Abstract
Seronegative hepatitis—non-A, non-B, non-C, non-D, non-E hepatitis—is poorly characterized but strongly associated with serious complications. We collected 92 sera specimens from patients with non-A–E hepatitis in Chongqing, China between 1999 and 2007. Ten sera pools were screened by Solexa deep sequencing. We discovered a 3,780-bp contig present in all 10 pools that yielded BLASTx E scores of 7e-05–0.008 against parvoviruses. The complete sequence of the in silico-assembled 3,780-bp contig was confirmed by gene amplification of overlapping regions over almost the entire genome, and the virus was provisionally designated NIH-CQV. Further analysis revealed that the contig was composed of two major ORFs. By protein BLAST, ORF1 and ORF2 were most homologous to the replication-associated protein of bat circovirus and the capsid protein of porcine parvovirus, respectively. Phylogenetic analysis indicated that NIH-CQV is located at the interface of Parvoviridae and Circoviridae. Prevalence of NIH-CQV in patients was determined by quantitative PCR. Sixty-three of 90 patient samples (70%) were positive, but all those from 45 healthy controls were negative. Average virus titer in the patient specimens was 1.05 e4 copies/µL. Specific antibodies against NIH-CQV were sought by immunoblotting. Eighty-four percent of patients were positive for IgG, and 31% were positive for IgM; in contrast, 78% of healthy controls were positive for IgG, but all were negative for IgM. Although more work is needed to determine the etiologic role of NIH-CQV in human disease, our data indicate that a parvovirus-like virus is highly prevalent in a cohort of patients with non-A–E hepatitis.
Keywords: hepatitis-associated aplastic anemia, liver failure, Solexa sequencing, virological diagnosis
Most viral hepatitis is secondary to infection by known hepatotropic viruses: hepatitis A virus (HAV) (1), hepatitis B virus (HBV) (2), hepatitis C virus (HCV) (3), hepatitis delta virus (HDV) (4), and hepatitis E virus (HEV) (5). Other hepatitis-associated viruses, including CMV (6), Epstein–Barr virus (EBV) (7), herpes simplex virus 1 and 2 (8), varicella-zoster virus, human herpesvirus 6 (9), human parvovirus B19V (B19V) (10), and adenoviruses (11), may cause liver injury, ranging from mild and transient elevation of aminotransferases to acute hepatitis and occasionally acute liver failure. Despite the number of hepatitis viruses and hepatitis-associated viruses, etiology cannot be determined in 10–20% of the cases of acute hepatitis (12), in 30% of cases of cryptogenic chronic liver diseases (13), in hepatitis-associated aplastic anemia (14), and in a large proportion of cases of acute liver failure (15). Patients with acute seronegative hepatitis have fewer parenteral risk factors, more severe bilirubin and transaminase elevations, and worse hepatocellular synthetic function than do patients with hepatitis C (16). Moreover, seronegative hepatitis is more strongly associated with serious complications, especially bone marrow failure (14) and childhood fulminant hepatitis (17).
Next-generation sequencing (NGS) technology has had an increasing impact on biological research and clinical diagnosis, because it provides high-resolution genome-scale data rapidly and reliably. The first application of NGS for pathogen discovery in a patient who died of a febrile illness after solid-organ transplantation led to the identification of an arenavirus (18). NGS-based platforms, such as 454 and Solexa deep sequencing, have been applied successfully to the investigation of emerging human pathogens, resulting in major discoveries of new human viruses (19–21).
In the present study, we established an experimental and analytic procedure for identifying virus identification in human specimens based on Solexa deep sequencing. We applied the method to screen for potential viral infection in human sera specimens and discovered a human virus in Chinese patients who had seronegative hepatitis. From its genome organization and phylogeny, the virus is unusual, because it is evolutionarily at the interface of the parvovirus and circovirus families, perhaps having emerged from interfamilial recombination. The virus was highly prevalent in a patient population, and active infection in seronegative hepatitis cases was suggested by the presence of viral DNA in the blood and serology. However, more work is needed to establish a firm disease association.
Results
Detection of a Virus in Patients with Seronegative Hepatitis.
Ten pools were made of sera from 92 patients with non-A–E hepatitis and were screened for potential pathogens by Solexa deep sequencing. After filter sterilization, the samples were digested with DNase and RNase to eliminate host nucleotide contamination, and the remaining nucleic acids were extracted using carrier RNA [synthetic poly(A)]. cDNA was synthesized from extracted viral nucleic acids with non-poly(A) random hexamers, which were designed specifically to block reverse transcription of the carrier RNA. With non-poly(A) random hexamers, redundant sequence tags derived from the carrier RNA were reduced to a level of <0.001%. High capacity enabled multiplex sequencing of more than 70 individual samples in a single Solexa run. Short DNA sequences were analyzed and filtered at the nucleotide level to exclude potential DNA contamination from human cells, mouse cells, and known bacteria. For detection of known viruses, nonredundant reads were mapped against virus genomes from the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) database using the short sequence alignment tool Bowtie. HAV and HCV sequences were detected in samples derived from sera pools 2 and 4, respectively. Individual samples from these two pools then were sequenced to clarify the source of the hepatitis viruses, and high copy numbers of HAV or HCV genomes were detected in one patient in each pool (these two samples were excluded from further analysis of seronegative hepatitis cases). Furthermore, sequences from Adenoviridae, Herpesviridae, Parvoviridae, and Anelloviridae were also commonly detected. For virus discovery, filtered short sequences were subjected to de novo assembly. Contigs from different samples were clustered based on nucleotide-level similarities. The resulting contigs were further compared with the NCBI nonredundant (NR) protein database using tBLASTx. A contig was predicted as viral origin if the best tBLASTx hit was from the kingdom of viruses, and a cluster was defined as a candidate if it contained a contig with viral origin. Contigs for each cluster were subjected to multiple alignments to define a consensus contig for later data analysis and experimental validation. By applying this pipeline to the 10 pooled samples, we identified a 3,780-bp contig that yielded BLASTx E scores of 7e-05–0.008 against parvoviruses (GenBank accession no.: KC617868).
