Abstract
ERK controls gene expression in development, but mechanisms that link ERK activation to changes in transcription are not well understood. We used high-resolution analysis of signaling dynamics to study transcriptional interpretation of ERK signaling during Drosophila embryogenesis, at a stage when ERK induces transcription of intermediate neuroblasts defective (ind), a gene essential for patterning of the nerve cord. ERK induces ind by antagonizing its repression by Capicua (Cic), a transcription factor that acts as a sensor of receptor tyrosine kinases in animal development and human diseases. A recent study established that active ERK reduces the nuclear levels of Cic, but it remained unclear whether this is required for the induction of Cic target genes. We provide evidence that Cic binding sites within the regulatory DNA of ind control the spatial extent and the timing of ind expression. At the same time, we demonstrate that ERK induces ind before Cic levels in the nucleus are reduced. Based on this, we propose that ERK-dependent relief of gene repression by Cic is a two-step process, in which fast reduction of repressor activity is followed by slower changes in nuclear localization and overall protein levels. This may be a common feature of systems in which ERK induces genes by relief of transcriptional repression.
Keywords: pattern formation, signal transduction, systems biology
ERK controls gene expression in countless developmental contexts (1–3). Most of what we know about transcriptional interpretation of ERK signaling in development is related to the E-twenty six family transcription factors (4–9). During the past decade, the high mobility group box transcription repressor Capicua (Cic) has been identified as a conserved sensor of ERK activation (10). ERK activation relieves gene repression by Cic, but the underlying mechanisms are not fully understood. During the terminal patterning of the fruit fly embryo, ERK-dependent relief of gene repression by Cic is preceded by strong reduction of Cic levels in the nucleus. Here, we show that ERK can relieve gene repression by Cic before reducing its nuclear localization. Based on this, we propose a two-tiered model in which fast antagonism of transcriptional repression is followed by slower reduction of nuclear levels.
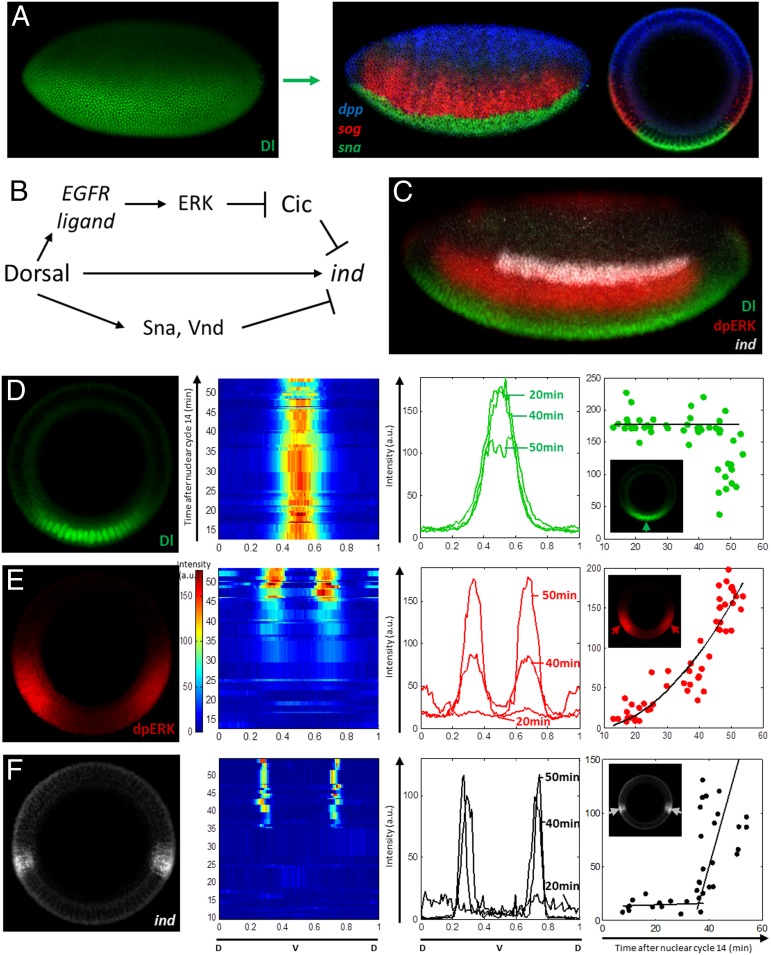
During the third hour of development, the Drosophila embryo is subdivided into three adjacent domains that give rise to the muscle, nerve, and skin cell types. One of these domains, the neural ectoderm, is further partitioned into three regions that map to the three columns of the future nerve cord (11). This patterning event depends on a Rel-family transcription factor Dorsal (Dl), which is distributed in a ventral-to-dorsal gradient of nuclear localization and controls gene expression along the dorsoventral (DV) axis of the embryo (Fig. 1A) (12–14). Dl patterns the DV axis directly by binding to enhancers of its target genes and indirectly through signaling and transcriptional cascades (15, 16). High levels of nuclear Dl directly activate Snail (Sna), a transcription factor, which represses neural cell fates in the ventral region of the embryo (Fig. 1 A and B) (17). At the same time, low levels are sufficient to directly repress decapentaplegic (Dpp), a bone morphogenetic protein ligand, which generates a signaling gradient that represses neural cell types in the dorsal region (18, 19). In addition to Dl and Dpp, the neural ectoderm is patterned by the epidermal growth factor receptor (EGFR), which establishes a gradient of ERK activation (20, 21). This gradient is also triggered by Dl through the localized production of EGFR ligands (Fig. 1 B and C) (22–24).
