Significance
Genotoxic attack results in temporary arrest of RNA synthesis. Mutations in the DNA repair factor Cockayne syndrome B gene product (CSB) that are responsible for the Cockayne syndrome phenotype lead to clinical features such as developmental and neurodegenerative defects and photosensitivity. In UV-irradiated CSB-deficient CS1AN cells, certain genes remain permanently repressed by the activating transcription factor 3, the product of a stress-response gene, which cannot be removed from promoter by the transcription machinery. We suggest that transcriptional defects observed in UV-irradiated CSB-deficient cells result from the permanent transcriptional repression of certain genes as well as from defects in DNA repair.
Abstract
Cockayne syndrome type B ATPase (CSB) belongs to the SwItch/Sucrose nonfermentable family. Its mutations are linked to Cockayne syndrome phenotypes and classically are thought to be caused by defects in transcription-coupled repair, a subtype of DNA repair. Here we show that after UV-C irradiation, immediate early genes such as activating transcription factor 3 (ATF3) are overexpressed. Although the ATF3 target genes, including dihydrofolate reductase (DHFR), were unable to recover RNA synthesis in CSB-deficient cells, transcription was restored rapidly in normal cells. There the synthesis of DHFR mRNA restarts on the arrival of RNA polymerase II and CSB and the subsequent release of ATF3 from its cAMP response element/ATF target site. In CSB-deficient cells ATF3 remains bound to the promoter, thereby preventing the arrival of polymerase II and the restart of transcription. Silencing of ATF3, as well as stable introduction of wild-type CSB, restores RNA synthesis in UV-irradiated CSB cells, suggesting that, in addition to its role in DNA repair, CSB activity likely is involved in the reversal of inhibitory properties on a gene-promoter region. We present strong experimental data supporting our view that the transcriptional defects observed in UV-irradiated CSB cells are largely the result of a permanent transcriptional repression of a certain set of genes in addition to some defect in DNA repair.
Gene expression is jeopardized by genotoxic attacks such as UV irradiation stress that challenge genome integrity. Several DNA repair factors are required to remove DNA lesions, indicating that connections between transcription and DNA repair orchestrate accurate gene expression.
UV-induced lesions that modify the DNA structure are eliminated through two subpathways of nucleotide excision repair (NER). Global genome NER removes DNA damage from the entire genome, whereas transcription-coupled NER (TCR) corrects DNA lesions located on the actively transcribed genes (1, 2). Cockayne syndrome type B (CSB) protein is involved in TCR; it is recruited to the stalled polymerase II (Pol II) and works as a coupling factor attracting histone acetyltransferase p300, NER proteins, and the Cockayne syndrome A (CSA)–damage-specific DNA binding protein 1 (DDB1) E3–ubiquitin ligase protein complex to remove the stalled transcription complex and induce chromatin remodeling to facilitate the repair of DNA lesions (3–5). CSB is a 168-kDa member of the Switch 2/Sucrose nonfermentable 2 (SWI2/SNF2) family of DNA-dependent ATPases and contains seven characteristic helicase motifs (6, 7), but no helicase activity has been demonstrated for CSB when using a conventional strand-displacement assay (8). It has been suggested that mutations in CSB prevent the recruitment of the repair machinery and the proper resumption of RNA synthesis. Experimental evidence also has indicated that CSB is involved in transcription as well as in DNA repair. The presence of CSB as the promoter of activated genes and its absolute requirement for reinitiating the transcription of undamaged genes after UV irradiation underline its function in promoting gene activation (5, 9, 10). CSB also stimulates RNA polymerase elongation (11–16).
Mutations in CSB and in Cockayne syndrome type A (CSA) result in Cockayne syndrome (CS), a rare inherited autosomal recessive disease with diverse clinical symptoms including severe growth failure, microcephaly, cachectic dwarfism, progressive neurological degeneration, white matter hypomyelination, lack of subcutaneous fat, cataracts, retinopathy, sensorineural deafness, and hypersensitivity to sunlight (17). Approximately two-thirds of the CS cases are caused by defects in the CSB gene (18, 19). The clinical features raise the question whether CS results solely from failure in DNA repair or if the severe CS phenotype has more complex causes.
Activating transcription factor 3 (ATF3) is a member of the ATF/cAMP response element (CRE) subfamily of basic-region leucine zipper (bZIP) proteins. The DNA binding of the longest isoform usually is associated with repression of its target genes (20). ATF3 is activated dramatically in various stress conditions in a variety of tissues (20, 21). The ATF3 transcriptional network itself is still poorly described and may vary depending on the cellular context (22–24).
In the present study we describe how the DNA-binding factor ATF3, the product of an immediate early gene (IEG), inhibits a panel of genes after a genotoxic stress. We found that ATF3 is recruited to its CRE/ATF-binding site, thus preventing the expression of the corresponding genes. Although in CSB-deficient cells the ATF3 target genes, including dihydrofolate reductase (DHFR), were unable to recover RNA synthesis after the genotoxic stress in CSB-deficient cells, we observed that transcription was restored rapidly in wild-type cells. In CSB cells, ATF3 remains bound at the promoter, thus preventing the recruitment of the RNA Pol II machinery. Silencing ATF3 restores RNA synthesis in UV-irradiated CSB cells suggesting that, in addition to its role in DNA repair, CSB also is required for regulating gene expression in response to a genotoxic attack.
Results
Transcription Recovery After UV Irradiation Depends on CSB Mutation.
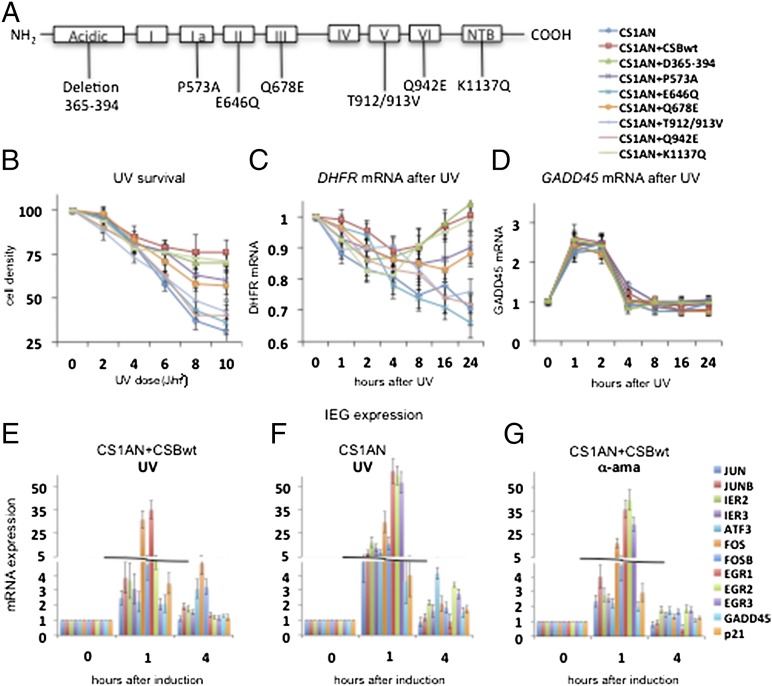
CSB contains seven helicase motifs that are conserved among the SWI2/SNF2 family. The 3D structure of SWI2/SNF2 proteins is organized into two main domains that are folded together to form a DNA-binding cleft harboring a composite ATP-binding site (8, 25). Domain 1 contains helicase motifs I, Ia, II, and III, and domain 2 contains motifs IV, V, and VI. To investigate the role of CSB in transcription, we initially performed a screen of cell lines expressing the following mutations in the CSB gene, five of which are located in the helicase motifs (Fig. 1A) (26): (i) a point mutation at P573 in motif Ia (also named the “Walker motif), which is involved directly in ATP binding and hydrolysis (27) and has a role in the transduction of energy from the ATPase domain to the DNA, because this motif is in close contact with DNA in crystallographic structures (28); (ii) a point mutation E646Q in the Walker B motif (motif II), that coordinates magnesium ions; this mutation completely inhibits the ATPase activity of CSB (28, 29); (iii) either a Q678E mutation in motif III or a Q942E mutation in motif VI; it has been suggested that motif III stabilizes the interaction between domains 1 and 2 by interacting with motif VI, which is located on the other side of the cleft (30, 31); (iv) a double T912/913V mutation in motif V that is located close to the ATP site and abolishes CSB ATPase activity (31); (v) in addition to cell lines with mutations in helicase motifs, we also used cell lines with either a deletion in the conserved acidic domain of amino acids 365–394 or a mutation at position K1137Q in the C-terminal nucleotide-binding (NTB) domain, both with unknown functions. CSB mutations then were introduced into a pcDNA3.1 vector and were stably transfected into a CSB-deficient cell line (CS1AN) to maintain the same genetic background (32). All mutant cell lines express CSB with the same efficiency (26).
