Abstract
The trans-translation pathway for protein tagging and ribosome release plays a critical role for viability and virulence in a wide range of pathogens but is not found in animals. To explore the use of trans-translation as a target for antibiotic development, a high-throughput screen and secondary screening assays were used to identify small molecule inhibitors of the pathway. Compounds that inhibited protein tagging and proteolysis of tagged proteins were recovered from the screen. One of the most active compounds, KKL-35, inhibited the trans-translation tagging reaction with an IC50 = 0.9 µM. KKL-35 and other compounds identified in the screen exhibited broad-spectrum antibiotic activity, validating trans-translation as a target for drug development. This unique target could play a key role in combating strains of pathogenic bacteria that are resistant to existing antibiotics.
Keywords: antibiotic target, tmRNA, non-stop translation
The increasing prevalence of antibiotic-resistant bacterial pathogens has spurred a search for new pathways that can be targeted for antibiotic development (1, 2). One pathway that has not been exploited is the trans-translation pathway, which resolves nonstop translation complexes. The components of trans-translation have been identified in every sequenced bacterial genome, and mutations in these components affect viability or virulence in a wide range of bacteria (3, 4), suggesting that inhibitors of trans-translation might be effective broad-spectrum antibiotics. In addition, the trans-translation pathway is not found in animals, so specific inhibitors are expected to have few side effects on the host.
The purpose of trans-translation is to remove nonstop translation complexes, i.e., translation reactions in which the ribosome has reached the 3′ end of the mRNA without terminating at a stop codon (4–6). These complexes are prevalent in bacteria because bacterial ribosomes do not require any information from the 3′ end of the mRNA to initiate translation, and bacteria lack most of the mechanisms for mRNA proofreading found in eukaryotes (7). Because hydrolysis of peptidyl-tRNA by release factors requires a stop codon in the A site, normal translation termination cannot occur when the ribosome reads to the 3′ end of the mRNA and there is no in-frame stop codon. Trans-translation resolves nonstop translation complexes using a ribonucleoprotein complex containing transfer-messenger RNA (tmRNA) and the small protein SmpB (4–6). tmRNA-SmpB recognizes nonstop translation complexes and enters the ribosomal A site mimicking a tRNA (8). The nascent polypeptide is transferred to tmRNA, and a specialized reading frame within tmRNA is inserted into the mRNA channel (4–6). Translation resumes using this sequence as a message and terminates at a stop codon at the end of the reading frame, releasing the ribosome and a tagged protein (4–6). The tag sequence is recognized by proteases, and the tagged protein is rapidly degraded (9–12). The net reaction of trans-translation is the removal of all components of the nonstop translation complex.
Trans-translation occurs with high frequency in bacteria—2–4% of translation reactions in Escherichia coli terminate with trans-translation (13)—suggesting that elimination of this pathway is likely to be detrimental. In fact, trans-translation is essential in some species, including Shigella flexneri, Helicobacter pylori, Neisseria gonorrheae, and Mycoplasma species (14–17). In addition, mutants lacking trans-translation activity are avirulent in Salmonella enterica, Yersinia pestis, Streptococcus pneumoniae, and Francisella tularensis (18–21), so inhibitors of the pathway could be effective against a wide array of pathogens. We identified compounds that inhibit trans-translation using a high-throughput screen (HTS) and demonstrated that these compounds have antibiotic activity. These results validate trans-translation as a target for antibiotic development.
Results
Development of a Cell-Based HTS Assay for Trans-Translation Activity.
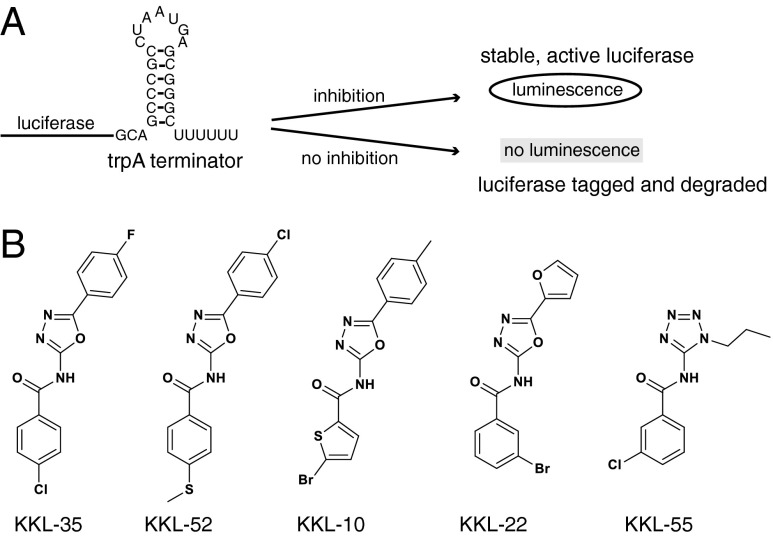
A luciferase-based reporter was constructed to monitor trans-translation activity in E. coli cells. A copy of the trpAt transcriptional terminator was inserted before the stop codon of luc, the gene encoding firefly luciferase, on a multicopy plasmid. Induction of luc expression from this reporter (luc-trpAt) produces luc mRNA with no stop codon, and translation of the nonstop luc mRNA in cells with an active trans-translation system results in tagging and proteolysis of the luciferase protein (Fig. 1A). Alternatively, if trans-translation is inhibited, active luciferase will be released and accumulate in the cells. Therefore, trans-translation activity can be monitored by assaying luciferase activity in cells expressing luc-trpAt.