Fig. 1.
Nuclear Dl, germ layers, and ERK signaling dynamics along the DV axis. (A) The nuclear Dl gradient controls the DV patterning of the embryo by activating or repressing transcription of its target genes in a concentration-dependent manner. The expression domains of three genes, sna (green), short gastrulation (sog) (red), and dpp (blue), correspond to different levels of nuclear Dl and mark the territories that give rise to the future muscle, nerve, and skin tissues. (B) Schematic representation of ind regulation by nuclear Dl. (C) Patterns of nuclear Dl (green), dpERK (red), and ind (white) visualized in the same embryo. (D–F) Dynamics of nuclear Dl, dpERK, and ind. Each embryo was assigned to a time within nuclear cycle 14, based on the extent of cellularization (as detailed in the text). Panels from left to right show (i) a representative DV pattern; (ii) pseudocolor plot of the spatiotemporal pattern along the DV axis; (iii) spatial patterns at 20, 40, and 50 min after cycle 14; and (iv) time course of expression level at a location corresponding to the maximum of the pattern. (D and E) A total of 51 embryos were stained for Dl, dpERK, and myosin. The pattern of nuclear Dl remains stable until the onset of gastrulation (∼50 min after cycle 14). dpERK pattern was dynamic throughout the third hour of development, and monotonic increase in amplitude was observed. (F) A total of 37 embryos were stained for ind mRNA by using FISH, and phase-contrast images were taken to visualize membrane. ind transcripts were observed after the activation of ERK pathway.
EGFR signaling in the neural ectoderm induces intermediate neuroblasts defective (ind), a gene that encodes a homeobox transcription factor required for patterning of the nerve cord (25–27). Expression of ind requires relief of its repression by Cic (28). Together with direct activation by Dl and Dl-dependent repression in the ventral side of the embryo (by Sna and ventral nervous system defective), EGFR establishes a thin lateral stripe of ind expression (Fig. 1C) (26, 27, 29, 30). Quantitative measurements of Cic dynamics in the early Drosophila embryo established that ERK activation reduces the nuclear residence time of Cic and, in this way, increases the rate of its degradation (31). Whether this effect is essential for induction of genes repressed by Cic was unclear. Here, we show that ind, a direct target of Cic, is induced before significant reduction of Cic levels in the nucleus. Thus, ERK relieves transcriptional repression by Cic via multiple mechanisms that operate on different time scales.
Results
ind Is Induced by Steady Dl and Dynamic Dual Phosphorylated ERK Signals.
We used a microfluidics approach for high-throughput orientation of embryos to study the patterns of nuclear Dl, dual phosphorylated ERK (dpERK), Cic, and transcriptional activity of the ind locus during the third hour of development. In these experiments, fixed and stained embryos are introduced into a microfluidics chip in which a fluid flow orients them vertically, enabling imaging of signaling and gene expression along the entire DV axis. Although each fixed embryo is arrested at one point of development, these points are different for different embryos. Hence, analysis of a large number of embryos provides an insight into developmental dynamics.