Fig. 1.
Arrest of RNA synthesis and induction of IEGs upon genotoxic stress in CS cells. (A) Schematic diagram of CSB secondary structure showing helicase motifs I–VI and the acidic and NTB domains. Cell lines used were transfected with pcDNA3.1 plasmid carrying point mutations with the denoted locations. (B) UV survival assay showing cell density 4 d after exposure to 0–10 J/m2 UV-C radiation. (C and D) DHFR mRNA (C) and GADD45 mRNA (D) after 10-J/m2 UV-C irradiation. Graphs show the average of three independent experiments. (E–G) A quantitative RT-PCR analysis of the set of IEG in CS1AN+CSBwt and CS1AN cells treated with 10 J/m2 of UV-C or 10 μg/mL of α-amanitin as indicated and harvested at different time points after UV-C irradiation. Genes listed from the top to the bottom at the right of G are shown from left to right in each histogram. ATP7A, ATPase, Cu++ transporting, alpha polypeptide DLD, Dihydrolipoamide dehydrogenase; NBN, Nibrin; RAB3GAP2, RAB3 GTPase activating protein subunit 2;RAD50, RAD50 homolog (S. cerevisiae). Other gene symbols are defined in the text. All results are presented as the ratio of the value obtained at each time point relative to that of the untreated cells at time t = 0. Each point represents the average of three independent experiments that were performed in triplicate.
We first determined the impact of the various CSB mutations on the sensitivity to UV light by measuring cell density 4 d after exposure to UV-C irradiation at 0–10 J/m2 (Fig. 1B). The mutation K1137A in the NTB domain or deletion of the N-terminal acidic domain (Δ365–394) did not substantially affect UV survival, which was similar to that of rescued wild-type (CS1AN+CSBwt) cells. Mutations in motifs Ia (P573A) and III (Q678E) caused a slight sensitivity to UV irradiation, with cell densities of ∼60% 4 d after treatment. However, mutations in helicase motif II (E646Q) and in motifs V (T912/913V) and VI (Q942E) resulted in a very severe UV sensitivity similar to that in CS1AN cells, demonstrating a gradient in the sensitivity toward UV irradiation depending on the location of the CSB mutation. Next, we investigated the recovery of RNA synthesis in wild-type cells (CS1AN+ CSBwt) and cells expressing mutated CSB (Fig. S1A and ref. 10). Although wild-type cells and cells with mutations in the NTB or the acidic domain recovered rapidly from the UV-induced inhibition of RNA synthesis, cell lines stably expressing mutations in helicase motif II or motifs V and VI, and cells not expressing full-length CSB were unable to recover transcription throughout the entire time course. Cells with mutations in helicase motifs Ia and III (33) displayed an intermediate ability to perform RNA synthesis after UV irradiation, similar to the pattern observed during UV survival.
We previously demonstrated that the absence of full-length CSB causes dysregulation in the transcription of the housekeeping gene DHFR in response to UV irradiation, whereas the expression of the protein 53 (p53)-dependent gene Growth arrest and DNA damage-inducible protein 45 (GADD45) remained unaltered (10). Therefore we investigated the expression of DHFR and GADD45 in cells carrying point mutations in the CSB gene. We irradiated the mutant cell lines with 10 J/m2 UV light and then collected the mRNA. We found that mutations in NTB (K1137Q) and the acidic domain (Δ365–394) did not affect the recovery of DHFR expression after UV irradiation (Fig. 1C). However, mutations in helicase motif Ia (P573A) and III (Q678E) caused a slight reduction in DHFR mRNA level 16–24 h after UV irradiation as compared with wild type. An even more prominent reduction in DHFR mRNA level was detected when helicase motifs II (E646Q), V (T912/913V), and VI (Q942E) were mutated, demonstrating that different mutations in the CSB gene can variably affect the transcriptional program. This result demonstrates that these motifs, most of which are associated with the CSB ATPase activity (34), are implicated in the expression of DHFR gene.
Interestingly, upon UV irradiation, GADD45 (Fig. 1D), as well as the IEGs, including Jun proto-oncogene (JUN), immediate early response 2 and 3 (IER2 and IER3), ATF3, FBJ murine osteosarcoma viral oncogene homolog (FOS), and Early growth response 1 (EGR1) (Fig. 1G), peak strongly in both wild-type and CSB-deficient cells (Fig. 1 E and F and Table S1). Their expression is not affected by mutations in NER factors (10, 35).
ATF3 Is Overexpressed in UV-Irradiated Cells.
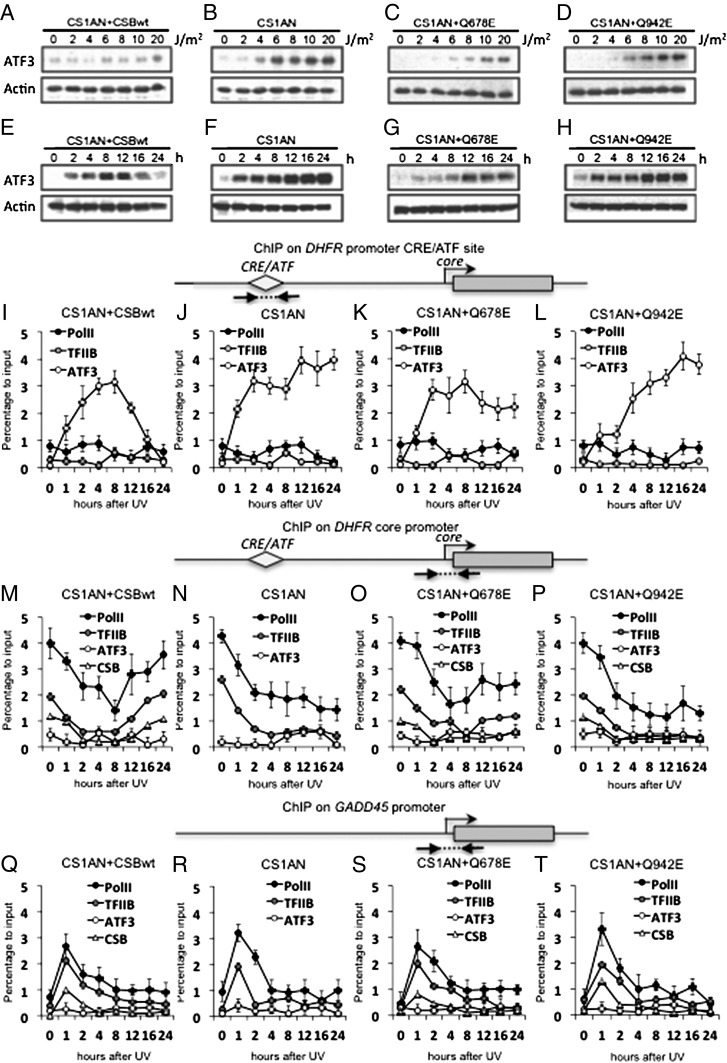
With the list of IEGs induced by UV irradiation in hand, we performed bibliographic studies and found that one of IEG, ATF3, is a repressor. Because ATF3 was expressed similarly several hours after the initial irradiation in both CSB and wild-type cells and because many housekeeping genes, including DHFR, exhibit a CRE/ATF-binding site upstream of their promoter, we considered ATF3 a good candidate and decided to analyze more deeply the potential role of this repressor in inhibiting transcription after a genotoxic attack (20, 36). The CRE (TGACGTATG) site is located at position −1686 relative to the DHFR transcription start site (TSS). We observed that mRNA synthesis of ATF3, as well as other IEGs, peaks a few hours after UV stress in both wild-type and CSB-deficient cells (Fig. 1 E and F). Thinking that ATF3 repressor overexpression could be a specific signature of UV stress response in an impaired CSB background, we further checked ATF3 protein levels in the CS1AN+CSBwt, CS1AN+Q678E, CS1AN+Q942E, and CS1AN cell lines. Increasing the UV dose from 0 to 20 J/m2 results in a strong accumulation of ATF3 in all the CSB-deficient cell lines as compared with wild-type cells (compare Fig. 2 A vs. B–D). Interestingly, at a UV dose of 4 J/m2 the ATF3 accumulation in CS1AN cells corresponded to the accumulation observed at 20 J/m2 in CS1AN+CSBwt cells (compare Fig. 2 B vs. A). The Q678E mutation in motif III resulted in a slightly lower accumulation of ATF3 than did the Q942E mutation in motif VI (Fig. 2 C and D). We also observed that ATF3 is recruited immediately at the chromatin in both CS1AN and CS1AN+CSBwt cells (Fig. S1 B and C). We next performed Western blotting after UV irradiation (10 J/m2) over a time course and found clear ATF3 accumulation in all four cells lines. In CS1AN+CSBwt cells, however, the induction of ATF3 peaked at 8 h and ceased at 16–24 h after treatment (Fig. 2 A and E), whereas the CS1AN, CS1AN+Q678E, and CS1AN+Q942E cell lines maintained a high level of ATF3 expression throughout the 24-h time course (Fig. 2 F–H).