Fig. 1.
Identification of trans-translation inhibitors by HTS. A cell-based assay with positive readout for trans-translation activity was used for HTS to identify inhibitors. (A) A schematic diagram of the HTS-compatible assay. Luciferase was made from a nonstop mRNA that has a strong transcriptional terminator (stem-loop sequence) before the stop codon. Translation of this mRNA when trans-translation is inhibited results in active luciferase and luminescence in the HTS assay. Conversely, when trans-translation is active, the luciferase is tagged and degraded, and there is no luminescence in the HTS assay. (B) Chemical structures of five compounds identified by HTS and characterized using secondary assays.
In preparation for HTS, the luc-trpAt reporter was tested in WT E. coli and a strain deleted for ssrA, the gene encoding tmRNA, using a 1,536-well format. WT cells produced little luciferase activity, even 2 h after induction, indicating that the luciferase was efficiently tagged and degraded. In contrast, luciferase activity was evident in ∆ssrA cells within 15 min after induction, and at 90 min, luciferase activity peaked at a level ∼40-fold higher than in WT. This assay had a Z′ = 0.8, indicating excellent statistics for HTS (Table S1).
Identification of Trans-Translation Inhibitors by HTS.
A library of 663,000 candidate compounds was screened for the ability to inhibit trans-translation in E. coli expressing luc-trpAt. In the primary screen, 0.2% of the compounds resulted in >1.5-fold activation of the reporter. When these initial hits were rescreened in duplicate using a dose–response format, 178 compounds produced greater than twofold activation of the reporter at a concentration of 10 µM (Table S1). Forty-six of these compounds were chosen for further investigation based on availability.
Characterization of Tagging and Proteolysis Inhibitors.
Compounds that inhibit either tagging of nascent luciferase or proteolysis of the tagged protein would pass the HTS. To eliminate any spurious positive hits and to determine whether each compound inhibits tagging or proteolysis, compounds were tested in cell-based fluorescent reporter assays. Compounds were first screened using two fluorescent reporters: gfp-trpAt, which produces green fluorescent protein (GFP) from a nonstop mRNA, and gfp-tag, which has the tmRNA tag peptide AANDENYALAA encoded at the 3′ end of the gfp gene. All of the GFP protein made from gfp-tag has the AANDENYALAA sequence regardless of trans-translation activity, so this reporter can be used to monitor proteolysis of tagged proteins. Inhibitors of tagging will make cells with gfp-trpAt fluorescent, but cells with gfp-tag will be dark; inhibitors of proteolysis will make cells with either gfp-trpAt or gfp-tag fluorescent (Fig. S1). All 46 compounds caused fluorescence when added to cells expressing gfp-trpAt, indicating that they are all trans-translation inhibitors and not spurious hits (Table S2). Nineteen compounds also caused fluorescence when added to cells expressing gfp-tag, indicating that they inhibit proteolysis of tagged proteins. The other 27 compounds did not cause fluorescence when added to cells expressing gfp-tag, indicating that they inhibit tagging and not proteolysis. Based on these assays, five of the compounds (KKL-35, KKL-52, KKL-10, KKL-22, and KKL-55) were selected for more detailed analysis (Fig. 1B).