The early embryo is a syncytium, a system in which nuclear divisions proceed without cytokinesis (32). Membranes enclosing nuclei into cells appear during the third hour of development, exactly the time window examined in this study. Membrane growth during cellularization follows highly reproducible kinetics, which can be used to assign a fixed embryo to a time window of ∼1 min (33, 34). By quantifying the length of the cellularization front in embryos stained for a specific component, such as dpERK, we can obtain a dynamic view of this component during the third hour of development. Thus, an ensemble of fixed embryos can be ordered in time to provide information about dynamics of dpERK activation and its transcriptional effects.
Fig. 1 D and E shows data from a representative experiment in which pattern formation was characterized in a group of 50 embryos stained with antibodies recognizing Dl, dpERK, and myosin regulatory light chain, which localizes to the cellularization front. In agreement with data from live imaging studies, we find that the levels of nuclear Dl within the ind expression domain remain essentially static during the third hour of development (Fig. 1D) (30). In contrast, the pattern of dpERK is dynamic and displays monotonic increase in amplitude (Fig. 1E). Fig. 1F shows data from a similar experiment, in which FISH was used to characterize the induction of ind. Consistent with the model in which ERK induces ind, we find that ind transcripts appear minutes after the ERK pathway is activated. These observations demonstrate that ind is induced by constant levels of nuclear Dl and increasing levels of dpERK.
Note that the positive input of Dl on ind is direct, through the Dl binding sites in the ind enhancer, and indirect, based on relief of ind repression by Cic. The indirect part of the mechanism relies on a signaling relay, in which Dl first induces the localized expression of EGFR ligand and ligand processor, which in turn stimulates ERK signaling that will relieve Cic repression of ind. This study focuses on the mechanisms underlying this relief of transcriptional repression.
Cic Binding Sites Control Spatial Extent and Timing of ind Expression.
The expression of ind is lost in embryos lacking the EGFR ligands (21, 27). To investigate the EGFR-dependent control of ind, we used a 1.4-kb transcriptional reporter, which drives neural ectoderm expression in a pattern that is very close to that of the ind gene and displays the same sensitivity to genetic perturbations (30). Ectopic expression of this reporter outside the neural ectoderm is not relevant for our analysis. For the rest of our presentation, we focus on expression only within the neural ectoderm.
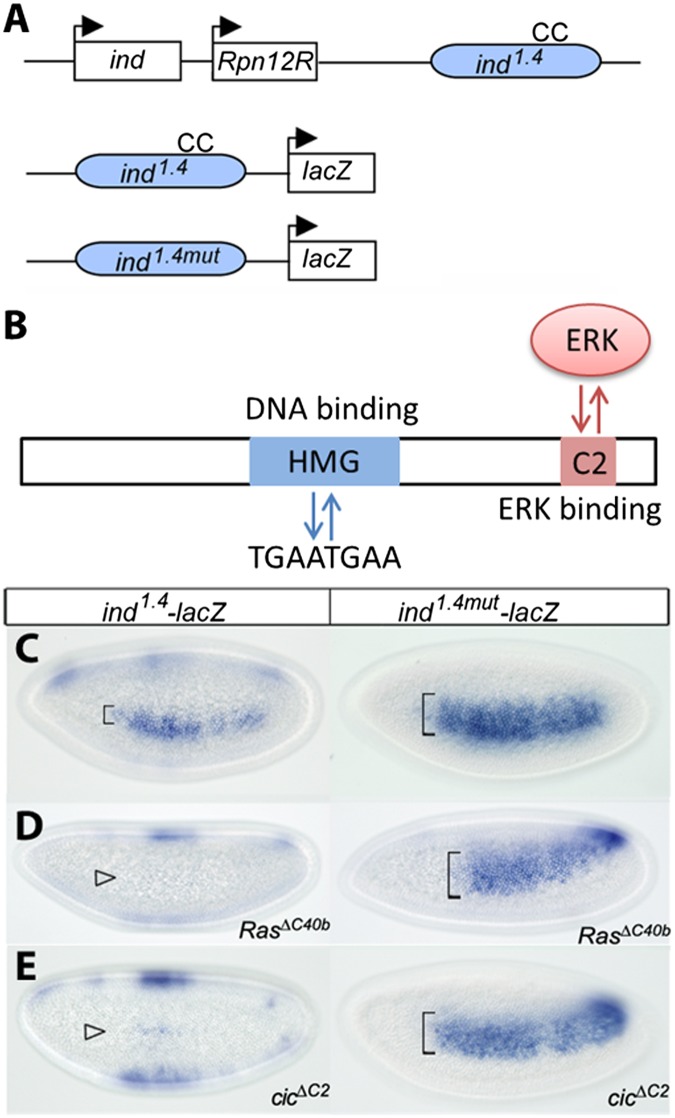
Studies with a shorter version of this reporter established that its sensitivity to EGFR depends on the DNA binding sites for Cic (Fig. 2 A and B) (28). We confirmed this with the ind1.4-lacZ construct, which serves as a more accurate reporter. Removal of Cic binding sites results in a clear expansion of the lacZ expression pattern (Fig. 2C). Importantly, removal of Cic binding sites overrules the need for receptor tyrosine kinase (RTK) signaling. Indeed, the activity of the WT reporter is dramatically reduced in embryos derived from mothers lacking Ras, a critical component of the EGFR signaling pathway. In contrast, the expression of ind1.4mut-lacZ lacking the Cic binding sites is essentially identical to that observed in embryos with unperturbed EGFR signaling (Fig. 2D).