Fig. 2.
Continuous ATF3 accumulation after UV irradiation in CS cells. (A–D) Western blots showing ATF3 expression 24 h after irradiation with 0, 2, 4, 6, 8, 10, and 20 J/m2 UV-C in CS1AN+CSBwt (A), CS1AN (B), CS1AN+Q678E (C), and CS1AN+Q942E (D) cells. (E–H) Western blots determining ATF3 accumulation in untreated cells and 2, 4, 8, 12, 16, and 24 h after exposure to 10 J/m2 UV-C in denoted cell lines. (I–L) ChIP determining Pol II, TFIIB, and ATF3 enrichment at the DHFR CRE/ATF site after exposure to 10 J/m2 UV-C in denoted cell lines. (M–P) ChIP at the DHFR core promoter. (Q–T) ChIP determining Pol II, TFIIB, ATF3, and CSB enrichment at the GADD45 promoter after exposure to 10 J/m2 UV-C in denoted cell lines. A schematic diagram of location of primers used in ChIP experiments is presented at the top of each set of ChIP. All the results are presented as percentage to input giving the respective percentage of enrichment in comparison with chromatin input. Each point represents the average of three real-time PCR reactions in three independent experiments.
Taken together, these data indicate that ATF3 gene expression and protein concentration are increased and accumulate after UV irradiation in CSB-deficient cells, suggesting a change in the ATF3 turnover rate.
ATF3 Remains Bound to the DHFR CRE/ATF Site in CSB-Deficient Cells.
We next performed ChIP to study whether ATF3 is recruited to the CRE/ATF site on the DHFR promoter after exposure to 10-J/m2 UV irradiation. In CS1AN+CSBwt cells, we discovered a high enrichment of ATF3 protein on the DHFR CRE/ATF site that peaked at 8 h after UV treatment and decreased 12–24 h later (Fig. 2I). Such ATF3 recruitment inversely parallels DHFR mRNA synthesis, which increases concomitantly with the release of ATF3 from its cognate site (Figs. 1C and 2I), and the recruitment of Pol II, CSB, and the basal transcription factor IIB (TFIIB) to the core promoter of DHFR (Fig. 2M). In CS1AN, CS1AN+Q678E, and CS1AN+Q942E CSB cells, ATF3 was recruited to the CRE/ATF site on the DHFR promoter immediately upon UV irradiation. However ATF3 protein enrichment at CRE/ATF did not cease in these cells after 12 h (as it did in wild-type cells) but instead remained recruited throughout the entire time course (compare Fig. 2 I vs. J–L). This result correlates well with the increased ATF3 accumulation in the corresponding cell lines (Fig. 2 F–H). The synchronization of the rate of ATF3 synthesis after UV irradiation and its presence in chromatin extracts in CS1AN+CSBwt and CS1AN cells (compare Fig. 2 E and F vs. Fig. S1 B and C) is noteworthy. The accumulation of ATF3 at the DHFR promoter in CSB-deficient cells parallels the substantial decrease in Pol II and TFIIB recruitment in response to UV, a decrease that was not restored even 24 h after treatment (Fig. 2 N–P).
In CS1AN, CS1AN+Q678E, and CS1AN+Q942E cells, CSB and Pol II were not recruited to the DHFR promoter after UV irradiation (Fig. 2 N–P). It should be noted that the decrease in Pol II and TFIIB enrichment seemed more prominent in CS1AN+Q942E cells than in CS1AN+Q678E cells (Fig. 2 O and P). This slight difference between the two cell lines correlates with the more pronounced ATF3 recruitment to CRE/ATF and the lower DHFR mRNA level after UV in CS1AN+Q942E cells and also with the lower UV survival and recovery of RNA synthesis in CS1AN+Q942E cells compared with CS1AN+Q678E cells (Fig. 1B and Fig. S1A).
In these four cell lines we did not detect any presence of ATF3 on the core promoter of GADD45, which does not contain a CRE/ATF site (Fig. 2 Q–T). However, antibodies against Pol II and TFIIB revealed a clear enrichment of these factors immediately after UV irradiation, peaking at 1 h after treatment and followed by a decrease that parallels the pattern of GADD45 expression (Fig. 1D). On both the DHFR core promoter and the GADD45 promoter, we detected the presence of CSB together with the transcription machinery in CS1AN+CSBwt, CS1AN+Q678E, and CS1AN+Q942E cells (Fig. 2 M, O–Q, S, and T). Because CS1AN cells do not express full-length CSB (26), we did not check for the presence of CSB on the DHFR and GADD45 core promoters (Fig. 2 N and R).
Taken together, these results suggest that f full-length CSB is needed to regulate ATF3-dependent DHFR expression and raise the question of whether CSB has a more general role of CSB in regulating gene expression.
ATF3 Inhibits Transcription of Its Direct Targets in CS Cells.
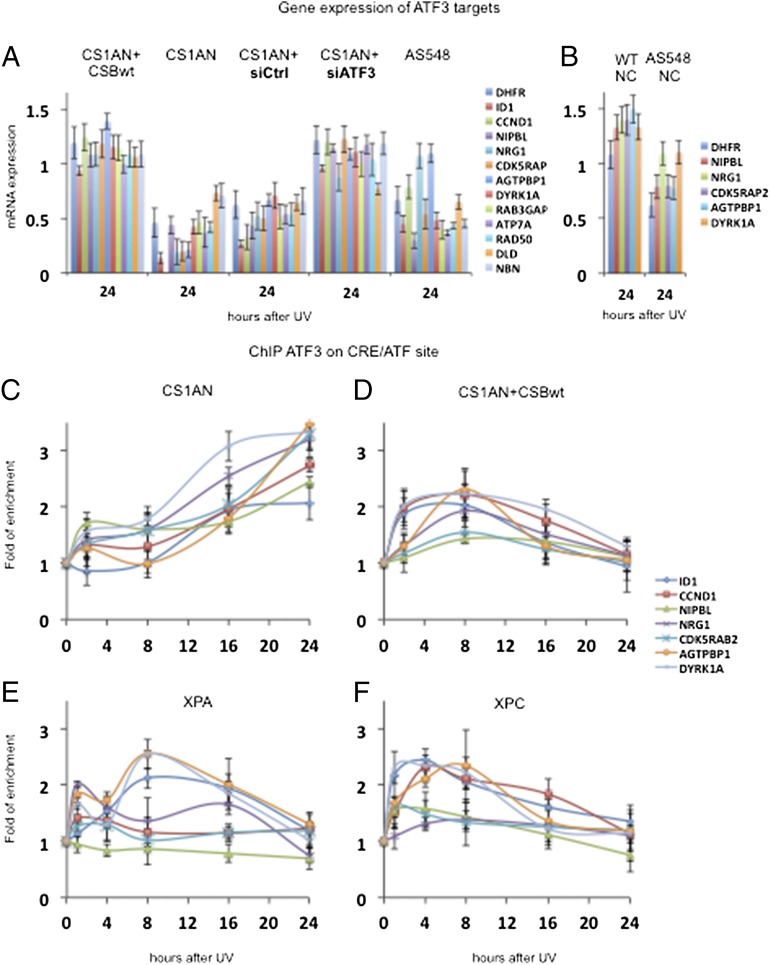
Although IEGs were induced similarly a few hours after UV treatment (Fig. 1 E and F), other genes were unable to recover normal RNA synthesis activity in the CS1AN+CSBwt, CS1AN, and AS548 cell lines as demonstrated by microarrays and quantitative RT-PCR (Fig. 3A, Fig. S1H, and Tables S1 and S2). We identified 1,217 genes that were down-regulated (by <0.5-fold, P < 0.001) (Materials and Methods, Fig. S1H, and Table S2) in the CS1AN, as compared with the CS1AN+CSBwt cell line, 24 h after UV treatment. Next, we aligned the mRNA expression profile together with the global ATF3 promoter occupancy (ChIP-Seq data) (24, 37). In the set of down-regulated genes we identified 334 genes (27%) that contain CRE/ATF-binding site candidates in close proximity to the corresponding TSS (Fig. S1H and Table S2). Among these were genes that previously had been identified as ATF3 transcriptional targets [e.g., Inhibitor of DNA binding 1 protein (ID1), Cyclin D1 (CCND1), Endothelin 1 (EDN1), Receptor-interacting serine-threonine kinase 2 (RIPK2), and AT rich interactive domain 5A (ARID5A)] (22, 33, 38) and others that had not. For a number of selected genes [ID1, CCND1, Nipped-B homolog (NIPBL), Neuregulin 1 (NRG1), CDK5 regulatory subunit associated protein2 (CDK5RAP2), ATP/GTP-binding protein 1 (AGTPBP1), and Dual-specificity tyrosine-phosphorylation kinase 1A (DYRK1A)], ChIP experiments show recruitment of ATF3 in CS1AN cells that remains bound at the CRE/ATF site throughout the entire time course and even longer (Fig. 3C). In wild-type cells, to the contrary, ATF3 remains recruited for a short period (Fig. 3D), after which RNA synthesis is restored (Fig. 1C and Fig. S1A). Interestingly, in Xeroderma pigmentosum, complementation group C (XPC)- and Xeroderma pigmentosum, complementation group A (XPA)-deficient cells, ATF3 is recruited at the promoter of the genes containing a CRE/ATF-binding site for a time period similar to that observed in wild-type cells (Fig. 3 E and F). These data strongly establish a correlation between the defective expression profile of genes (other than DHFR) and the presence of ATF3 on their respective promoters in CSB-deficient cells.