Characterization of a Human Virus Genome.
The 3,780-bp contig was composed of three major ORFs (Fig. 1 and Fig. S1). Protein Blast (BLASTp) searches showed that ORF1 encoded a 45-kDa protein, which contained a domain (GxxxxGK[T/S]) (22) for a phosphate-binding loop (P-loop) that is conserved among different DNA viruses (Table S1); ORF1 was homologous to the replication-associated protein (Rep) of bat circovirus with E values of 7e-04. ORF2 encoded a 55-kDa protein which was homologous to Parvovirus coat protein 1 (VP1) of porcine parvovirus (PPV) and goose parvovirus, with E scores of 2e-05. Because of low homology, only the first 87 amino acid residues encoded by ORF2 could be aligned reliably to the first 100 amino acids of VP1 of PPV. The first 100 amino acids of the N-terminal region of the PPV VP1 region constituted an active phospholipase A2 (PLA2), which is highly conserved among the members of the Parvoviridae and is essential for parvovirus infection (23). The putative capsid protein (CP) encoded by ORF2 included a PLA2-like motif that is highly conserved among the members of the Parvoviridae (Table S2). ORF3 was located on the left side of the genome on the viral negative strand and encoded a 15-kDa protein. There was no homology at the nucleic acid level between the 3,780-bp contig and known viruses in GenBank.
Fig. 1.

Schematic diagram of the NIH-CQV genome. The putative ORFs are diagramed in boxes, and the arrows indicate the orientation of the ORFs. The conserved P-loop is shown as a shaded box in rep, and the conserved PLA2-like motifs are shown as shaded boxes with the Ca2+ binding loop in black and the catalytic residues in gray.
Sequence analysis also revealed inverted terminal repeats (ITR) at both ends of the genome (Fig. S1). The ITR was 156 nucleotides long. The first 75 nucleotides complemented nucleotides 82–156, allowing the formation of a stem of a hairpin, whereas nucleotides 76–81 were mismatched and could form a loop. By sequence analysis of the intergenic region between ORF1 and ORF3, we found multiple direct repeats and TATA boxes (Fig. S1). In silico promoter prediction suggested that consensus sequences for a bidirectional eukaryote promoter were present in this region (www.fruitfly.org/seq_tools/promoter.html) and that the NIH-CQV might have an ambisense genome.
To confirm the sequence of the de novo-assembled 3,780-bp contig, eight pairs of PCR primers (Table S3) were designed, based on the sequence predicted and were used to amplify overlapping DNA fragments from the pooled patient samples. Eight amplified DNA fragments were of the expected lengths (Fig. S2), and their sequences could be assembled into a 3,629-bp contig. Sequencing analysis revealed that the 3,629-bp contig, composed of the entire coding region plus about half of the ITRs from both 5′ and 3′ termini, aligned exactly with that of the de novo-assembled 3,780-bp contig, indicating that the contig contained the nearly complete virus genome, was indeed present in the patient samples, and was not an artifact generated by misassembly. The virus was provisionally designated NIH-CQ virus (NIH-CQV), because the samples were collected in a hospital in Chongqing and the laboratory experiments were conducted at the National Institutes of Health (NIH).
Because ORF1 of NIH-CQV encodes a putative Rep protein homologous to circovirus Rep, we speculated that the virus might have a circular form of its genome. To test this hypothesis, we performed inverse PCR with a primer pair (Table S3) oriented outwardly with respect to each other. Amplification with the inverted-primer pair generated an amplicon of 116 bp from two of three patient samples tested. Sequencing and alignment of the inverse PCR product showed a junction region between the 5′ and 3′ termini in a head-to-tail orientation. In comparison with the liner genome of NIH-CQV, a 419-bp region composed of both 5′ and 3′ ITRs present in the linear virus genome was absent in the inverse PCR product (Fig. S3), indicating a closed circular form of the virus genome of 3,361 bp present in the patient samples. Circularization of the viral genome resulted in an extension of the ORF3 at the left end of the genome, from 369 bp to 435 bp, with predicted encoding of a 17-kDa protein. The circular form of NIH-CQV was designated NIH-CQV-Co.
Phylogenetic and Evolutionary Analyses of NIH-CQV.