Fig. 2.
Regulation of ind expression by Cic. (A) Diagram of the ind locus. The coding region and its 1.4-kb enhancer (in blue) are separated by the unrelated regulatory particle non-ATPase 12-related (Rpn12R) gene. “CC” indicates a pair of conserved Cic DNA binding sites (TGAATGAA). Reporter genes ind1.4-lacZ and ind1.4mut-lacZ containing WT and mutated CC sites are represented. (B) Schematic representation of the Cic protein highlighting its functional domains. The high mobility group box region binds to the octameric TGAATGAA sites in target gene enhancers, resulting in their transcriptional repression. The C2 motif functions as a docking site for ERK and mediates down-regulation of Cic in response to RTK-dependent ERK activation. Expression of ind-lacZ reporters. In situ hybridization of ind1.4-lacZ and ind1.4mut-lacZ in WT (C), RasΔC40b (D), and cicΔC2 (E) embryos. All images correspond to lateral surface views taken at mid- to late nuclear cycle 14. (C) In the WT background, ind1.4mut-lacZ expression is expanded compared with ind1.4-lacZ expression. (D and E) ind1.4-lacZ appears strongly repressed in RasΔC40b and cicΔC2 embryos (open arrowheads), whereas ind1.4mut-lacZ is insensitive to Cic and EGFR signaling.
A recent proteomics study established that Cic binding sites in the ind1.4 sequence interact with other transcription factors that may provide additional links between EGFR activation and ind expression (35). To test this idea, we assayed the activity of ind1.4-lacZ in embryos expressing a variant of Cic protein that lacks a motif necessary for interaction with ERK and is thus largely insensitive to the EGFR-dependent down-regulation of gene repression by Cic (CicΔC2) (36). If EGFR induces ind mainly by relieving its repression by Cic, expression of CicΔC2 should prevent this control and lead to dominant silencing of the reporter. Consistent with this scenario, the CicΔC2-expressing embryos show ind1.4-lacZ activity in only a few cells. In contrast, CicΔC2 does not repress the ind1.4mut enhancer that lacks the Cic binding sites (Fig. 2E). Based on this, we conclude that EGFR induces ind mainly through Cic.