Fig. 3.
ATF3 binding to CRE/ATF sites and subsequent transcriptional repression of target genes. (A) Quantitative RT-PCR analysis of the ATF3 target genes (shown in Fig. S1H and Table S2) in CS1AN+CSBwt cells, CS1AN cells, CS1AN cells transfected with either siCtrl or siATF3, and AS548 cells (19) 24 h after 10-J/m2 UV-C treatment. (B) Quantitative RT-PCR analysis of the newly identified ATF3 target genes in wild-type (CRL-2097) and AS548 neuronal cultures 24 h after treatment with 10-J/m2 UV-C. Cells were harvested at the 0- and 24-h time points after UV-C treatment. Gene expression at 24 h was normalized to 0 h. (C–F) ChIP assays showing enrichment of ATF3 binding at CRE/ATF sites of selected gene promoters. CS1AN (C), CS1AN+CSBwt (D), XPA (XP12RO) (E), and XPC (GM14867) (F) cells were treated with 10 J/m2-UV-C and harvested at different time points within 24 h. The genes that are named from the top to the bottom at the right in A and B are shown from left to right in each histogram. All the results are presented as fold recruitment, the ratio of the value obtained at each time point relative to that of the untreated cells at time t = 0. Each point represents the average of three real-time PCR reactions of three independent experiments.
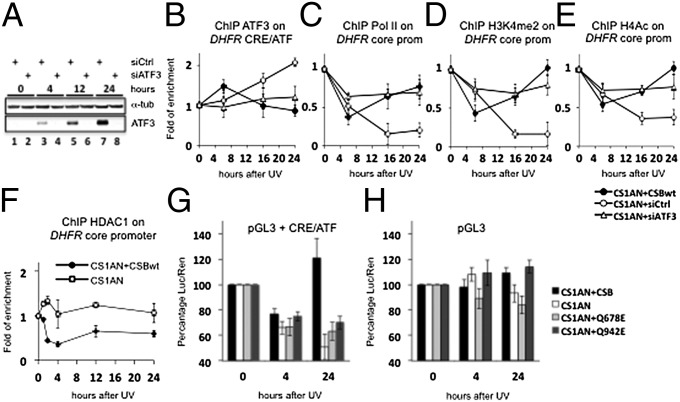
We next investigated the regulatory function of ATF3 in CSB-deficient cells. CS1AN cells were treated with either siRNA against ATF3 (siATF3) or control siRNA without a target (siCtrl) and were exposed to UV irradiation. As compared with siCtrl, the siRNA-mediated knockdown of ATF3 was very significant even 24 h after UV treatment as demonstrated by Western blot (Fig. 4A). In our experimental conditions there were no significant differences in UV survival in CS1AN+siATF3 and CS1AN+siCtrl cells 48 h after 10-J/m2 UV irradiation (Fig. S1E). As a consequence, we observed that all the genes tested thus far that were repressed in UV-treated CS1AN+siCtrl cells recovered their RNA synthesis activity in CS1AN+siATF3 cells (Fig. 3A). In those cells, the level of DHFR mRNA synthesis reached that of CS1AN+CSBwt cells, but DHFR mRNA synthesis was not recovered in CS1AN+siCtrl cells. As expected, ChIP assay next showed that at 24 h after irradiation the recruitment of ATF3 on the DHFR CRE/ATF-binding site was strongly reduced in CS1AN+siATF3 cells as compared with CS1AN+siCtrl cells (Fig. 4B). Under these conditions the level of Pol II on the DHFR core promoter in CS1AN+siATF3 cells was similar to that observed in CS1AN+CSBwt cells and was much higher than in CS1AN+siCtrl cells 24 h after UV irradiation (Fig. 4C), thus demonstrating that transcription did occur (Fig. 3A).
Fig. 4.
siRNA-mediated ATF3 down-regulation abolishes the repression of ATF3-dependent genes in CS cells. (A) Western blot analysis of ATF3 protein in CS1AN cells transfected with siCtrl or siATF3 constructs and harvested at different time points after UV-C treatment. (B–E) ChIP experiments showing enrichment of ATF3 on the DHFR CRE/ATF site (B), Pol II on the DHFR core promoter (C), H3K4me2 (D), and acetylated histone H4 on the DHFR core promoter (E) after exposure to 10-J/m2 UV-C in CS1AN+CSBwt, CS1AN+siCtrl, and CS1AN+siATF3 cells. (F) ChIP assay on the DHFR core promoter in CS1AN+CSBwt and CS1AN cell lines showing the stable presence of HDAC1 over a time course of 24 h after UV-C (10 J/m2) treatment in CS1AN and CSi1AN+CSBwt cells. (G and H) Luciferase assay in untreated CS1AN, CS1AN+CSBwt, CS1AN+Q678E, and CS1AN+Q942E cells and 4 and 24 h after 10-J/m2 UV-C irradiation. These cells were transfected with luciferase plasmid with (G) or without (H) a CRE/ATF site in front of the SV40 promoter. All results are presented as fold recruitment, which represents the ratio of the value obtained at each time point relative to that of the untreated cells at time t = 0. Each point represents the average of three real-time PCR reactions of three independent experiments.
Active transcription often is associated with heterochromatin landmarks such as di/tri methylation of histone H3 (H3K4me2/3) and histone H3/Histone H4 (H3/H4) acetylation. We also observed that silencing ATF3 in CS1AN restores the H3K4 dimethylation as well as H4/H3 acetylation, two chromatin landmarks already observed around the DHFR promoter in CS1AN+CSBwt (Fig. 4 D and E). In CS1AN+siCtrl, this decrease in histone modification lasted up to 24 h after UV irradiation. ATF3 promotes transcriptional repression by recruiting histone deacetylases (HDACs) (22). HDACs have a critical role in chromatin remodeling by participating in the acetylation/deacetylation cycle of histones: Acetylation relaxes the chromatin structure, thereby allowing access of transcription factors to DNA; conversely, deacetylation alters the chromatin structure to limit access of transcription factors. Thus, it was not surprising to observe the permanent presence of HDAC1 at the DHFR promoter in CS1AN cells (Fig. 4F).