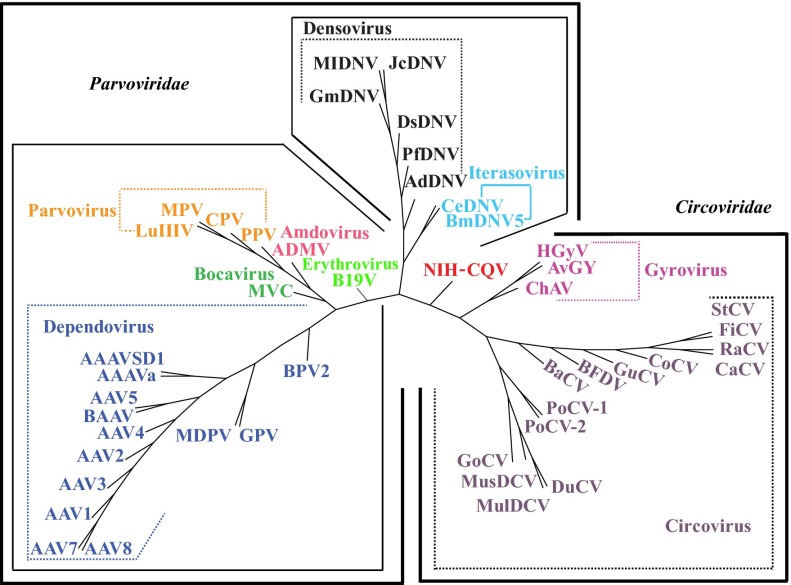
Because ORF1 and ORF2 of NIH-CQV were homologous to different viral families, phylogenetic analysis of the two ORFs was performed separately by comparison with members of Circoviridae and Parvoviridae. Although BLASTp searches showed that the amino acid sequences encoded by ORF1 were homologous to Rep of bat circovirus, by gene tree analysis NIH-CQV was not closely related to any known circoviruses (Fig. 2A). For ORF2, the amino acid similarity between NIH-CQV and known parvoviruses was below 20%. Because of low homology, only the first 87 amino acid residues encoded by ORF2 could be aligned reliably with the VP1 of porcine parvovirus and goose parvovirus, and in this region amino acid identity was ∼31%. NIH-CQV was not closely related to any known parvoviruses and thus appeared to be deeply rooted by lineage between human and animal parvoviruses and parvoviruses identified in arthropods (Fig. 2B). To confirm further the phylogenetic location of NIH-CQV, whole-proteome phylogeny analysis of NIH-CQV with 14 circoviruses and 21 prototypes of parvoviruses was conducted using a dynamic language model (24), which correctly divided the 35 selected viruses into two families, Parvoviridae and Circoviridae, and placed NIH-CQV at the interface of Parvoviridae and Circoviridae (Fig. 3).
Fig. 2.
Phylogenetic analysis of Rep and CP of NIH-CQV. (A) Phylogenetic relationship of the Rep of NIH-CQV and other related circoviruses based on an alignment of amino acid sequences. (B) Phylogenetic relationship of the CP of NIH-CQV and other related parvoviruses based on an alignment of amino acid sequences. The phylogenetic trees were constructed by neighbor joining (NEIGHBOR program from PHYLIP) based on alignments generated with CLUSTAL V, and 1,000 bootstrap replications were performed. All sequences were downloaded from GenBank and SwissProt with a BioPerl script. Detailed information is available in Dataset S1A.
Fig. 3.
Whole-proteome tree of NIH-CQV and members of Parvoviridae and Circoviridae. A total of 18 circoviruses and 28 parvoviruses are included in the analysis. The tree was constructed by neighbor joining based on protein sequences using dynamic language method for K = 4. The different viral genera are color-coded to the branches of the tree based on the taxonomic partitions. Detailed information regarding abbreviations and accession numbers are available in Dataset S1B.
Deep-sequencing data showed that NIH-CQV exhibited remarkable intrapatient genetic heterogeneity, suggesting the presence of closely related variants or quasispecies in the patients infected with NIH-CQV. To assess the dynamics of circulating quasispecies, we measured the ratios of nonsynonymous (Ka) versus synonymous (Ks) substitutions across the entire rep and cp of NIH-CQV from patients with acute (n = 13) or chronic (n = 9) non-A–E hepatitis. A higher value of Ka/Ks reflects selection favoring amino acid mutation (positive selection), whereas lower values are consistent with selection against nonsynonymous variants (negative selection). Positive selection results in the dominance of virus variants with a survival advantage, whereas negative selection acts to eliminate variants with a diminished capacity to complete the virus-replication cycle. The average Ka/Ks ratio of the cp of the virus was greater in patients with chronic hepatitis than in patients with acute hepatitis (0.98 ± 0.27 versus 0.71 ± 0.17; P < 0.01) (Fig. 4A). There were three regions in cp, ranging from nucleotides 2,360–2,448, 2,562–2,732, and 2,969–3,036 of the NIH-CQV genome, in which Ka/Ks ratios were greater than 1 and were significantly higher than in other regions (P < 0.05) (Fig. 4B), suggesting positive selection acting on specific regions of the cp gene. In contrast, although the average Ka/Ks ratio of the rep from patients with chronic hepatitis was greater than that for patients with acute hepatitis [0.47 ± 0.08 versus 0.39 ± 0.08 (P < 0.05)] (Fig. 4C), the overall Ka/Ks ratio was much lower (0.42), and there was no evidence for positive selection of this gene (Fig. 4D).
Fig. 4.
Quasispecies of NIH-CQV in patients with acute and chronic hepatitis. (A and C) Scatterplot presentations of Ka/Ks ratios of cap and rep genes of NIH-CQV from patients with acute or chronic hepatitis. The average values of Ka/Ks ratios of cap or rep of NIH-CQV from 22 individual patients (acute: n = 13; chronic: n = 9) are indicated. (B and D) Distribution of Ka/Ks ratios throughout the cp and rep. The y-axis is the averaged values of Ka or Ks within a sliding window of 100 amino acids. The average values of Ka/Ks across the whole gene region of NIH-CQV from the 22 patients are indicated. Genomic regions with significant high Ka/Ks (P < 0.05; binomial test) are shown in gray.