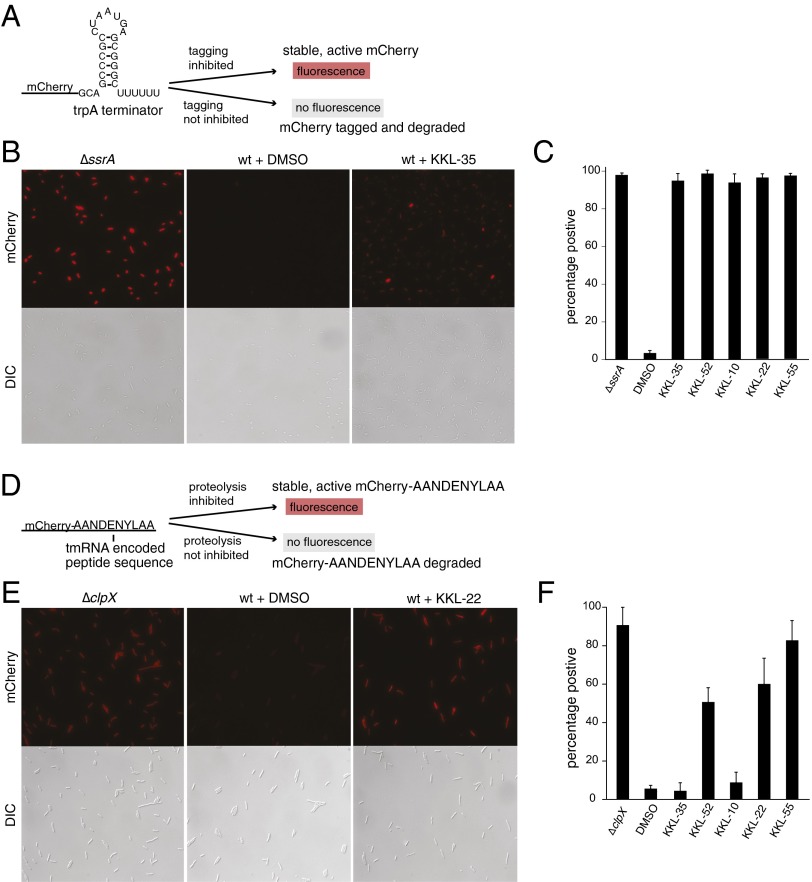
The effects of the five compounds on trans-translation were quantified using mCherry-trpAt and mCherry-tag reporters that are directly analogous to gfp-trpAt and gfp-tag (Fig. 2). Incubation of KKL-35, KKL-52, KKL-10, KKL-22, or KKL-55 with cells expressing mCherry-trpAt resulted in a three- to fivefold increase in the average fluorescence signal per cell (Fig. 2B; Fig. S2A). Greater than 95% of the cells in the treated populations had a fluorescence signal >2 SDs above the mean for the DMSO-treated control population (Fig. 2C). A similar analysis using E. coli expressing mCherry-tag confirmed that KKL-52, KKL-22, and KKL-55 inhibit the proteolysis of tagged proteins, but KKL-35 and KKL-10 do not affect proteolysis (Fig. 2 E and F; Fig. S2B). Collectively, these in vivo reporter assays demonstrate that the HTS successfully identified inhibitors of the tagging and proteolysis steps of the trans-translation pathway.
Fig. 2.
Characterization of inhibitors of tagging and proteolysis. Two mCherry-based reporters were used to determine which compounds inhibited tagging of nascent polypeptides and which inhibited proteolysis of tagged proteins. Schematic diagrams of the mCherry-trpAt reporter (A), and mCherry-tag reporter (D) indicate the conditions that produce fluorescent cells. Epifluorescence (mCherry panels) and DIC micrographs were used to measure the fluorescence in ΔssrA cells, ΔclpX cells, or in WT cells treated with DMSO only or with one of the compounds. Representative micrographs for mCherry-trpAt (B) and mCherry-tag (E) are shown. The fluorescence intensity in >330 individual cells was measured, and the cells were scored as positive if the intensity was >2 SDs higher than the average intensity for the DMSO-treated control. The percentage of positive cells for mCherry-trpAt (C) and mCherry-tag experiments (F) is shown. Compounds were added at 100 μM, with the exception of KKL-52 in C, which was added at 10 μM. Error bars indicate SDs.
KKL-35 Inhibits Trans-Translation In Vitro.
In vivo assays indicated that KKL-35 and KKL-10 inhibit trans-translation at some step before proteolysis of tagged proteins. In principle, these compounds could block tagging by directly interfering with the activity of tmRNA-SmpB on nonstop translation complexes, or they could act indirectly to decrease tagging. To test whether the compounds act directly on the tagging reaction, trans-translation activity was assayed in vitro. A gene encoding dihydrofolate reductase (DHFR) with no stop codon (DHFR-ns) was used as template in a coupled in vitro transcription/translation reaction with E. coli components. In vitro expression of DHFR-ns resulted in full-length DHFR protein. When tmRNA-SmpB was included in the reaction, the larger, tagged DHFR was produced (Fig. 3A). Control reactions that expressed a DHFR gene containing a stop codon (DHFR-stop) had little tagged DHFR even when tmRNA-SmpB was added (Fig. 3A). KKL-35 and KKL-55 inhibited tagging of DHFR-ns (Fig. 3B). Dose–response experiments showed that KKL-35 inhibited tagging with an IC50 = 0.9 µM (Fig. 3 C and D). Addition of DMSO or the proteolysis inhibitors KKL-52 or KKL-22 had no effect on tagging in vitro (Fig. 3B). A large amount of untagged DHFR was produced in reactions with the highest concentrations of KKL-35, indicating that KKL-35 does not inhibit translation. In agreement with this result, KKL-35 did not inhibit production of DHFR-stop in in vitro translation assays (Fig. 3 E and F). These results demonstrate that KKL-35 specifically inhibits trans-translation and not translation.
