI Materials and Methods.
SAXS Measurements and Modeling.
SAXS data for solutions of Cµ4tp were measured for several solute concentrations in the range from 1 to 10 mg/mL. The structure of the Cμ4tp hexamer was modeled using the program CORAL (39) as described in SI Materials and Methods. The model of the IgM Fc hexamer was generated in CORAL using the structures of (i) Cµ2, (ii) Cµ3, (iii) the Cµ4 hexameric ring as determined based on SAXS data, (iv) the disulfide linkages (C414 and C575), and (v) the Cµ3–Cµ4 interface derived from NMR chemical shift titrations as input. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
The mouse IgM cDNA was a kind gift from Roberto Sitia (San Raffaele Scientific Institute, Milan). We thank the staff of the Swiss Lightsource for their assistance in X-ray data collection. We are grateful to Matthias Feige and Moritz Marcinowski for helpful discussion and technical support and Thomas Kriehuber for assistance during MALS measurements. We thank the Bavarian NMR Centre for NMR measurement time and Anton Paar GmbH for access to the SAXSess mc2 in-house SAXS instrument. This work was supported by grants from the Swiss National Foundation (to R.M.), DFG (to M.S., M.G., and J.B.), the Emmy Noether Program Grant MA 5703/1-1 (to T.M.), the Helmholtz Association, the Bavarian Ministry of Sciences, Research and the Arts, Bavarian Molecular Biosystems Research Network (to T.M.), and the Austrian Academy of Sciences (Austrian Programme for Advanced Research and Technology Fellowship, to T.M.). M.A.G. was supported by the Peter und Traudl Engelhornstiftung and J.P. by the Studienstiftung des deutschen Volkes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4JVU (Cµ2 domain), 4BA8 (Cµ3 domain), 4JVW (Cµ4 domain), and 4BLE (C-alpha coordinates of the Cµ4tp hexamer of dimers small angle X-ray scattering model).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300547110/-/DCSupplemental.
References
- 1.Wannemuehler MJ, Galvin JE. Bacterial immunogens and protective immunity in swine. Vet Immunol Immunopathol. 1994;43(1-3):117–126. doi: 10.1016/0165-2427(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 2.Ohanian SH, Schlager SI. Humoral immune killing of nucleated cells: mechanisms of complement-mediated attack and target cell defense. Crit Rev Immunol. 1981;1(3):165–209. [PubMed] [Google Scholar]
- 3.Boes M. Role of natural and immune IgM antibodies in immune responses. Mol Immunol. 2000;37(18):1141–1149. doi: 10.1016/s0161-5890(01)00025-6. [DOI] [PubMed] [Google Scholar]
- 4.Thomas HI, Morgan-Capner P. The avidity of specific IgM detected in primary rubella and reinfection. Epidemiol Infect. 1990;104(3):489–497. doi: 10.1017/s095026880004749x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor B, et al. C1q binding properties of monomer and polymer forms of mouse IgM mu-chain variants. Pro544Gly and Pro434Ala. J Immunol. 1994;153(11):5303–5313. [PubMed] [Google Scholar]
- 6.Brewer JW, Corley RB. Late events in assembly determine the polymeric structure and biological activity of secretory IgM. Mol Immunol. 1997;34(4):323–331. doi: 10.1016/s0161-5890(97)00029-1. [DOI] [PubMed] [Google Scholar]
- 7.Sidman C. B lymphocyte differentiation and the control of IgM mu chain expression. Cell. 1981;23(2):379–389. doi: 10.1016/0092-8674(81)90133-1. [DOI] [PubMed] [Google Scholar]
- 8.Sitia R, et al. Developmental regulation of IgM secretion: The role of the carboxy-terminal cysteine. Cell. 1990;60(5):781–790. doi: 10.1016/0092-8674(90)90092-s. [DOI] [PubMed] [Google Scholar]
- 9.Feinstein A, Munn EA. Conformation of the free and antigen-bound IgM antibody molecules. Nature. 1969;224(5226):1307–1309. doi: 10.1038/2241307a0. [DOI] [PubMed] [Google Scholar]
- 10.Perkins SJ, Nealis AS, Sutton BJ, Feinstein A. Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modelling. A possible mechanism for complement activation. J Mol Biol. 1991;221(4):1345–1366. doi: 10.1016/0022-2836(91)90937-2. [DOI] [PubMed] [Google Scholar]
- 11.Wan T, et al. The crystal structure of IgE Fc reveals an asymmetrically bent conformation. Nat Immunol. 2002;3(7):681–686. doi: 10.1038/ni811. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Shopes B, Holowka D, Baird B. Conformations of IgE bound to its receptor Fc epsilon RI and in solution. Biochemistry. 1991;30(38):9125–9132. doi: 10.1021/bi00102a002. [DOI] [PubMed] [Google Scholar]