To investigate further the role of ATF3 in repressing RNA synthesis in UV-irradiated CSB cells, we designed two luciferase reporter constructs, one containing a CRE/ATF site upstream of a simian virus 40 (SV40) promoter (pGL3+CRE/ATF), and one without the CRE/ATF site (pGL3). These plasmids were transfected into the CS1AN, CS1AN+CSBwt, CS1AN+Q678E, and CS1AN+Q942 cell lines that then were UV irradiated. In response to UV irradiation, the luciferase activity was decreased significantly in all cell lines at 4 h after treatment when CRE/ATF was inserted upstream of the SV40 promoter (Fig. 4G). However, CS1AN+CSBwt cells recovered luciferase activity24 h after UV treatment, correlating with the down-regulation of ATF3 expression observed 24 h after treatment in this cell line (Fig. 2E). At that time, CS1AN, CS1AN+Q678E, and CS1AN+Q942E cells were unable to restore luciferase activity (Fig. 4G). The three different mutated or truncated CSB cell lines as well as the restored CSB wild-type cell lines transfected with a plasmid lacking the CRE/ATF-binding site allow proficient luciferase activity in response to UV irradiation (Fig. 4H), demonstrating that the CRE/ATF site indeed is responsible for the ATF3-mediated repression of transcription.
Taken together, these results demonstrate that with UV irradiation there is a repression of ATF3-responsive genes in CSB-deficient cells that is circumvented when its CRE/ATF target site is deleted and/or when ATF3 expression itself is silenced. Additionally, these results suggest that CSB, as well as ATF3, is implicated in the acetylation/deacetylation process of histones.
Inhibition of Pol II Transcription Promotes ATF3 Induction.
Having shown that ATF3 is induced by UV and binds to CRE/ATF sites within promoters and prevents the recruitment of Pol II either temporarily (in CSB wild-type cells) or permanently (in CSB-deficient cells), we asked whether the ATF3-induced repression is caused by the unavailability of Pol II to join the DHFR promoter. To induce a slight inhibition of Pol II activity in wild-type cells, CS1AN+CSBwt cells were incubated for 1 h with 10 μg/mL of α-amanitin, a concentration 2,500 times lower than usually used to inhibit mRNA synthesis completely (39, 40). When interacting with the bridge helix in RNA pol II, α-amanitin prevents its binding to the DNA and/or constrains its mobility and hence slows the translocation of the polymerase and the rate of synthesis of the RNA molecule (41).
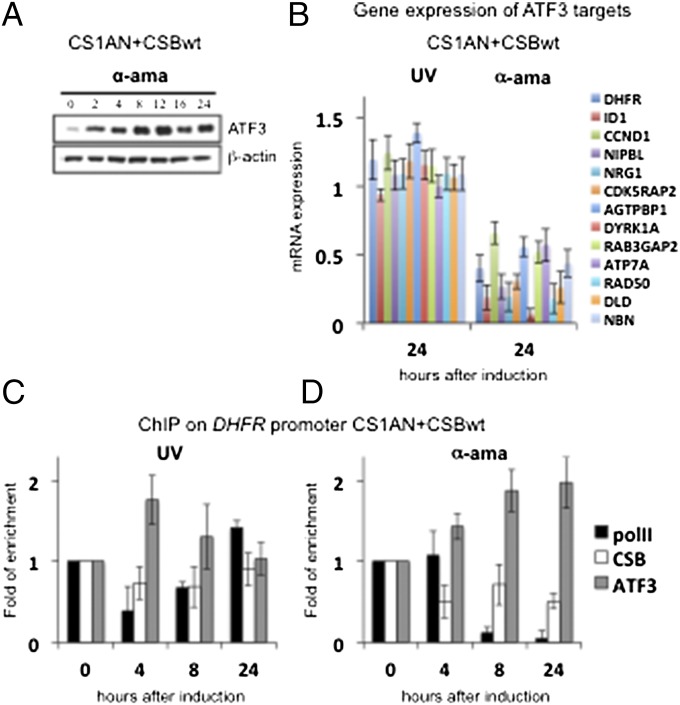
First, we quantitatively compared the mRNA expression levels of different IEGs in CS1AN+CSBwt and CS1AN cells after treatment with UV-C (Fig. 1 E and F) with the expression levels in CS1AN+CSBwt cells treated with α-amanitin (Fig. 1G). In all three settings we observed an almost similar induction of these IEGs, including ATF3 (Fig. 1 E–G and Table S1). Twenty-four hours after α-amanitin administration the accumulation of ATF3 protein in the treated CS1AN+CSB cells (Fig. 5A) was similar to that in CSB-deficient CS1AN cells after UV-C treatment (Fig. 2F). As a consequence, mRNA expression of DHFR still is abrogated considerably 24 h after α-amanitin treatment in CS1AN+CSBwt cells (Fig. 5B), similar to the previously observed down-regulation in UV-irradiated CS1AN cells (Figs. 1C and 3A). We also analyzed quantitatively the mRNA expression levels of the direct ATF3-target genes that we had monitored previously (Fig. 5B). ChIP experiments next showed that ATF3 protein remains recruited throughout the entire time course on the DHFR promoter region in CS1AN+CSBwt cells treated with α-amanitin (Fig. 5D), in clear contrast to what occurs in UV-treated cells (Fig. 5C; also see Fig. 2 I and M). The recruitment of CSB and Pol II is diminished significantly at 8 and 24 h after treatment with α-amanitin, explaining the defect in RNA synthesis (Fig. 5D). These data from the CS1AN+CSBwt cell line clearly demonstrate that α-amanitin treatment induces the expression of the stress-response gene ATF3, thereby causing transcriptional effects comparable to those seen after UV-C treatment. That is, the recruitment of ATF3 protein at the DHFR promoter region matches its recruitment in UV-C–irradiated CS1AN cells, showing that the transcriptional arrest observed in UV-C–irradiated CS1AN cells is caused not only by defective TCR but also by disturbed regulation of transcription.
Fig. 5.
α-Amanitin treatment mimics UV-induced stress and causes continued ATF3 accumulation. (A) Western blot with antibody against ATF3 and β-actin as loading control in CS1AN+CSBwt cells incubated with 10 μg/mL α-amanitin for 1 h. Cells were collected at indicated time points after treatment. The genes that are listed from the top to the bottom at the right of Fig. 3 A and B are shown from left to right in each histogram. (B) Quantitative RT-PCR analysis of direct ATF3 target genes (shown in Fig. S1H and Table S2) in CS1AN+CSBwt cells: (i) 24 h after UV-C irradiation (10 J/m2) and (ii) 24 h after administration with 10 μg/mL α-amanitin for 1 h. (C and D) ChIP assay showing the enrichment of Pol II, CSB, and ATF3 at the DHFR promoter at 0, 4, 8, and 24 h after UV (C) or α-amanitin (D) treatment. All results are presented as fold recruitment, which represents the ratio of the value obtained at each time point relative to that of the untreated cells at time t = 0. Each point represents the average of three real-time PCR reactions of three independent experiments.
Discussion
In response to UV irradiation, a panel of genes does not recover RNA synthesis in CSB-deficient cells, although the IEGs are activated whether or not CSB is mutated. The developmental consequences of CSB mutations shown by the very severe clinical features observed in patients with CS (19) may be explained by the defective expression of a certain set of genes. This study provides mechanistic insights into the role of CSB in regulating the expression of a certain set of genes after genotoxic stress and also provides explanations for the selective down-regulation of ATF3-dependent genes and the relationship of this down-regulation to CS clinical features.
In wild-type cells, CSB is recruited cyclically at the chromatin upon either UV irradiation or ligand induction, but such recruitment is abolished in CSB-deficient cells (3, 9, 10, 34). In both CBS-proficient and -deficient cells, ATF3 expression is induced with UV irradiation, and further ATF3 is recruited to the CRE/ATF-binding site located in the vicinity of the promoter of numerous genes, including DHFR, to repress transcription (Figs. 2 I–L and 3 A–D; see also refs. 36 and 42). Although in wild-type cells the overexpression and the binding of ATF3 to its target sites peaks at 8 h and disappears completely at 24 h when RNA synthesis restarts, in CSB-deficient cells ATF3 accumulates and is maintained at the promoter of several genes (compare Figs. 2 I vs. J and 3 C vs. D). In this latter case, it is likely that the half-life of ATF3 is increased. The recruitment of ATF3 by the various target genes parallels the removal of Pol II together with CSB, and during that period transcription is arrested (Fig. 2).
We further demonstrate that the arrest of RNA synthesis is caused by the binding of ATF3 itself to its native target sites: (i) expression of luciferase from a construct that contains one CRE/ATF-binding site in front of the SV40 promoter is abolished in UV-treated cells with a CSB-deficient background, and deleting the ATF/CRE site allows luciferase expression (Fig. 4 G and H); and (ii) siRNA-mediated knockdown of ATF3 releases the repression of direct targets of ATF3 in UV-irradiated cells, thereby circumventing some of the deleterious effects of CSB mutations (Fig. 3A).