Fig. 3.
In vitro assays for inhibition of trans-translation and translation. (A) DHFR genes with or without a stop codon were expressed in vitro in the absence or presence of additional tmRNA and SmpB. The locations of DHFR, as determined in control reactions, and of the lower-mobility tagged DHFR protein are indicated. (B) In vitro trans-translation reactions were performed after addition of 1.67% DMSO or 10 μM compound. Representative reactions are shown. The intensity of the DHFR and tagged DHFR bands were measured, and the tagging efficiency was calculated as the percentage of total DHFR protein in the tagged DHFR band. The average tagging efficiency with SD for at least three repeats is shown. To determine the dose–response behavior, in vitro trans-translation reactions performed after addition of KKL-35 at different concentrations, and the tagging efficiencies were calculated. (C) A representative experiment is shown. (D) Data from three repeats were averaged, graphed, and fit with a sigmoidal function to determine the IC50. Whiskers indicate SD for each point. (E) In vitro translation reactions were performed after addition of DMSO, 100 μM chloramphenicol (chlor), or 100 μM compound, and a representative experiment is shown. The amount of DHFR in each lane was measured as a percentage of the amount in the DMSO-treated control, and the average value for three experiments was graphed with whiskers indicating the SD. (F) Inhibitor-treated reactions did not result in significantly different translation activity relative to the DMSO-treated control; P values from one-way ANOVA tests are indicated.
KKL-55 inhibited tagging in vitro even though it inhibited proteolysis of tagged proteins in vivo, suggesting that KKL-55 inhibits both steps in the pathway. These results also indicate that KKL-35 and KKL-55 act differently in vivo even though they both inhibit tagging in vitro. Conversely, KKL-10, which inhibited trans-translation in vivo, did not inhibit tagging in vitro at 10 μM. A decrease in tagging was observed when KKL-10 was added at 100 µM (Fig. S3). It is possible that KKL-10 is active in vivo but not in vitro because it is modified in the cell or targets trans-translation indirectly by interacting with a cellular component that is not present in the in vitro assays.
KKL-35 Prevents Growth of Cells That Require Trans-Translation.
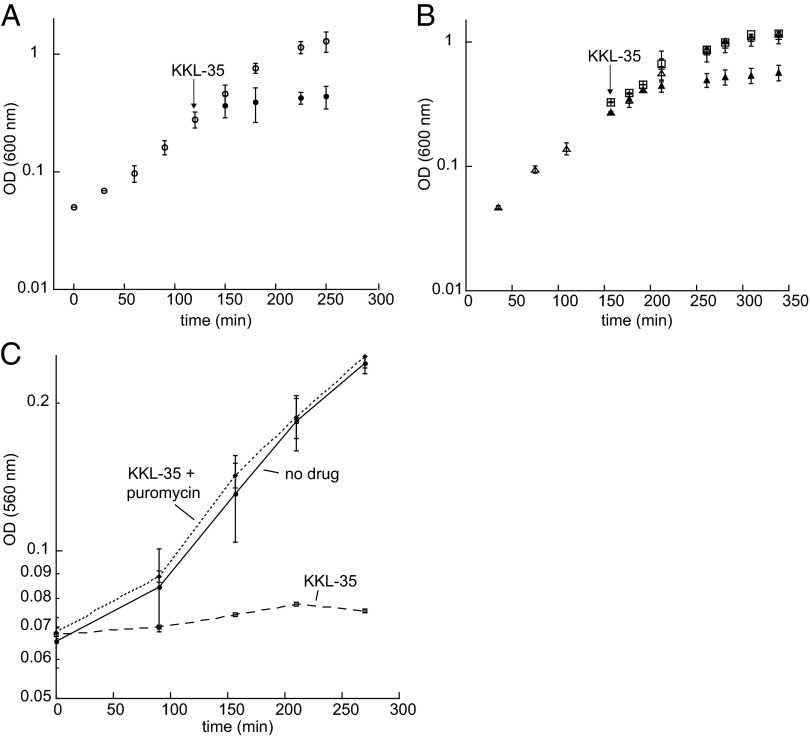
Because KKL-35 specifically inhibits trans-translation, it should kill bacteria under conditions where trans-translation is required for viability but have little effect if trans-translation is dispensable for growth. A good system to test this prediction is S. flexneri, a close relative of E. coli (22). ssrA is essential in S. flexneri, but can be deleted in E. coli because E. coli has an alternative release factor, ArfA, that can resolve nonstop translation complexes in the absence of trans-translation (14, 23). When E. coli ArfA is expressed in S. flexneri, ssrA can be deleted (14). Broth microdilution assays showed that KKL-35 prevented growth of WT S. flexneri with a minimum inhibitory concentration (MIC) of 6 µM (Table 1), and addition of KKL-35 to a growing culture of S. flexneri stops growth (Fig. 4A). In an S. flexneri strain expressing ArfA and deleted for ssrA, addition of KKL-35 has little effect on viability or growth rate (Fig. 4B). These data are consistent with KKL-35 inhibiting trans-translation and preventing growth of cells that have no other mechanism to resolve nonstop translation complexes. These results also indicate that KKL-35 does not inhibit ArfA activity in vivo, and in vitro assays confirmed that ArfA activity is not inhibited by KKL-35 (Fig. S4).