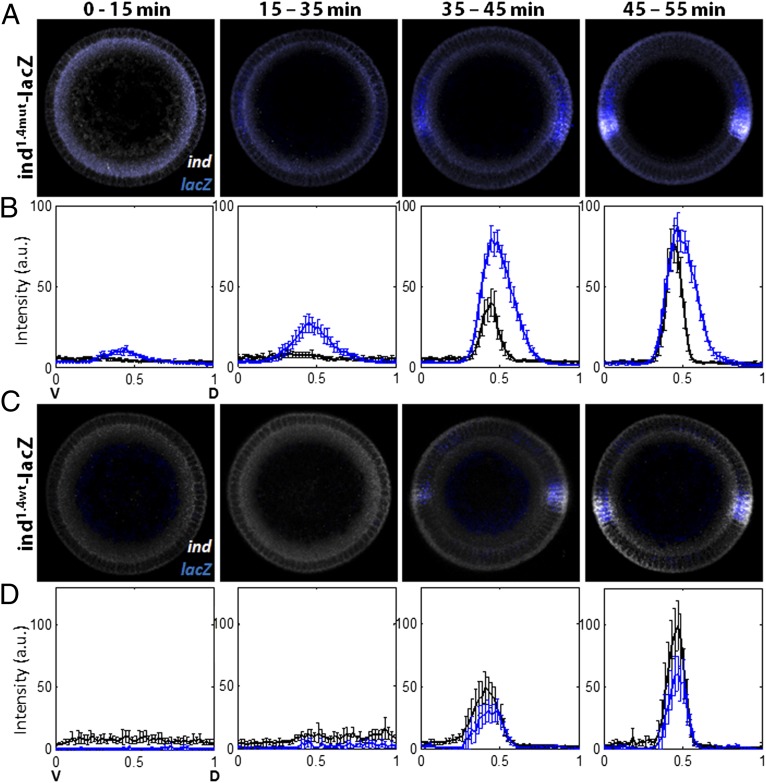
We found that Cic binding sites affect the timing of ind expression. This conclusion is based on the results of experiments in which we used multiplex FISH and temporal staging of embryos to compare the dynamics of the ind and lacZ transcripts. As shown in Fig. 3 A and B, transcriptional activity of the ind1.4mut-lacZ reporter is detected ∼20 min earlier than the ind transcripts.
Fig. 3.
Cic binding sites control the spatial pattern and timing of ind expression. Duplex FISH images show expression patterns of the transcripts of ind and lacZ, driven by the mutant (A and B) or WT ind (C and D) enhancers. ind1.4mut-lacZ (A and B) and ind1.4WT-lacZ (C and D) embryos were costained for ind (white) and lacZ (blue), and average signals were quantified for four temporal classes. Phase-contrast images were used to quantify membrane ingression length, which corresponds to a time point in cycle 14. Expression patterns are plotted from ventralmost (x = 0) to dorsalmost (x = 1) point. (A and B) Expression of ind1.4mut-lacZ is dorsally expanded relative to the ind pattern. Note the premature activation of ind1.4mut-lacZ expression during early/mid-cycle 14 (∼20 min after cycle 14, and before ERK pathway activation, based on Fig. 1). Totals of 19, 10, 13, and 13 embryos were examined for each class, respectively, and error bars correspond to SEM. (C and D) Expression of ind1.4WT-lacZ and that of endogenous ind pattern matches in its spatial and temporal pattern. In both cases, the expression is first observed ∼40 min after start of cycle 14, which is after the activation of ERK pathway. The numbers of embryos used for each class are 7, 5, 15, and 8, respectively. Error bars correspond to SEM.
In theory, this result could reflect a greater sensitivity in detecting lacZ vs. ind transcripts. However, two lines of evidence argue against this possibility. First, both transcripts display similar signal intensities at late cellularization stages (45–55 min after initiation of nuclear cycle 14; Fig. 3 A and B), implying a different time course for reaching these signals. Second, the WT ind1.4-lacZ reporter did not show premature activation compared with endogenous ind (Fig. 3 C and D).
Thus, the Cic binding sites control the spatial domain and the timing of ind expression: in the absence of these sites, lacZ is expressed prematurely and in a wider domain. The observed expansion of the lacZ domain (Fig. 3 A and B) is caused by a shift in the dorsal border of expression, which reflects the spatial range of the Dl morphogen gradient (37). In contrast, the ventral border of the lacZ pattern remains unaffected (Fig. 3B), reflecting regulation by transcriptional repressors that are activated by higher levels of nuclear Dl (29).
ind Is Induced Without Significant Reduction of Cic Levels in the Nucleus.
ERK signaling in the presumptive neural ectoderm is activated shortly after the initial phase of ERK signaling at the anterior and posterior poles of the embryo. This earlier phase of ERK signaling depends on Torso, another RTK, which is activated by ligand produced at the poles. ERK activation at the poles induces the expression of several genes with Cic binding sites in their regulatory DNA (38).
A recent live imaging study established that genes regulated by Torso are induced after a significant reduction of Cic levels in the nucleus, suggesting that this down-regulation may be required for RTK-dependent antagonism of Cic repression. On the contrary, studies of EGFR signaling in cultured human cells led to a model that does not rely on Cic degradation. Instead, RTK-dependent phosphorylation of Cic was proposed to interfere with Cic binding to DNA (39).