These findings raise questions about the role of CSB in the arrest of RNA synthesis in addition to its function at the sites of DNA lesions with stalled transcription machinery (1, 3, 43, 44).
Interplay Between CSB and ATF3 Controls the Expression of ATF3 Direct Targets.
To explain further the arrest of RNA synthesis in UV-irradiated cells, a scenario was proposed in which the appearance of genotoxic stress originated by UV irradiation induces activation of both the ATF3 early immediate response and the p53 pathways (Fig. 1 E–G). Activation of the p53 branch mediates the transcription of genes such as GADD45, whose products can be involved in cell-cycle regulation and apoptosis. ATF3 activation, which is p53 independent, would act as a pivotal transcription factor that represses genes (Figs. 3 A and B and 5B), including those involved in the cell cycle and in apoptosis as well (45–47). Such a defense mechanism allowing cellular arrest could be developed to recover normal activity; then Pol II rejoins the promoter to reinitiate RNA synthesis as observed in CSB-proficient cells. It seems that the presence of CSB and ATF3 at the promoter is mutually exclusive (Fig. 2 I–P). ATF3 neither interacts with the CSB ATPase in a protein–protein interaction nor allows its removal from DNA upon the addition of ATP (at least in our in vitro EMSA experiment) (Fig. S1D).
An initial hypothesis suggested that arrest of transcription by ATF3 allows the repair of damaged DNA (48) and the recycling of the phosphorylated elongating Pol II so that normal gene transcriptions can resume (1, 3, 10). ATF3 could be involved in transcriptional arrest and thus stimulate DNA repair (46). In this scenario, CSB is recruited to the stalled Pol II upstream of a lesion and works to recruit further chromatin remodeling and NER proteins consecutively to facilitate the repair of DNA lesions (3–5, 10, 11). Mutations in CSB are unable to initiate the formation of a TCR complex, thus preventing the reinitiation of RNA synthesis, because the elongating Pol II is blocked upstream of the lesion (2) and therefore cannot be recycled for further reinitiation. Because 25 J/m2 of UV introduced around two DNA lesions per 10 kb of DNA, another hypothesis suggested that damaged genes are defective in transcription (49). In this scenario, the CSB-initiated repair of DNA damage and the further availability of Pol II are required to restart RNA synthesis.
However, these assumptions are insufficient or at least are incomplete. Indeed we demonstrate that silencing ATF3 and/or deleting its CRE/ATF target site circumvents transcription arrest and allows transcription (at least of the ATF3-dependent genes) in UV-irradiated CSB cells (Figs. 3A and 4 G and H), suggesting that the presence of unrepaired DNA lesions is not the sole cause of the defect in RNA synthesis. This result also demonstrates that active Pol II is still available for RNA synthesis, even if it is partially blocked by DNA lesions in CSB-deficient cells. Thus it seems that the processes of transcription arrest and DNA repair can be separated and that eliminating DNA lesions does not initiate the restart of transcription. The perturbation of Pol II activity by very low concentrations of α-amanitin also causes ATF3 induction and increases binding to the CRE/ATF site in the absence of extrinsic damage in a numerous set of genes, with resulting persistent shutdown of the expression of these genes even in wild-type cells. However, we cannot exclude the possibility that inhibition of Pol II (by α-amanitin and/or stalled in front of a DNA lesion) provides a CSB-independent signal for ATF3-dependent repression of a large set of genes, as also observed after UV treatment. Such signal in response to UV damage could be resolved after TCR in normal cells but persists in CS cells because they cannot perform TCR, with resulting persistent repression of the class of ATF3-responsive genes (Figs. 3 A and B and 5B). Therefore, we conclude that the arrest of RNA synthesis is not caused exclusively by the defect in repairing damaged DNA.
Previously it has been shown that CSB is recruited at the promoter of activated genes (Fig. 2M) (9), but questions remain about the role of CSB in removing the repressor from its cognate site. Our results suggest that not all of Pol II is blocked at DNA lesions, but active Pol II alone is not sufficient to restore transcription in UV-irradiated cells unless wild-type CSB is present. Thus, in addition to its role in DNA repair, CSB is a key component in the transcription arrest that occurs with genotoxic stress. CSB’s key role in regulating transcriptional arrest upon UV stress also is seen in cells with an XPC or XPA NER-defective background that otherwise are CSB proficient. In such cases CSB may be required to remove ATF3, although the presence at the promoter is mutually exclusive.
CSB was defined as an ATP-dependent chromatin remodeler belonging to the SWI/SNF family that uses ATP as energy to alter DNA–histone interactions. ChIP analysis suggests that ATF3 may silence DHFR by recruiting HDACs but did not tell us what role CSB plays in relieving ATF3 bound to its native CRE/ATF site. However, we have observed that the ATPase activity of CSB is absolutely required for its recruitment at chromatin (3, 9, 10, 34) and likely participates in the removal of ATF3 and restoration of optimal gene expression after genotoxic attack. Indeed mutations in CSB that affect its ATPase activity as well as mutations in the helicase motifs (28, 29) prevent its recruitment at chromatin and consequently prevent the removal of ATF3 from its target sites (Fig. 2 O and P). The CSB/K1137Q and CSB/P1042L mutations located at the C terminus and the CSB/P573A mutation, which does not affect its ATPase activity, do not prevent the restart of RNA synthesis after UV treatment (Fig. S1 A and C) (2, 10, 15, 34). Our data suggest that the energy provided by CSB contributes, in collaboration with Pol II, to remove ATF3 repressor from its binding site, suggesting that the helicase activity might have a role in modifying DNA–histone as well as DNA–transcription factor contacts (50). Further investigation will be needed to analyze the interconnections between CSB, ATF3, and various HDACs, all of which seem to be implicated in the acetylation/deacetylation process of histones and thus in the chromatin remodeling of promoter regions of CSB-sensitive downstream genes.
ATF3 Overexpression and CS Clinical Features.
The cellular or genotoxic stress experienced during a lifespan is managed efficiently in wild-type cells. In a CSB-deficient background such stress results in continuous up-regulation of ATF3 that changes the transcriptional landscape. We propose that such down-regulation of ATF3 target genes might be responsible, at least in part, for the clinical features seen in patients with CS.
Focusing on the neurodegenerative aspects of CS, we selected a set of genes that become down-regulated in CS1AN cells 24 h after UV treatment (51). Among these candidates, we selected those that offer ATF3 binding close to their corresponding TSSs and that previously have been shown to be associated with multiple neurodegenerative or hypomyelination disorders (Fig. S1H and Tables S2 and S3). A selected set of these genes (NIPBL, NRG1, CDK5RAB2, AGTPBP1, and DYRK1A) was confirmed to be true ATF3 targets in CS1AN cells (Fig. 3 A, C, and D). Additionally, the same pattern of gene down-regulation was confirmed in another cell line of fibroblasts derived from a CS patient (Fig. 3A) with a different CSB mutation (AS548) and with phenotypes of varying severity (19).
To mimic a cell type-specific content of affected tissue, we differentiated the wild-type and AS548 human induced pluripotent stem cells (hiPSCs) into neuronal cultures (52, 53) and measured the expression of direct ATF3 targets 24 h after UV treatment (Fig. 3B). We compared the expression of ATF3 directly in UV-induced and noninduced populations in wild-type and AS548 neuronal cultures. Interestingly, three newly described targets, NIPBL, CDK5RAP2, and AGTPBP1, were down-regulated in neuronal cultures of CS AS548 cells after UV treatment. NIPBL is a homolog of the Drosophila melanogaster Nipped-B gene product and fungal sister chromatid cohesion type 2 (Scc2) proteins. Mutations in NIPBL were found to be responsible for Cornelia De Lange syndrome (54, 55), a disorder characterized by dysmorphic facial features, growth delay, limb reduction defects, and mental retardation. Interestingly, down-regulation of NIPBL also may impair adipogenesis (56). CDK5RAP2 is a CDC2-like kinase that is a key regulator of centrosomal maturation. Mutations in this gene are associated with autosomal primary recessive microcephaly, a disorder characterized by small brain size caused by deficient neuron production in the developing cerebral cortex (57–59). AGTPBP1 is a zinc carboxypeptidase that initially was cloned from spinal motor neurons undergoing axon regeneration. Interestingly, AGTPBP1 was found to be deleted inside a Purkinje cell degeneration (PCD) allele (60). The PCD phenotype is associated with changes in nuclear chromatin architecture and function (61, 62).