Table 1.
Antibacterial activity of small molecule inhibitors
| Inhibitor | S. flexneri | B. anthracis | M. smegmatis | E. coli ∆tolC | E. coli ∆tolC ∆ssrA | |||||
| MIC* | MBC† | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| KKL-35 | 6.0 (0) | S‡ | 3.0 (2.6) | S | 1.3 (0.4) | 1.3 (0.4) | 0.3 (0.1) | ND | 0.3 (0.1) | ND |
| KKL-52 | 50 (0) | S | 1.5 (0) | 6.0 (0) | 100 (0) | 100 (0) | 4.0 (0) | ND | 4.0 (0) | ND |
| KKL-10 | 7.8 (5.5) | S | 2.3 (0.3) | 7.5 (6.4) | 1.5 (0) | 1.5 (0) | 0.4 (0) | ND | 0.4 (0) | ND |
| KKL-22 | 19 (11) | S | 10 (3.5) | 25 (0) | 5.0 (1.7) | 5.0 (1.7) | 1.5 (0) | ND | 1.5 (0) | ND |
| KKL-55 | 13 (9.0) | S | 25 (0) | 50 (0) | 25 (0) | 25 (0) | 16 (7.5) | ND | 16 (7.5) | ND |
ND, not determined.
Mean (SD) values from at least three broth microdilution assays (μM).
Mean (SD) values from at least three plating assays (μM).
Bacteriostatic at all tested concentrations.
Fig. 4.
KKL-35 inhibits cell growth by preventing resolution of nonstop translation complexes. (A) Growth curves of S. flexneri with (●) and without (○) addition of KKL-35. The arrow indicates when KKL-35 was added. (B) ArfA suppresses the effect of KKL-35 on cell growth. Growth curves of S. flexneri ssrA::kan pCA24N-His6-ArfA cells were similar when ArfA expression was induced before addition of KKL-35 (□), when ArfA expression was induced and no KKL-35 was added (+), and when ArfA expression was not induced and no KKL-35 was added (△). Growth was slower when ArfA expression was not induced before addition of KKL-35 (▲). The arrow indicates when KKL-35 was added. (C) Puromycin antagonizes the effect of KKL-35 on culture growth. Growth curves of untreated E. coli ΔtolC cells (no drug), cells treated with 3.3 µM KKL-35 (10× MIC), and cells treated with 3.3 µM KKL-35 and 0.1 µg/mL puromycin. Error bars indicate SDs.
KKL-35 also prevents the growth of S. flexneri cells that have both ArfA and trans-translation activity, but interpretation of this result is complicated by the fact that ArfA production is regulated by trans-translation (24, 25). When trans-translation is functional, ArfA is tagged and degraded and does not accumulate in the cell. Therefore, when KKL-35 is added to cultures of WT cells expressing ArfA, the steady-state levels of ArfA are extremely low, and inhibition of trans-translation will prevent growth (for a more detailed explanation, see SI Results and Figs. S5 and S6). Likewise, KKL-35 inhibits the growth of E. coli ∆tolC, which is deficient in small molecule efflux, with an MIC = 0.3 µM (Table 1). These results are consistent with KKL-35 preventing growth of both E. coli and S. flexneri strains by inhibiting trans-translation under conditions where there are no other functional mechanisms to resolve nonstop translation complexes.
Pharmacological experiments also suggest that the effects of KKL-35 on cell growth are due to inhibition of trans-translation. The drug puromycin, which hydrolyzes peptidyl-tRNA in the ribosome, can rescue growth of E. coli ∆arfA cells during depletion of tmRNA (23), presumably because it releases some of the nonstop translation complexes that accumulate. Puromycin could also rescue growth of E. coli ∆tolC in the presence of KKL-35. Addition of 0.1 µg/mL puromycin rescued normal growth, even when KKL-35 was added at concentrations >10-fold above the MIC (Fig. 4C). The antagonism between puromycin and KKL-35 indicates that growth inhibition caused by KKL-35 is due to accumulation of nonstop translation complexes and not a secondary target in the cell. Taken together, the in vivo effects of KKL-35 demonstrate that this compound interferes with release of nonstop translation complexes by trans-translation.