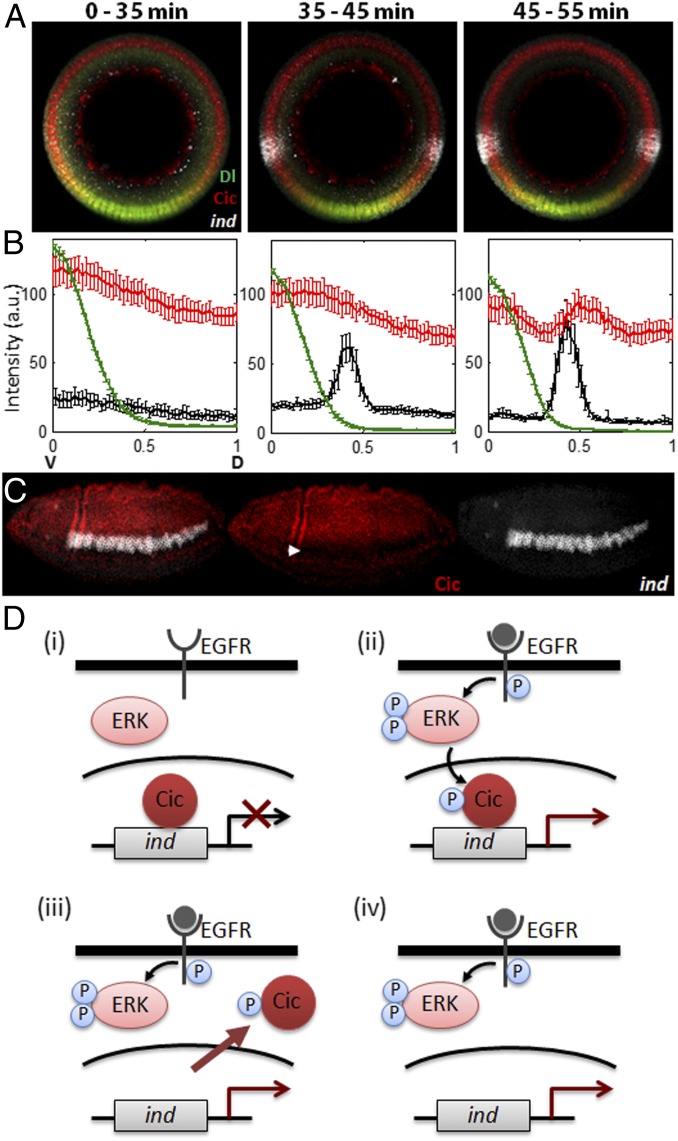
We reasoned that ERK signaling in the neural ectoderm can be used to investigate temporal control of Cic by RTKs. The half-life of Cic protein in the early embryo is approximately 15 min (31), which is comparable to the duration of the first phase of EGFR signaling that triggers ind transcription (Fig. 1 E and F). Based on this, we predicted that the EGFR-dependent derepression of ind does not require down-regulation of Cic levels. To test this, we stained embryos for ind mRNA, dpERK, and Cic protein. We found that the onset of ind expression is first detected during midcycle 14 and is closely associated with the onset of EGFR activation, visualized by the pattern of dpERK (Fig. 1E). On the contrary, nuclear levels of Cic within the ind expression domain appear uniform when ind is activated (Fig. 4 A and B).
Fig. 4.
Induction of ind precedes down-regulation of Cic nuclear levels. (A and B) WT embryos were costained for Dl (green), Cic (red), and ind mRNA (white), and average signals (y axis) were quantified along the DV axis (x-axis) for three distinct temporal classes. Phase-contrast images were taken to measure membrane ingression length, and each embryo was given a time point based on its length. Zero corresponds to the ventralmost point of the embryo. Before ind derepression, Cic nuclear levels are uniform (Left). Cic nuclear levels appear uniform at the stage when ind is derepressed (Center). Cic level begins to decrease only around the time of gastrulation (Right). The numbers of embryos in each temporal class are 14, 15, and 12, respectively, and error bars correspond to SEM. (C) Expression of Cic (red) and ind (white) in a gastrulating embryo (∼3 h 10 min after egg laying). Note the significant decrease of Cic nuclear levels in dpERK and ind expressing domain (solid arrowhead). (D) Schematic representation of the two-tiered model of RTK-dependent antagonism of gene repression by Cic. EGFR-mediated activation of ERK phosphorylates Cic (i), which immediately relieves its repression on ind (ii). Phosphorylated Cic is then exported to cytoplasm (iii) and subject to degradation (iv).