Prevalence of the NIH-CQ Virus in Clinical Specimens.
We designed a quantitative PCR (qPCR) assay for the rep region of NIH-CQV and generated a standard curve using a synthetic DNA rep fragment. Sensitivity of detection was <10 copies. We conducted screening of the 90 sera specimens from the patients with seronegative hepatitis and 45 sera specimens from healthy blood donors. Seventy percent (63/90) of the patient samples were positive, but all the samples from 45 healthy controls were negative. The average virus titer in the patient specimens was 1.05 e4 copies/µL, and the highest was 3.1 e4 copies/µL (Fig. 5). No significant difference in NIH-CQV DNA levels was observed between patients with acute or chronic hepatitis (Table S4). The two patients with cryptic HAV and HCV infection were negative for NIH-CQV by qPCR.
Fig. 5.
Detection of NIH-CQV DNA by qPCR. Scatterplot showing virus load per microliter of sera specimens obtained from patients (n = 90) and healthy controls (n = 45). Each dot represents one specimen. Bars show the average copy numbers of virus genome.
Detection of Specific Antibody Against NIH-CQV in Patients with Non-A–E Hepatitis.
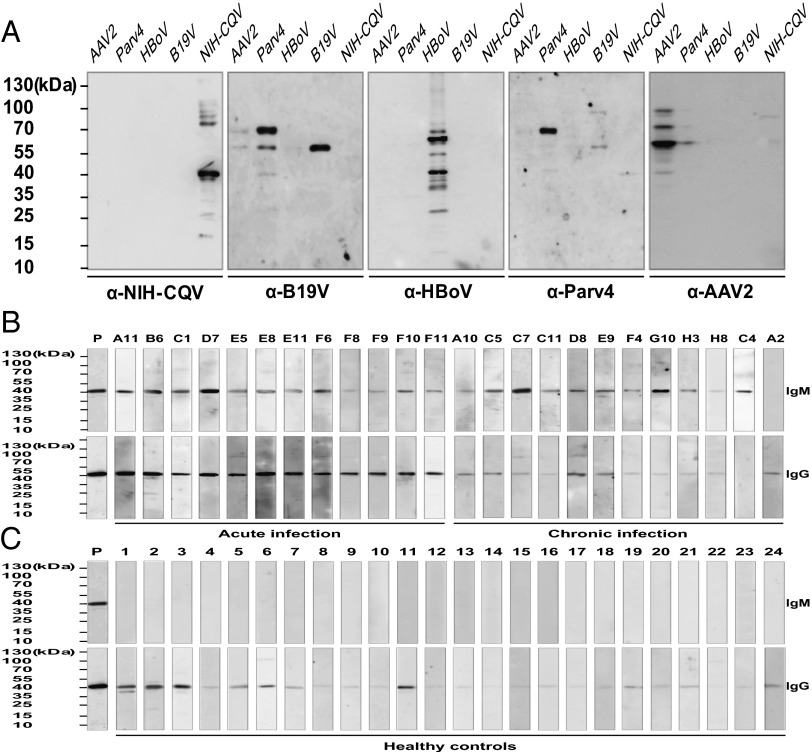
To investigate the immune response against NIH-CQV, a synthetic DNA fragment encoding a 184-amino acid polypeptide of the C-terminal portion of CP was cloned into an expression vector, and the affinity-purified recombinant CP (rCP) was used for serological studies. Cross-reactivities between NIH-CQV and other human parvoviruses, including B19V, human parvovirus 4 (Parv4), human bocavirus (HBoV), and adeno-associated virus 2 (AAV2), were tested by immunoblotting. No cross-reactivity was seen between CP of NIH-CQV and other major human parvoviruses (Fig. 6A). Immunoreactivity of rCP with a total of 135 human sera specimens was examined. By immunoblotting, 84% (76/90) of the patients with seronegative hepatitis were positive for NIH-CQV IgG, and 31% (28/90) were positive for IgM, suggestive of recent infection. In contrast, 78% (35/45) of healthy controls were positive for IgG, but all were negative for IgM, consistent with past NIH-CQV exposure (Fig. 6B, Fig. S4 A and B, and Table 1). There was a significant difference in NIH-CQV–specific IgM, but not in IgG, between the patients with acute and chronic hepatitis (Table S4).
Fig. 6.
Immunoblotting of sera from patients with non-A–E hepatitis and from healthy controls using rCP of NIH-CQV. (A) Specificity test for the rCP of NIH-CQV. The cell lysates derived from the cells transfected with the plasmids that expressed capsid proteins of AAV2, Parv4, HBoV, and B19V and purified rCP were subjected to SDS/PAGE and then were transferred to a nitrocellulose membrane. After blocking with 5% (wt/vol) nonfat milk for 2 h, the membrane was incubated with respective antisera at 1:1,000 dilution. (B and C) Detection of specific antibodies against the rCP of NIH-CQV. Samples subjected to SDS/PAGE consisted of 25 ng of affinity-purified rCP. These proteins were transferred to a nitrocellulose membrane and incubated with a 1:1,000 dilution of patient sera. The numbers at the top are patient identification numbers. Only representatives result are shown; complete data are shown in Fig. S4 A and B. Mouse monoclonal antibody against the polyhistidine tag was used as a positive control. The numbers on the left indicate molecular masses in kilodaltons based on the PageRuler Prestained Protein Ladder (Fermentas).