Our findings suggest that such stress-dependent down-regulation of these genes is coupled to the neurological features of CS patients, because dysfunction in these genes may account for mental retardation (NIPBL), microcephaly (CDK5RAP2), and Purkinje cell degeneration in the cerebella cortex (AGTPBP1) (19).
Materials and Methods
Cell Lines.
CS1AN, CS1AN+CSBwt, CS1AN+Q678E, and CS1AN+Q942E SV40 transformed fibroblasts were grown in Dulbecco/HamF10 medium containing 10% FCS and 40 mg/mL gentamycin. Primary fibroblasts from CS patients AS548 (19) and GM14867 and XP12RO were grown in MEM containing 10% (vol/vol) or 15% (vol/vol) FCS and 40 mg/mL gentamycin.
Generation and Culture of Human iPS Cells.
The human wild-type (CRL-2097; American Type Culture Collection) and AS548 fibroblasts were transduced with concentrated retroviruses of Yamanaka mixture (Vectalys) (63). After a week of culture in DMEM/FBS 10% (vol/vol) medium, fibroblasts were passaged onto mouse embryonic fibroblasts (MEFs) and were maintained and cultured in human ES cell medium: DMEM/F12 containing 20% (vol/vol) Knockout Serum Replacement (KSR; Invitrogen), 10 ng/mL basic FGF (PeproTech), 1 mM l-glutamine, 100 μM nonessential amino acids, 100 μM β-mercaptoethanol, 50 U/mL penicillin, and 50 mg/mL streptomycin. Several hiPSC clones were selected and expanded in each cell line.
Directed Differentiation of hiPSCs.
hiPSCs were dissociated using TrypLE (Invitrogen) and were preplated on gelatin-coated dishes in human ES cell medium supplemented with 10 μΜ Rho-kinase (ROCK) inhibitor (Tocris) for 1 h to eliminate MEFs,. A suspension of nonadherent iPSCs then was plated on Matrigel-coated dishes (BD Bioscience). Confluent hiPSC cultures were induced for neurogenesis by switching to N3 medium (53) and addition of 10 μM of SB431542 (Tocris) and 5 μg/mL of dorsomorphin (Tocris) for 10 d. Then neural rosettes were transferred and maintained as neuroepithelial cultures on polyornithine/laminin-coated dishes in N3/FGF2 medium. Further differentiation into neuronal cultures was promoted in N3 medium by the removal of FGF2.
Western Blotting.
Cells were grown to subconfluency on 15-cm dishes, exposed to UV-C radiation when noted, harvested, and suspended in radioimmunoprecipitation assay (RIPA) buffer. After 15 min sonication, 30 μg of cell lysates was run on Invitrogen NuPAGE 4–12% Bis-Tris gel and were blotted onto PVDF membranes. Antibodies used were ATF3 (sc-188; Santa Cruz), β-actin (sc-1615; Santa Cruz), and α-tubulin (ab15246; Abcam).
Cellular Compartment Fractionation.
To determine the translocation of ATF3 to chromatin, cells were seeded in 15-cm dishes and grown to subconfluency before irradiation with 10-J/m2 UV-C. At various time points after radiation, cells were lysed on ice in lysis buffer (20 mM Hepes, 150 mM NaCl, 0.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 10% glycerol), and fractions were separated and visualized according to the procedures in ref. 34, using antibodies against ATF3 (sc-188; Santa Cruz), GAPDH, and H4 [Institute of Genetics and Molecular and Cellular Biology (IGBMC) Antibody Facility, Strasbourg, France].
ChIP.
Cells were crosslinked with 1% (vol/vol) formaldehyde solution for 10 min at room temperature. Crosslinking was stopped by adding glycine to a final concentration of 125 mM. Samples were sonicated to generate DNA fragments smaller than 500 bp. For immunoprecipitations, 1 mg of chromatin extract was precleared for 2 h with 50 mL of a 50% slurry of protein A/G-Sepharose mix (50:50) before the addition of the indicated antibodies. Then 2 mg of each antibody was added to the reactions and was incubated over night at 4 °C in the presence of 50 mL of protein A/G beads. After serial washings, the immunocomplexes were eluted twice for 10 min at 65 °C, and crosslinking was reversed by adjusting to 200 mM NaCl and overnight incubation at 65 °C. Further proteinase K digestion was performed for 2 h at 42 °C. DNA was purified using Qiagen columns (Qiagen QIAquick PCR Purification Kit). Immunoprecipitated DNA was quantified by real-time quantitative PCR (Qiagen QuantiTect SYBR Green PCR Kit). Antibodies used for ChIP assay were ATF3 (sc-188; Santa Cruz), Pol II (sc-9001; Santa Cruz), H3K4me2 (31209; Cell Signaling), CSB, H4Ac, and TFIIB (IGBMC Antibody Facility). Primer sequences are available upon request.
RNAi.
A pool of four RNA oligonucleotides (Dharmacon) forming a 19-base duplex core, specifically designed to target ATF3 mRNA (siATF3) was transfected in CS1AN cells at a concentration of 50 nM. A pool of RNA oligonucleotides without any target mRNA (siCtrl) was used as control. RNA transfection was performed using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions.
Determination of Global RNA Synthesis.
Cells in log phase were grown in the presence of 14C-thymidine (0.02 mCi/mL) for 2 d to label the DNA uniformly. The irradiated cells (10 J/m2 UV-C) and cells treated with α-amanitin for 1 h (when noted) were pulse-labeled with 5 μCi/mL of 3H-Uridine for 30 min at different time points. The cells were collected and washed once with ice-cold PBS and were lysed in buffer containing 0.5% SDS and 100 μg/mL Proteinase K for 2h at 37 °C. After trichloroacetic acid [TCA, 10% (vol/vol)] precipitation, the samples were spotted onto fiberglass discs (Whatman).Then the filters were washed sequentially in 5% TCA and 70% (vol/vol) ethanol/acetone, and their radioactivity was counted.
EMSA Assay.
For the gel-shift assay ATF3-GST and CSB-His were purified from bacteria or CF21 cells, respectively. The DNA fragment containing the ATF3-binding site, a part of CDK5RAP2 promoter, was amplified using the primers GCTATTTGGAAGTTGGTTTTCC and CATCTGACTGCAAGCTACTTCC for the wild-type ATF3-binding site and CCTAGAGGattCGTCACTACCACC and GGTGGTAGTGACG–aatCCTCTAGG for the mutated ATF3-binding site. Both wild-type and mutated CDK2RAP2 fragments were cloned into a pCG blunt cloning vector and were verified by sequencing. For the binding reaction, [32P]ATP-labeled wild-type or mutated fragments (100,000 cpm), purified ATF3-GST and CSB-His, and 2.5 μg of poly(deoxyinosinic-deoxycytidylic) acid [poly(dI-dC)] were added to a solution containing 20 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.2 mM EDTA, 1 mM MgCl2, 1 mM DTT, and 5% glycerol in a final assay volume of 25 μL, in the presence or absence of 4 mM ATP. The binding assay was carried out at room temperature for 30 min, and DNA–protein complexes were separated by gel electrophoresis as previously described (64).
Luciferase Reporter Assay.
Cells were double-transfected with Renilla control plasmid (Promega) and either pGL3 promoter vector (Promega) or pGL3 promoter vector containing the CRE/ATF repressor site using JetPei transfection reagent according to the manufacturer’s instructions. After 24 h cells were irradiated with 10 J/m2 UV-C, harvested at denoted time points, and screened for Renilla and luciferase activity using the Promega Dual-Luciferase Reporter Assay System. Values were calculated as the ratio of luciferase to Renilla (Luc/Ren), and unirradiated cells were set at 100%.
Affymetrix Microarray.
At each time point after treatment, total mRNA samples from two independent experiments were isolated and hybridized further to Affymetrix Human 1.0 ST Array chips according standard protocols. The quality of mRNA hybridization was very similar in each sample within replicates and time points, as measured via internal quality controls of the Affymetrix chips. We also measured the correlation of signal intensity between two replicas of each time point. All samples shared a high degree of similarity (P = 0.005%).
Extraction and Alignment of ATF3 ChIP-Seq Data.