Thus, the EGFR-dependent relief of Cic repression does not require Cic down-regulation. As proposed by studies in human cells, RTKs can first counteract Cic-dependent repression at the level of DNA binding (39). This mechanism does not depend on changes in protein levels and can be effective minutes after pathway activation. At later times, repression may be further attenuated by changes in Cic subcellular localization and stability. Indeed, Cic protein levels within the ind-expression domain are significantly reduced during gastrulation (Fig. 4C). We propose that RTKs control Cic through a mechanism that combines fast relief of transcriptional repressor function and slower reduction of nuclear localization and protein levels (Fig. 4D).
Discussion
CNS development in flies (1, 5, 40), vulval patterning in nematodes (2, 41), and DV patterning in amphibians (3) are just a few examples of systems in which ERK-dependent control of gene expression is important for normal development. Understanding how ERK activation is converted to changes in gene expression requires analysis of ERK signal transduction as a function of space, time, and cellular context.
Spatial control of ERK activation and its effects on target genes can be studied in vivo, although at low temporal resolution. At this point, most of our knowledge about temporal aspects of ERK-dependent gene expression comes from experiments with cultured cells (42–49). Our work establishes the neural ectoderm in the early Drosophila embryo as a powerful experimental system in which ERK-dependent transcription can be studied with spatial resolution of a single cell and temporal resolution of minutes.
We used the developed data collection and analysis tools to examine how ERK activation relieves gene repression by Cic, a transcriptional repressor with important roles in animal development and human diseases (38, 50, 51). Our analyses of signaling dynamics establish that ind is activated by a time-dependent ERK signal and that the spatial domain and kinetics of ind expression are controlled by specific Cic binding sites in the regulatory DNA of ind.
Induction of ind depends on ERK and is detected before any significant decrease in the nuclear levels of Cic. In contrast, during terminal patterning of the Drosophila embryo, which also depends on ERK, the induction of Cic-target genes is preceded by a clear decrease in Cic levels (31). Based on previous studies with receptor tail swaps, we propose that these differences do not reflect differences in the RTKs used by the terminal and DV systems (52). Instead, a time delay between ERK activation and gene transcription in the terminal system may result from the lack of transcriptional activators of Cic target genes at the embryonic termini when ERK signaling is initiated. Some of these genes are positively regulated by the nuclear Dl gradient and Zelda, a uniformly expressed activator of the zygotic genome (53). Nuclear levels for both of these signals increase during the time preceding the expression of the terminal genes (54). The same activators play a role in the induction of ind, but because they are already present when ERK is activated in the presumptive neural ectoderm, there is no delay between ERK activation and ind induction. Cic degradation is observed on longer time scales, when additional layers of control become important for robust response. This may be a common feature of systems in which ERK controls its target genes by relief of transcriptional repression (45, 55).
What is the molecular basis for fast ERK-dependent relief of transcriptional repression by Cic? One possibility is that phosphorylated Cic dissociates from DNA. This mechanism was put forward on the basis of three results (39). First, human Cic is phosphorylated on sites close to the DNA binding domain. Second, phosphorylation of Cic promotes binding to 14-3-3 proteins, which function as key regulators of phosphoproteins. Third, 14-3-3 proteins interfere with binding of phosphorylated Cic to DNA in vitro. Analysis of this potential mechanism in flies will require (i) determination of the ERK-dependent phosphorylation sites in Drosophila Cic, (ii) testing of the role of 14-3-3 proteins in regulating Cic embryonic functions, and (iii) measurement/quantification of the time scales on which Cic DNA binding could be affected in vivo.
Alternatively, or in addition to the DNA-binding mechanism, phosphorylation of Cic could disrupt binding of Cic to a cofactor required for repressor activity. Addressing this hypothesis will involve the characterization of Cic interactome and its sensitivity to ERK activation. Given the conserved association between ERK signaling and Cic-target gene derepression, the early Drosophila embryo offers an ideal system for elucidating the mechanisms of Cic regulation in vivo. The experimental approach developed in this study paves the way for testing these mechanisms at high temporal, spatial, and molecular resolution.
Materials and Methods
Fly Stocks and Transgenes.