Table 1.
Results of real-time PCR and immunoblot analysis
| qPCR | Seronegative hepatitis patients | Healthy controls | ||||||
| IgM | IgG | IgM | IgG | |||||
| + | - | + | - | + | - | + | - | |
| + | 27.8% (25/90) | 42.2% (38/90) | 70.0% (63/90) | 0 (0/90) | 0 (0/45) | 0 (0/45) | 0 (0/45) | 0 (0/45) |
| - | 3.3% (3/90) | 26.7% (24/90) | 14.4% (13/90) | 15.6% (14/90) | 0 (0/45) | 100% (45/45) | 77.8% (35/45) | 22.2% (10/45) |
| Total | 31.1% (28/90) | 68.9% (62/90) | 84.4% (76/90) | 15.6% (14/90) | 0 (0/45) | 100% (45/45) | 77.8% (35/45) | 22.2% (10/45) |
Discussion
We report the identification of a virus, NIH-CQV, in patients with non-A–E hepatitis. Phylogenetically NIH-CQV appears to lie at the interface of parvoviruses and circoviruses. The genome of NIH-CQV has no homology to any known viruses in GenBank. Although comparative analysis revealed that the Rep and CP proteins of NIH-CQV have limited homologies with circoviruses and parvoviruses, respectively, the overall genome organization of NIH-CQV has the basic characteristics of viruses in Parvoviridae.
NIH-CQV has a small, compact linear DNA genome with two tandemly arranged major ORFs encoding Rep and CP proteins, respectively, and a pair of ITRs at the left and right ends of the genome. In silico promoter prediction suggested that transcription of the rep and cp was regulated by a single promoter upstream of the rep, because there was no apparent promoter sequence immediately upstream of cp. A pre-mRNA encoded by a single promoter that is processed by alternative splicing and alternative polyadenylation to generate multiple mRNAs had been reported for other parvoviruses, such as B19V (25) and Aleutian mink disease virus (26). By amino acid sequence analysis, CP protein of NIH-CQV is minimally homologous to porcine and goose parvovirus VP1 (27). The homologous region is mainly in the N terminus of CP and contains the conserved PLA2-like motif, which is present in the N-terminal extension of the VP1 unique region of members of Parvoviridae (23). The phylogenetic tree constructed using amino acid sequences of CP of NIH-CQV and VP1 of other representative viruses in Parvoviridae showed that NIH-CQV represents a deeply rooted lineage between two groups: (i) human and animal parvoviruses and (ii) insect parvoviruses. Thus, NIH-CQV appears to be a parvovirus-like virus. However, NIH-CQV also has features shared by members of the Circoviridae family (28). First, a BLASTp search revealed that the Rep of NIH-CQV is more related to circoviral Rep than to those of other viruses. Second, NIH-CQV appears to have an ambisense genome because there are bidirectional putative promoters in the intergenic region between the rep and 15-kDa protein and because rep and 15-kDa protein arranged head-to-head flank the intergenic region, which is a feature of circoviruses. Finally, sequencing and alignment of the inverse PCR products indicated the existence of the circular genome of NIH-CQV in the clinical samples. The circular form of NIH-CQV genome might represent a subviral DNA or an intermediate structure of viral replication, as reported for other parvoviruses (29). Therefore, NIH-CQV appears to be a “hybrid” virus, perhaps formed by interfamilial recombination between a parvovirus and a circovirus. This idea was supported further by whole-proteome phylogenetic analysis, which showed NIH-CQV located between Parvoviridae and Circoviridae.
Both recombination and a high mutation rate are often cited as key in evolutionary innovation. For many viruses, recombination allows single-step acquisition of multiple genetic changes, and a high mutation rate enhances adaptation to new hosts (30). These processes are critical driving forces in viral evolution and lead to viral emergence, host-switching, and adaptation, which often result in disease outbreaks (31, 32). Recent studies had demonstrated that small eukaryotic ssDNA viruses have high rates of nucleotide substitutions and genetic recombination, which may surpass rates seen in RNA viruses (33, 34). These observations may explain the extensive diversity seen in ssDNA viruses. Parvoviruses (family Parvoviridae) and circoviruses (family Circoviridae) are both ssDNA viruses that infect a wide variety of animal species, including mammals, birds, and insects. Although horizontal gene transfer and high mutation rates have been documented in these viruses, cross-species viral transfers resulting from interfamilial recombination and leading to a infection in the new host species are not common.
A high rate of viral evolution has been reported for emerging parvoviruses (35). Canine parvovirus (CPV) is one well-characterized example (35). CPV emerged from feline panleukopenia parvovirus or a closely related virus, differing at several amino acid residues. Sequences of the viruses collected at time points before and after host-switching suggest that the emergence of CPV was dependent on a high mutation rate and positive selection of the major capsid gene. Our deep-sequencing data showed that NIH-CQV has a high substitution mutation rate throughout its genome. However, the impact of selective pressures, as measured by the Ka/Ks ratio, appeared to vary depending on location. The cp gene contained three regions in which the Ka/Ks ratio exceeded 1, suggesting positive selection. These substitutions would be presumed to enhance fitness. In contrast, selective pressure on the rep of NIH-CQV appeared to be dominated by purifying selection, because the average Ka/Ks ratio for the rep from 14 patients was only 0.42. The Ka/Ks ratios for the regions encoding the PLA2 motif in the CP protein or P-loop nucleoside triphosphatase (NTPase) in the Rep proteins were lower than in other regions, as is consistent with strong purifying selection and functional importance of this region in viral replication.