KKL-35 Has Broad-Spectrum Antibacterial Activity.
In addition to preventing growth of S. flexneri and E. coli ∆tolC, broth microdilution experiments showed that KKL-35 prevented growth of B. anthracis and M. smegmatis with MIC values ≤6 µM (<2 µg/mL; Table 1). Plating assays to determine the minimum bactericidal concentration (MBC) showed that KKL-35 was bactericidal at the MIC against M. smegmatis but was bacteriostatic against other species (Table 1). The effects of KKL-35 on B. anthracis and M. smegmatis indicate that release of nonstop translation complexes is required for growth of each of these species and suggest that compounds such as KKL-35 could be developed as broad-spectrum antibiotics. Several other compounds identified in the HTS, including KKL-10, KKL-22, KKL-55, KKL-06, KKL-07, and KKL-24, also exhibited broad-spectrum antibiotic activity (Table 1; Table S3). Disk diffusion assays confirmed the broad-spectrum antibiotic activity, but for some compounds, diffusion appeared to be limited (Fig. S7).
Discussion
The HTS described here identified specific inhibitors of the trans-translation pathway. Inhibitors of both tagging and proteolysis of tagged proteins were recovered, and several of the compounds had broad-spectrum antibiotic activity. The ability of the compounds identified by HTS to cross the bacterial membrane and function in vivo is not surprising, because the screen was designed with a cell-based assay dependent on a positive readout. Compounds that are not cell permeable or are not active inside a bacterial cell would not score as positive hits. In principle, the screen described here could be used with a wide variety of compound libraries to identify other inhibitors of the pathway.
KKL-35 is one of the most potent inhibitors isolated in the HTS, and several lines of evidence indicate that the in vivo effects of KKL-35 are caused by inhibition of the release of nonstop translation complexes by trans-translation. KKL-35 inhibits trans-translation in vivo and in vitro and prevents growth of S. flexneri strains that require trans-translation. The correlation between inhibition of trans-translation and growth is supported by genetic and pharmacological experiments showing that alternative mechanisms to release nonstop translation complexes relieve the growth suppression of KKL-35.
What is the molecular target of KKL-35? The target must be required for trans-translation but not for translation or ArfA activity. In principle, candidate steps include binding of tmRNA and SmpB, charging of tmRNA, association of tmRNA-SmpB with the ribosome, and conformational changes of the ribosome that are required for trans-translation but not translation. KKL-35 had little effect on binding of tmRNA and SmpB as measured by filter-binding assays (Kd = 1.3 ± 0.13 nM with 100 µM KKL-35 vs. 1.6 ± 0.35 nM with DMSO only), suggesting that KKL-35 is likely to affect one of the subsequent steps in trans-translation. Despite repeated attempts, no spontaneous or UV-induced mutants of E. coli or S. flexneri that were resistant to KKL-35 or KKL-55 could be recovered. One possible explanation for this observation is that mutations in the targets of KKL-35 and KKL-55 that would lead to resistance are highly detrimental for growth in culture. Further biochemical and structural studies of KKL-35 will be required to identify the mechanism of action.
KKL-35 inhibits growth of very distantly related bacteria, suggesting that it may have antibiotic activity against a broad spectrum of species. KKL-35 is expected to prevent growth of other bacteria known to require trans-translation, including Neisseria gonorrheae, Haemophilus influenza, Helicobacter pylori, Mycoplasma genitalium, and Mycoplasma pneumonia, assuming there are no problems with bioavailability in these species. Intriguingly, KKL-35 was also identified in a HTS for molecules that prevent growth of Mycobacterium tuberculosis (26). Based on the data presented here, we suggest that KKL-35 kills M. tuberculosis by inhibiting trans-translation. The requirement for trans-translation in M. tuberculosis has not been rigorously tested, but trans-translation has been implicated as a target for pyrazinamide (PZA) (27), a frontline drug for anti-tuberculosis therapy. Although it is not yet clear if KKL-35 has the clinical properties required for use as an antibiotic, the activity of KKL-35 and other compounds described here indicate that trans-translation is a promising target for antibiotic development.
Methods
Bacterial Strains and Plasmids.
All strains (Table S4) were grown at 37 °C in lysogeny broth supplemented with 100 μg/mL ampicillin, 30 μg/mL kanamycin, or 20 μg/mL chloramphenicol as appropriate. For detailed descriptions of strain and plasmid construction, see SI Methods.
HTS.
A library of 663,000 small molecule compounds was screened using the luc-trpAt reporter. An overnight culture of LC1083 grown at 37 °C was diluted 10-fold, and 3 µL diluted overnight culture was dispensed per well in 1,536-well white solid plates. To induce luc-trpAt expression, 1 µL 4 mM isopropyl ß-D-1-thiogalactopyranoside (IPTG) was added per well, and the plates were incubated at room temperature for 90 min. Four microliters Bright-Glo reagent (Promega) was added per well, the plates were incubated at room temperature for 30 min, and the luminescence signal was measured using a CLIPR system (Molecular Devices).