- 13.Czajkowsky DM, Shao Z. The human IgM pentamer is a mushroom-shaped molecule with a flexural bias. Proc Natl Acad Sci USA. 2009;106(35):14960–14965. doi: 10.1073/pnas.0903805106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11(9):2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Borrok MJ, Jung ST, Kang TH, Monzingo AF, Georgiou G. Revisiting the role of glycosylation in the structure of human IgG Fc. ACS Chem Biol. 2012;7(9):1596–1602. doi: 10.1021/cb300130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brewer JW, Randall TD, Parkhouse RME, Corley RB. IgM hexamers? Immunol Today. 1994;15(4):165–168. doi: 10.1016/0167-5699(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 18.Davis AC, Shulman MJ. IgM—molecular requirements for its assembly and function. Immunol Today. 1989;10(4):118–122, 127–128. doi: 10.1016/0167-5699(89)90244-2. [DOI] [PubMed] [Google Scholar]
- 19.Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem Sci. 2010;35(4):189–198. doi: 10.1016/j.tibs.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feige MJ, et al. The structure of a folding intermediate provides insight into differences in immunoglobulin amyloidogenicity. Proc Natl Acad Sci USA. 2008;105(36):13373–13378. doi: 10.1073/pnas.0802809105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beale D, Feinstein A. Studies on the reduction of a human 19S immunoglobulin M. Biochem J. 1969;112(2):187–194. doi: 10.1042/bj1120187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milstein CP, Richardson NE, Dieverson EV, Feinstein A. Interchain disulphide bridges of mouse immunoglobulin M. Biochem J. 1975;151(3):615–624. doi: 10.1042/bj1510615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis AC, Roux KH, Pursey J, Shulman MJ. Intermolecular disulfide bonding in IgM: effects of replacing cysteine residues in the mu heavy chain. EMBO J. 1989;8(9):2519–2526. doi: 10.1002/j.1460-2075.1989.tb08389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiersma EJ, Shulman MJ. Assembly of IgM. Role of disulfide bonding and noncovalent interactions. J Immunol. 1995;154(10):5265–5272. [PubMed] [Google Scholar]
- 25.Saphire EO, et al. Crystal structure of a neutralizing human IGG against HIV-1: A template for vaccine design. Science. 2001;293(5532):1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]
- 26.McDonnell JM, et al. The structure of the IgE Cepsilon2 domain and its role in stabilizing the complex with its high-affinity receptor FcepsilonRIalpha. Nat Struct Biol. 2001;8(5):437–441. doi: 10.1038/87603. [DOI] [PubMed] [Google Scholar]
- 27.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426(6968):891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 28.Anelli T, et al. Sequential steps and checkpoints in the early exocytic compartment during secretory IgM biogenesis. EMBO J. 2007;26(19):4177–4188. doi: 10.1038/sj.emboj.7601844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, et al. Crystal structure of human ERp44 shows a dynamic functional modulation by its carboxy-terminal tail. EMBO Rep. 2008;9(7):642–647. doi: 10.1038/embor.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lalla C, Fagioli C, Cessi FS, Smilovich D, Sitia R. Biogenesis and function of IgM: the role of the conserved mu-chain tailpiece glycans. Mol Immunol. 1998;35(13):837–845. doi: 10.1016/s0161-5890(98)00073-x. [DOI] [PubMed] [Google Scholar]
- 31.Cortini M, Sitia R. ERp44 and ERGIC-53 synergize in coupling efficiency and fidelity of IgM polymerization and secretion. Traffic. 2010;11(5):651–659. doi: 10.1111/j.1600-0854.2010.01043.x. [DOI] [PubMed] [Google Scholar]
- 32.Randall TD, Brewer JW, Corley RB. Direct evidence that J chain regulates the polymeric structure of IgM in antibody-secreting B cells. J Biol Chem. 1992;267(25):18002–18007. [PubMed] [Google Scholar]
- 33.Davis AC, Roux KH, Shulman MJ. On the structure of polymeric IgM. Eur J Immunol. 1988;18(7):1001–1008. doi: 10.1002/eji.1830180705. [DOI] [PubMed] [Google Scholar]
- 34.Randall TD, King LB, Corley RB. The biological effects of IgM hexamer formation. Eur J Immunol. 1990;20(9):1971–1979. doi: 10.1002/eji.1830200915. [DOI] [PubMed] [Google Scholar]
- 35.Bolen DW, Santoro MM. Unfolding free energy changes determined by the linear extrapolation method. 2. Incorporation of delta G degrees N-U values in a thermodynamic cycle. Biochemistry. 1988;27(21):8069–8074. doi: 10.1021/bi00421a015. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szyperski T, Neri D, Leiting B, Otting G, Wüthrich K. Support of 1H NMR assignments in proteins by biosynthetically directed fractional 13C-labeling. J Biomol NMR. 1992;2(4):323–334. doi: 10.1007/BF01874811. [DOI] [PubMed] [Google Scholar]
- 38.Coggins BE, et al. Structure of the LpxC deacetylase with a bound substrate-analog inhibitor. Nat Struct Biol. 2003;10(8):645–651. doi: 10.1038/nsb948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89(2):1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.