The global ATF3 occupancy was extracted from ATF3 ChIP-Seq data available at the ENCODE database (http://genome.ucsc.edu/cgi-bin/hgFileUi?db=hg19&g=wgEncodeHaibTfbs) (24, 37). Specifically, the Bed files were extracted and proceed through GPAT software (http://bips.u-strasbg.fr/GPAT/) to retrieve genomic annotations of ATF3 peak positions through the promoter window. The extracted ATF3 peak data were aligned to the microarray dataset of the CS1AN+CSBwt and CS1AN UV-treated cells, using Galaxy software (https://main.g2.bx.psu.edu) (Fig. S1H and Table S2).
Supplementary Material
Acknowledgments
We thank R. Velez Cruz, A. Zadorin, and E. Compe for fruitful discussions; Charlotte Saint-André for helping with antibody production and protein expression; and the Institute of Genetics and Molecular and Cellular Biology Cell Culture Facility. This study was supported by grants from the European Research Council Advanced Scientists (to J.-M.E.), the Institut National du Cancer Grant INCA-2008-041 (to F.C.), and l’Association de la Recherche contre le Cancer (ARC) Grant 3153. U.K, was supported by a JME (Jean-Marc Egly) Advanced European Research Council grant and by an ARC fellowship for young scientists. M.-A.R. was supported by the French Ministère des Affaires Etrangères (Séjour Scientifique de Haut Niveau).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220071110/-/DCSupplemental.
References
- 1.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 2.Lainé J-P, Egly J-M. When transcription and repair meet: A complex system. Trends Genet. 2006;22(8):430–436. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LHF. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23(4):471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 4.Bregman DB, et al. UV-induced ubiquitination of RNA polymerase II: A novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA. 1996;93(21):11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citterio E, et al. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20(20):7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23(14):2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troelstra C, et al. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71(6):939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272(3):1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 9.Le May N, et al. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell. 2010;38(1):54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Proietti-De-Santis L, Drané P, Egly J-M. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25(9):1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Boom V, et al. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J Cell Biol. 2004;166(1):27–36. doi: 10.1083/jcb.200401056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradsher J, et al. CSB is a component of RNA pol I transcription. Mol Cell. 2002;10(4):819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 13.Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA. 1997;94(9):4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997;94(21):11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dianov GL, Houle JF, Iyer N, Bohr VA, Friedberg EC. Reduced RNA polymerase II transcription in extracts of cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 1997;25(18):3636–3642. doi: 10.1093/nar/25.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. 1997;17(12):6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nance MA, Berry SA. Cockayne syndrome: Review of 140 cases. Am J Med Genet. 1992;42(1):68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 18.Mallery DL, et al. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet. 1998;62(1):77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laugel V, et al. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31(2):113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 20.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: Activating transcription factor proteins and homeostasis. Gene. 2001;273(1):1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 21.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7(4-6):321–335. [PMC free article] [PubMed] [Google Scholar]
- 22.Gilchrist M, et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441(7090):173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka Y, et al. Systems analysis of ATF3 in stress response and cancer reveals opposing effects on pro-apoptotic genes in p53 pathway. PLoS ONE. 2011;6(10):e26848. doi: 10.1371/journal.pone.0026848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 25.Dürr H, Körner C, Müller M, Hickmann V, Hopfner K-P. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121(3):363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 26.Tuo J, et al. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J Biol Chem. 2001;276(49):45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 27.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall MC, Matson SW. Helicase motifs: The engine that powers DNA unwinding. Mol Microbiol. 1999;34(5):867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 29.Christiansen M, et al. Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res. 2003;31(3):963–973. doi: 10.1093/nar/gkg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt JF, et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297(5589):2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 31.Sharma V, et al. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA. 2003;100(5):2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horibata K, et al. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci USA. 2004;101(43):15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J Biol Chem. 2006;281(15):10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 34.Lake RJ, Geyko A, Hemashettar G, Zhao Y, Fan HY. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell. 2010;37(2):235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vélez-Cruz R, Zadorin AS, Coin F, Egly JM. Sirt1 suppresses RNA synthesis after UV irradiation in combined xeroderma pigmentosum group D/Cockayne syndrome (XP-D/CS) cells. Proc Natl Acad Sci USA. 2013;110(3):E212–E220. doi: 10.1073/pnas.1213076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfgang CD, Chen BP, Martindale JL, Holbrook NJ, Hai T. gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol Cell Biol. 1997;17(11):6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields S. Molecular biology. Site-seeing by sequencing. Science. 2007;316(5830):1441–1442. doi: 10.1126/science.1144479. [DOI] [PubMed] [Google Scholar]
- 38.Kang Y, Chen C-R, Massagué J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11(4):915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- 39.Kedinger C, Gniazdowski M, Mandel JL, Jr, Gissinger F, Chambon P. Alpha-amanitin: A specific inhibitor of one of two DNA-pendent RNA polymerase activities from calf thymus. Biochem Biophys Res Commun. 1970;38(1):165–171. doi: 10.1016/0006-291x(70)91099-5. [DOI] [PubMed] [Google Scholar]
- 40.Lindell TJ, Weinberg F, Morris PW, Roeder RG, Rutter WJ. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970;170(3956):447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- 41.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: Alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc Natl Acad Sci USA. 2002;99(3):1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen BP, Liang G, Whelan J, Hai T. ATF3 and ATF3 delta Zip. Transcriptional repression versus activation by alternatively spliced isoforms. J Biol Chem. 1994;269(22):15819–15826. [PubMed] [Google Scholar]
- 43.Lainé JP, Egly JM. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 2006;25(2):387–397. doi: 10.1038/sj.emboj.7600933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tornaletti S. Transcription arrest at DNA damage sites. Mutat Res. 2005;577(1-2):131–145. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 45.Turchi L, et al. Hif-2alpha mediates UV-induced apoptosis through a novel ATF3-dependent death pathway. Cell Death Differ. 2008;15(9):1472–1480. doi: 10.1038/cdd.2008.74. [DOI] [PubMed] [Google Scholar]
- 46.Turchi L, et al. ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ. 2009;16(5):728–737. doi: 10.1038/cdd.2009.2. [DOI] [PubMed] [Google Scholar]
- 47.Yang G, Zhang G, Pittelkow MR, Ramoni M, Tsao H. Expression profiling of UVB response in melanocytes identifies a set of p53-target genes. J Invest Dermatol. 2006;126(11):2490–2506. doi: 10.1038/sj.jid.5700470. [DOI] [PubMed] [Google Scholar]
- 48.Chiganças V, Lima-Bessa KM, Stary A, Menck CF, Sarasin A. Defective transcription/repair factor IIH recruitment to specific UV lesions in trichothiodystrophy syndrome. Cancer Res. 2008;68(15):6074–6083. doi: 10.1158/0008-5472.CAN-07-6695. [DOI] [PubMed] [Google Scholar]
- 49.Mathonnet G, et al. UV wavelength-dependent regulation of transcription-coupled nucleotide excision repair in p53-deficient human cells. Proc Natl Acad Sci USA. 2003;100(12):7219–7224. doi: 10.1073/pnas.1232161100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Hippel PH. Biochemistry. Completing the view of transcriptional regulation. Science. 2004;305(5682):350–352. doi: 10.1126/science.1101270. [DOI] [PubMed] [Google Scholar]
- 51.Hunt D, Raivich G, Anderson PN. Activating transcription factor 3 and the nervous system. Front Mol Neurosci. 2012;5:7. doi: 10.3389/fnmol.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee G, Chambers SM, Tomishima MJ, Studer L. Derivation of neural crest cells from human pluripotent stem cells. Nat Protoc. 2010;5(4):688–701. doi: 10.1038/nprot.2010.35. [DOI] [PubMed] [Google Scholar]
- 53. Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ (2012) Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci 15(3):477–486. [DOI] [PMC free article] [PubMed]
- 54.Krantz ID, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36(6):631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36(6):636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 56.Kawauchi S, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5(9):e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37(4):353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 58.Megraw TL, Sharkey JT, Nowakowski RS. Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol. 2011;21(8):470–480. doi: 10.1016/j.tcb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fong KW, Choi YK, Rattner JB, Qi RZ. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell. 2008;19(1):115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernandez-Gonzalez A, et al. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 2002;295(5561):1904–1906. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- 61.Baltanás FC, et al. Purkinje cell degeneration in pcd mice reveals large scale chromatin reorganization and gene silencing linked to defective DNA repair. J Biol Chem. 2011;286(32):28287–28302. doi: 10.1074/jbc.M111.246041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125(4):801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 64.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2(8):1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.