NIH-CQV exhibited remarkable genetic heterogeneity within patients, indicating the presence of closely related variants or quasispecies in NIH-CQV–infected individuals. The Ka/Ks ratios of both viral cp and rep were greater in patients classified as having chronic hepatitis than in those with acute hepatitis, suggesting greater variation or more diversity of quasispecies of NIH-CQV over time. For hepatitis viruses C and E, diversification of quasispecies during infection is driven by the interaction between the viruses and host immune response and may be associated with different clinical outcomes (36, 37). With a limited number of serial samples from a single individual, we were unable to conclude whether quasispecies dynamics of NIH-CQV in patients correlated with disease.
Despite technological advances in molecular biology, an etiology cannot be determined in as many as 20% of hepatitis cases. As of yet, no causative agent may be defined in a significant proportion of patients with chronic liver disease and cirrhosis, whose disease therefore is characterized as “cryptogenic.” Additional hepatitis agents have been suspected. Over several decades, several human viruses, such as GB virus C (38), Torque teno virus (39), and SEN virus (40), have been putatively claimed to be hepatitis viruses, but subsequent investigation revealed no conclusive association between these viruses and liver disease. Therefore, the etiology of non-A–E hepatitis remains unresolved. In this study, we applied NGS methodology to virus discovery and indentified a parvovirus-like virus in the sera samples collected from a cohort of patients with non-A–E hepatitis. Our results showed that 28% of patients were positive for both NIH-CQV–specific IgM and viral DNA, suggesting recent infection in patients. Most patients and healthy controls were positive for virus-specific IgG, suggesting that exposure was common in the population studied. The patients who yielded positive specimens suffered from symptoms of hepatitis with a wide range of liver dysfunction, including acute and chronic hepatitis. There was no difference between patients diagnosed with acute versus or chronic hepatitis with respect to detection of NIH-CQV DNA, but viral DNA was not detected in healthy controls. Although these findings are suggestive of an etiologic relationship, our study has limitations. By electron microscopy, we observed virus-like particles in NIH-CQV qPCR-positive sera which cosedimented with viral DNA, but we were unable to confirm their identity by immunogold labeling with our antisera. In addition, because of the limited availability of samples, we had insufficient material for viral propagation and isolation in cell culture, and efforts are in progress to detect viral proteins in a few liver biopsy tissue blocks. Additionally, with the limited number of samples available, we cannot exclude the possibility of reactivation of a latent virus in the course of liver inflammation.
In general, parvoviruses cause systemic infection, but clinical manifestations of the infection are varied and may depend on specific tissue tropisms as well as on host immunologic competency. For two human pathogenic parvoviruses, B19V (10) and HBoV (41), clinical symptoms range from none to acute systemic manifestations involving skin and joints to subtle persistence of infection; in immunocompromised populations, B19V chronic infection causes pure red cell aplasia. In addition, coinfection of parvoviruses with other viruses, especially related DNA viruses, can lead to synergy and more severe clinical symptoms in the affected host. Because of the complexity of disease associations, more comprehensive epidemiology and in vitro studies are needed to establish a pathogenic role of NIH-CQV in hepatitis.
Materials and Methods
Study Subjects.
We studied a total of 92 sera samples from patients with non-A–E hepatitis who were admitted to the Institute of Infectious Disease of Southwest Hospital, Third Military Medical University, China, between 1999 and 2007. According to the criteria of the Chinese Society of Infectious Disease and Parasitology, and the Chinese Society of Hepatology (42), 33 patients were diagnosed with acute hepatitis by clinical and biochemical parameters. Fifty-nine patients had chronic aggressive hepatitis confirmed by biopsy, and 10 of them had cirrhosis; eight outpatients with chronic persistent hepatitis did not have liver biopsies performed. The patients had a median age of 42 y (range: 12–73 y); among of them, 55 males had a median age of 41 y (range: 12–73 y), and 37 females had median age of 43 y (range: 15–70 y). On admission, mean liver function tests were as follows: total bilirubin 174 ± 130 μmol/L (range: 14–735 μmol/L), alanine aminotransferase 421 ± 623 IU/L (range: 5–3,150 IU/L). Serological tests for anti-HAV IgM (HAV IgM, ELISA Kit; Wantai Biological Pharmacy), HBV markers (HBsAg and HBeAg; Roche Diagnostic Systems) anti-HCV antibodies (HCV ELISA Kit; Wantai Biological Pharmacy), anti-HDV IgM (HDV IgM ELISA Kit; Wantai Biological Pharmacy), anti-HEV IgM (HAV IgM ELISA Kit; Wantai Biological Pharmacy), HCV RNA tests (HCV Quantitative RT-PCR Kit; Kehua Bio-engineering), anti-HIV-1 and 2 antibodies (HIV ELISA Kit; Wantai Biological Pharmacy), HIV viral load test (HIV Quantitative RT-PCR Kit; Roche Diagnostic Systems), and serological tests for anti-CMV IgM and (CMV IgM ELISA Kit; Wantai Biological Pharmacy) and anti-EBV IgM (EBV IgM Kit; YHLO Biotech) were all negative. Additional tests for antinuclear antibody, rheumatoid factor, anti-mitochondrial antibody (Microplate ELISA Test Systems for Autoimmunity; EUROIMMUN), as well as blood cultures, urine cultures, and throat swabs for bacteria were negative. The research protocol was approved by the Human Bioethics Committee of the Third Military Medical University, and all participants provided written informed consent.