Compounds.
KKL-35, KKL-52, KKL-10, KKL-22, and KKL-55 were purchased from Life Chemicals in multiple batches, dissolved in 100% (vol/vol) DMSO, and stored at −80 °C. KKL-55 was also synthesized as previously published (28).
Fluorescent Reporter Assays.
Overnight cultures of strains KCK161, LC1031, LC1131/32, LC1189/90, NSR259, and NSR262 were diluted to OD600 = 0.02 and grown with aeration at 37 °C to OD600 = 0.2. IPTG was added to 1 mM final concentration, and the cells were transferred to culture tubes, and DMSO [2% (vol/vol) final concentration] or compound was added. Cells were observed after 20 min (strains containing gfp-tag or mCherry-tag) or 4 h (strains containing gfp-trpAt or mCherry-trpAt) on agarose pads using epifluorescence microscopy (Nikon Eclipse 90i microscope, 60× TIRF NA 1.4 objective, Nikon CoolSNAP HQ CCD camera). Exposure times were 0.01 s for differential interference contrast (DIC), 0.5 s for mCherry, and 2 s for GFP. The intensity in >330 cells per sample was determined using Simple PCI software (Hamamatsu Photonics). The percentage of positive cells was calculated by determining the number of cells in the population that had a fluorescent intensity >2 SDs above the average intensity of the DMSO-treated control.
MIC and MBC Assays.
MIC assays were performed according to Clinical and Laboratory Standards Institute guidelines for determining the antimicrobial activity of the compounds (29). Overnight cultures of S. flexneri 2a 2457T, B. anthracis, M. smegmatis, JW5503, and NSR253 were grown in lysogeny broth (LB). Cultures were diluted to a final inoculum of ∼5 × 105 cfu/mL in 96-well microtiter plates, compounds were added at appropriate concentrations, and growth was observed after 24-h incubation at 37 °C. The MIC was determined by the concentration of the compound in the last well that showed no bacterial growth. For MBC assays, 10 μL from wells containing the MIC, 2× MIC, and 4× MIC of each compound was diluted 10-fold, spotted on LB plates, and grown overnight at 37 °C. An inhibitor was scored as bactericidal if it resulted in 1,000-fold reduction in colony-forming units per milliliter from the original inoculum.
In Vitro Translation and Trans-Translation.
Translation assays were performed using the PURExpress (New England Biolabs) protein synthesis kit with the control DHFR template and [35S]-methionine, according to the manufacturer’s instructions, with inhibitors or chloramphenicol added to 100 μM final concentration or with DMSO added to 6.7% (vol/vol). After 2- to 3-h incubation at 37 °C, reactions were precipitated with acetone, separated on 12% (wt/vol) SDS polyacrylamide gels, and visualized by exposure to a phosphor screen (GE Healthcare). Relative translation activity of each inhibitor or chloramphenicol was determined with respect to the DMSO-treated control, for at least three repeats. A one-way ANOVA test was conducted to determine whether relative translation activities were significantly different from the DMSO-treated control value.
For in vitro trans-translation assays, the following modifications were made to the in vitro translation reaction. DHFR-ns and DHFR-stop template DNA was made by PCR amplification of the DHFR control plasmid using T7 universal forward primer and either DHFR-ns or DHFR-stop reverse primer. tmRNA and SmpB were added to 2 μM final concentration. Inhibitors were added to 10 μM final concentration. Anti-ssrA oligonucleotide (5 μM) (30) was added to reactions without tmRNA and SmpB to inhibit background trans-translation activity contributed by tmRNA-SmpB in the kit. Tagging efficiency was calculated as the ratio of tagged DHFR to total DHFR (tagged + untagged) from at least three repeats. For dose-dependent inhibition experiments with KKL-35, tagging efficiency at each concentration was graphed and fit with a sigmoidal function (Kaleidagraph; Synergy Software) to obtain the IC50 value.
Growth Curves.
Saturated cultures of S. flexneri ssrA::kan pCA24N-His6-ArfA were diluted 1:50 into LB, grown with shaking at 37 °C, induced with 1 mM IPTG at an OD600 = 0.1, and grown for 30–45 min before KKL-35 was added to 10 μM final concentration. Cultures were sampled every 20–30 min, and the OD600 values determined were plotted as a function of time. Doubling times were obtained by fitting curves to a single exponential function.
For growth in the presence of puromycin, E. coli tolC::kan cultures were grown at 25 °C in 96-well plates in the presence of 0–10 µM KKL-35 and 0–1 µg/mL puromycin, and culture growth was monitored by reading the optical density at 560 nm.