Solexa Deep Sequencing and Phylogenetic and Evolutionary Analysis.
Details of deep sequencing and phylogenetic and evolutionary analysis are provided in SI Material and Methods.
Overlapping PCR and Reverse PCR.
To verify the sequence of the viral genome assembled from the Solexa data, eight sets of overlapping primer pairs were designed (Table S3). Viral DNA was extracted from sera as described in the Solexa deep sequencing. Extracted DNA (5 μL) was used as template for the PCR. The 50-μL reaction mix consisted of 1× Platinum Taq PCR Buffer (Invitrogen), 1.5 mM MgCl2, each dNTP at 0.2 mM, and 25 pmol each of the primers. After 2 min at 94 °C, 35 cycles of amplification (94 °C for 1 min, 54 °C for 1 min, and 72 °C for 1 min) were performed. To detect circularized viral DNA, inverse PCR with a primer pair (Table S3) that oriented outwardly with respect to each other was used for amplification. PCR was performed as described above, and amplified products were visualized on an agarose gel. All PCR products were sequenced.
Diagnostic qPCR.
For quantitative PCR, DNA was extracted by the QIAamp MinElute Virus Kit (Qiagen), and 5 µL of the resulting DNA was used for analysis with the NIH-CQV rep primers set (NIH-CQV_Rep-1295F and NIH-CQV_Rep-1470R), and NIH-CQV_Rep probe was used for qPCR (Table S3). All reactions were performed using the Chromo4 real-time detector (Bio-Rad). The reaction started with activation of the polymerase at 95 °C for 15 min, followed by 45 cycles of 15 s at 94 °C and 1 min at 60 °C. The quantitation of amplicon was performed by interpolation with the standard curve to the synthesized rep gene (ORF1) with serial dilutions.
Immunoblotting.
To investigate the immune response against NIH-CQV, a synthetic DNA fragment encoding a 184-amino acid polypeptide of the C-terminal portion of CP was cloned into an expression vector, and the affinity-purified rCP was used for immunoblotting. Cross-reactivities between NIH-CQV and other human parvoviruses, including B19V, Parv4, HBoV, and AAV2, were tested by immunoblotting. Details are provided in SI Material and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jun Zhu at the DNA Sequencing and Genomics Core of the National Heart, Lung, and Blood Institute (NHLBI) for assistance in deep-sequencing and data analysis; Dr. Amit Kapoor of Columbia University for technical advice; and Genoveffa Franchini of the National Cancer Institute, Leonid Margolis of the National Institute of Child Health and Development, Cynthia Dunbar of the NHLBI, and Mike Fang in our laboratory for their careful reading of the manuscript. This work was supported by the National Institutes of Health Intramural Research Program.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KC617868).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303744110/-/DCSupplemental.
References
- 1.Cohen JI. Hepatitis A virus: Insights from molecular biology. Hepatology. 1989;9(6):889–895. doi: 10.1002/hep.1840090617. [DOI] [PubMed] [Google Scholar]
- 2.Theilmann L, et al. HBV DNA and other hepatitis B virus markers in sera from long-term hemodialysis and kidney transplant patients. Hepatogastroenterology. 1988;35(4):147–150. [PubMed] [Google Scholar]
- 3.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 4.Rizzetto M, et al. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977;18(12):997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes GR, et al. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247(4948):1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 6.Lemonovich TL, Watkins RR. Update on cytomegalovirus infections of the gastrointestinal system in solid organ transplant recipients. Curr Infect Dis Rep. 2012;14(1):33–40. doi: 10.1007/s11908-011-0224-6. [DOI] [PubMed] [Google Scholar]
- 7.Crum NF. Epstein Barr virus hepatitis: Case series and review. South Med J. 2006;99(5):544–547. doi: 10.1097/01.smj.0000216469.04854.2a. [DOI] [PubMed] [Google Scholar]
- 8.Riediger C, et al. Herpes simplex virus sepsis and acute liver failure. Clin Transplant. 2009;23(Suppl 21):37–41. doi: 10.1111/j.1399-0012.2009.01108.x. [DOI] [PubMed] [Google Scholar]
- 9.Razonable RR, Zerr DM. AST Infectious Diseases Community of Practice HHV-6, HHV-7 and HHV-8 in solid organ transplant recipients. Am J Transplant. 2009;9(Suppl 4):S100–S103. doi: 10.1111/j.1600-6143.2009.02899_2.x. [DOI] [PubMed] [Google Scholar]
- 10.Young NS, Brown KE. Parvovirus B19. N Engl J Med. 2004;350(6):586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 11.Lynch JP, 3rd, Fishbein M, Echavarria M. Adenovirus. Semin Respir Crit Care Med. 2011;32(4):494–511. doi: 10.1055/s-0031-1283287. [DOI] [PubMed] [Google Scholar]
- 12.Alter MJ, et al. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med. 1992;327(27):1899–1905. doi: 10.1056/NEJM199212313272702. [DOI] [PubMed] [Google Scholar]
- 13.Kodali VP, Gordon SC, Silverman AL, McCray DG. Cryptogenic liver disease in the United States: Further evidence for non-A, non-B, and non-C hepatitis. Am J Gastroenterol. 1994;89(10):1836–1839. [PubMed] [Google Scholar]
- 14.Brown KE, Young NS. Human parvovirus B19: Pathogenesis of disease. In: Anderson LJ, Young NS, editors. Human parvovirus B19. Basel: Karger; 1997. pp. 105–119. [Google Scholar]
- 15.Ferraz ML, et al. Fulminant hepatitis in patients undergoing liver transplantation: Evidence for a non-A, non-B, non-C, non-D, and non-E syndrome. Liver Transpl Surg. 1996;2(1):60–66. doi: 10.1002/lt.500020110. [DOI] [PubMed] [Google Scholar]
- 16.Rochling FA, et al. Acute sporadic non-A, non-B, non-C, non-D, non-D, non-E hepatitis. Hepatology. 1997;25(2):478–483. doi: 10.1002/hep.510250237. [DOI] [PubMed] [Google Scholar]
- 17.Lee WS, McKiernan P, Kelly DA. Etiology, outcome and prognostic indicators of childhood fulminant hepatic failure in the United kingdom. J Pediatr Gastroenterol Nutr. 2005;40(5):575–581. doi: 10.1097/01.mpg.0000158524.30294.e2. [DOI] [PubMed] [Google Scholar]
- 18.Palacios G, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358(10):991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- 19.Briese T, et al. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5(5):e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yozwiak NL, et al. Human enterovirus 109: A novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J Virol. 2010;84(18):9047–9058. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319(5866):1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momoeda M, Wong S, Kawase M, Young NS, Kajigaya S. A putative nucleoside triphosphate-binding domain in the nonstructural protein of B19 parvovirus is required for cytotoxicity. J Virol. 1994;68(12):8443–8446. doi: 10.1128/jvi.68.12.8443-8446.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zádori Z, et al. A viral phospholipase A2 is required for parvovirus infectivity. Dev Cell. 2001;1(2):291–302. doi: 10.1016/s1534-5807(01)00031-4. [DOI] [PubMed] [Google Scholar]
- 24.Yu ZG, et al. Whole-proteome phylogeny of large dsDNA viruses and parvoviruses through a composition vector method related to dynamical language model. BMC Evol Biol. 2010;10:192. doi: 10.1186/1471-2148-10-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JM, Green SW, Shimada T, Young NS. A block in full-length transcript maturation in cells nonpermissive for B19 parvovirus. J Virol. 1992;66(8):4686–4692. doi: 10.1128/jvi.66.8.4686-4692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu J, Cheng F, Pintel D. The abundant R2 mRNA generated by aleutian mink disease parvovirus is tricistronic, encoding NS2, VP1, and VP2. J Virol. 2007;81(13):6993–7000. doi: 10.1128/JVI.00244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tijssen P, et al. Family Parvoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Family Parvoviridae Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic; 2011. pp. 405–425. [Google Scholar]
- 28.Biagini P, et al. Family Circoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Family Circoviridae Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevler Academic; 2011. pp. 343–349. [Google Scholar]
- 29.Kapoor A, et al. Bocavirus episome in infected human tissue contains non-identical termini. PLoS ONE. 2011;6(6):e21362. doi: 10.1371/journal.pone.0021362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woolhouse ME, Haydon DT, Antia R. Emerging pathogens: The epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20(5):238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, et al. Animal origins of the severe acute respiratory syndrome coronavirus: Insight from ACE2-S-protein interactions. J Virol. 2006;80(9):4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Bueno A, Villarreal LP, Almendral JM. Parvovirus variation for disease: A difference with RNA viruses? Curr Top Microbiol Immunol. 2006;299:349–370. doi: 10.1007/3-540-26397-7_13. [DOI] [PubMed] [Google Scholar]
- 34.Martin DP, et al. Recombination in eukaryotic single stranded DNA viruses. Viruses. 2011;3(9):1699–1738. doi: 10.3390/v3091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shackelton LA, Parrish CR, Truyen U, Holmes EC. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci USA. 2005;102(2):379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farci P, et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288(5464):339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 37.Lhomme S, et al. Hepatitis E virus quasispecies and the outcome of acute hepatitis E in solid-organ transplant patients. J Virol. 2012;86(18):10006–10014. doi: 10.1128/JVI.01003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linnen J, et al. Molecular cloning and disease association of hepatitis G virus: A transfusion-transmissible agent. Science. 1996;271(5248):505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 39.Nishizawa T, et al. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241(1):92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka Y, et al. Genomic and molecular evolutionary analysis of a newly identified infectious agent (SEN virus) and its relationship to the TT virus family. J Infect Dis. 2001;183(3):359–367. doi: 10.1086/318091. [DOI] [PubMed] [Google Scholar]
- 41.Jartti T, et al. Human bocavirus-the first 5 years. Rev Med Virol. 2012;22(1):46–64. doi: 10.1002/rmv.720. [DOI] [PubMed] [Google Scholar]
- 42.Chinese Society of Infectious D & Parasitology Chinese Society of Hepatology. Management scheme of diagnostic and therapeutic criteria of viral hepatitis. Zhonghua Gan Zang Bing Za Zhi. 2000;8:324–329. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.