Supplementary Material
Acknowledgments
We thank Dr. Peter Schultz for guidance and support of high-throughput screening. We are grateful to the laboratories of Sarah Ades (Pennsylvania State University), Chris Hayes (University of California, Santa Barbara), Jeff Cox (University of California, San Francisco), and Chuck Turnbough (University of Alabama at Birmingham) for gifts of strains and plasmids. This work was supported by National Institutes of Health Grant GM68720 (to K.C.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302816110/-/DCSupplemental.
References
- 1.Leung E, Weil DE, Raviglione M, Nakatani H. World Health Organization World Health Day Antimicrobial Resistance Technical Working Group The WHO policy package to combat antimicrobial resistance. Bull World Health Organ. 2011;89(5):390–392. doi: 10.2471/BLT.11.088435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Infectious Diseases Society of America The 10 x ’20 Initiative: Pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis. 2010;50(8):1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 3.Gueneau de Novoa P, Williams KP. The tmRNA website: Reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32(Database issue):D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- 5.Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 6.Keiler KC, Ramadoss NS. Bifunctional transfer-messenger RNA. Biochimie. 2011;93(11):1993–1997. doi: 10.1016/j.biochi.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131(4):660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 8.Neubauer C, Gillet R, Kelley AC, Ramakrishnan V. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science. 2012;335(6074):1366–1369. doi: 10.1126/science.1217039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choy JS, Aung LL, Karzai AW. Lon protease degrades transfer-messenger RNA-tagged proteins. J Bacteriol. 2007;189(18):6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman C, Thévenet D, Bouloc P, Walker GC, D’Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 1998;12(9):1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keiler KC, Sauer RT. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J Biol Chem. 1996;271(5):2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12(9):1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito K, et al. Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS ONE. 2011;6(12):e28413. doi: 10.1371/journal.pone.0028413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramadoss NS, Zhou X, Keiler KC. tmRNA is essential in Shigella flexneri. PLoS ONE. 2013;8(2):e57537. doi: 10.1371/journal.pone.0057537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C, Wolfgang MC, Withey J, Koomey M, Friedman DI. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000;19(5):1098–1107. doi: 10.1093/emboj/19.5.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchison CA, et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286(5447):2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 17.Thibonnier M, Thiberge JM, De Reuse H. Trans-translation in Helicobacter pylori: Essentiality of ribosome rescue and requirement of protein tagging for stress resistance and competence. PLoS ONE. 2008;3(11):e3810. doi: 10.1371/journal.pone.0003810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J Bacteriol. 2000;182(6):1558–1563. doi: 10.1128/jb.182.6.1558-1563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okan NA, Mena P, Benach JL, Bliska JB, Karzai AW. The smpB-ssrA mutant of Yersinia pestis functions as a live attenuated vaccine to protect mice against pulmonary plague infection. Infect Immun. 2010;78(3):1284–1293. doi: 10.1128/IAI.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann B, et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 2012;8(7):e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svetlanov A, Puri N, Mena P, Koller A, Karzai AW. Francisella tularensis tmRNA system mutants are vulnerable to stress, avirulent in mice, and provide effective immune protection. Mol Microbiol. 2012;85(1):122–141. doi: 10.1111/j.1365-2958.2012.08093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei J, et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun. 2003;71(5):2775–2786. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadani Y, et al. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol Microbiol. 2010;78(4):796–808. doi: 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- 24.Garza-Sánchez F, Schaub RE, Janssen BD, Hayes CS. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol Microbiol. 2011;80(5):1204–1219. doi: 10.1111/j.1365-2958.2011.07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaub RE, Poole SJ, Garza-Sánchez F, Benbow S, Hayes CS. Proteobacterial ArfA peptides are synthesized from non-stop messenger RNAs. J Biol Chem. 2012;287(35):29765–29775. doi: 10.1074/jbc.M112.374074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds RC, et al. High throughput screening of a library based on kinase inhibitor scaffolds against Mycobacterium tuberculosis H37Rv. Tuberculosis (Edinb) 2012;92(1):72–83. doi: 10.1016/j.tube.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi W, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333(6049):1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGillivray SM, et al. Pharmacological inhibition of the ClpXP protease increases bacterial susceptibility to host cathelicidin antimicrobial peptides and cell envelope-active antibiotics. Antimicrob Agents Chemother. 2012;56(4):1854–1861. doi: 10.1128/AAC.05131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. National Committee for Clinical Laboratory Standards (1999) Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline, Vol. M26-A (National Committee for Clinical Laboratory Standards, Wayne, PA) [Google Scholar]
- 30.Schaffitzel C, Hanes J, Jermutus L, Plückthun A. Ribosome display: An in vitro method for selection and evolution of antibodies from libraries. J Immunol Methods. 1999;231(1-2):119–135. doi: 10.1016/s0022-1759(99)00149-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.