Oregon-R (OreR), spaghetti squash (Sqh)-green fluorescent protein (GFP), cicΔC2, RasΔC40b, ind1.4-lacZ, ind1.4mut-lacZ stocks were used in this study (28, 36, 53). ind1.4 enhancer sequences were obtained by PCR using primers ind1 (5 AAT GAA TTC AAA CGT TTT GTT ATA ATC 3) and ind3 (5 TAT AGA TCT GGG CCT TCG GTC CGA AAA TG 3). The ind1.4mut enhancer has mutated Cic binding sites [CACACGCA (underlining indicates the nucleotides being mutated in the ind1.4mut construct)] generated by recombinant PCR. WT and mutant ind1.4-lacZ reporters were assembled in pCaSpeR-hs43-lacZ and transformed into flies by standard methods. Sqh-GFP flies were used to visualize the membrane invagination.
Staining Procedures.
Antibody staining and FISH were performed as described before (56). For FISH, embryos were hybridized overnight at 60 °C with digoxigenin and biotin-labeled antisense probes. Labeled embryos were visualized by using standard immunofluorescence techniques. Monoclonal rabbit anti-dpERK (1:100; Cell Signaling), rabbit anti-Cic (1:2,000; gift from Celeste Berg, University of Washington, Seattle, WA), mouse anti-dorsal (1:100; DSHB), sheep anti-digoxigenin (1:125; Roche), and mouse anti-biotin (1:125; Jackson Immunoresearch) were used as primary antibodies. DAPI (1:10,000; Vector Laboratories) was used to stain nuclei. Alexa Fluor conjugates (1:500; Invitrogen) were used as secondary antibodies.
Imaging.
A Nikon A1 confocal microscope was used for imaging. For lateral imaging of embryos, a Nikon 20× Plan-Apo objective was used. Images were collected from the focal plane in the midsagittal cross section of embryos. For upright imaging, a Nikon 60× Plan-Apo oil objective was used, and images were collected at the focal plane ∼90 μm from the anterior or posterior pole of an embryo. End-on imaging was performed by using the microfluidics device described previously (57). Spatial patterns of Dl, dpERK, and Cic proteins, and of ind and lacZ mRNAs, were automatically extracted from raw confocal images as described elsewhere (58). To quantify nuclear Dl and Cic signal, we used DAPI staining as nuclear masks, which provided nuclei positions to be quantified. For end-on imaging, the maximum of the quantified nuclear Dl gradient was used to find the ventralmost point of the embryo (59).
Temporal Staging of Embryos.
The progression of membrane invagination and nuclear elongation during cellularization was used to assign time point to each embryo in third hour of development. During the first 30 min of nuclear cycle 14 (2 h 10 min to 2 h 40 min after fertilization), membranes that will divide future cells begin to form. During the remaining 15 min before gastrulation, the length of internuclear membranes reaches approximately 30 μm (33). We measured the membrane length by costaining embryos with a membrane marker (myosin-GFP) or by taking phase-contrast images. We used a correlation curve between the membrane ingression length and the development time to assign a fixed embryo to a time window of approximately 1 min.
Acknowledgments
The authors thank Celeste Berg, Eric Wieschaus, and Anna Sokac for fly stocks, reagents, and data; B.L. thanks Yoosik Kim, Lily Cheung, and Jitendra Kanodia for generous help during the early stages of this project; B.L. and S.Y.S. thank Victoria Sanchez-Zini, Oliver Grimm, Eric Wieschaus, Trudi Schupbach, Mike Levine, Ze’ev Paroush, Alexey Veraksa, Amanda Simcox, James Skeath, Ilaria Rebay, and Benny Shilo for helpful discussions; and N.S. and G.J. thank Ainhoa Olza for assistance with Drosophila injections. G.J. was supported by Institució Catalana de Recerca i Estudis Avançats (ICREA), Ministerio de Ciencia e Innovación Grants BFU2008-01875 and BFU2011-23611 and Generalitat de Catalunya Grant 2009SGR-1075; B.L. and S.Y.S. were partially supported by the National Institutes of Health Grant R01GM086537 and National Science Foundation (NSF) Grant Emerging Frontiers in Research and Innovation (EFRI) 1136913; and H.L. and C.R. were partially supported by NSF Grant EFRI 1136913.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122(1):223–230. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- 2.Yoo AS, Bais C, Greenwald I. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science. 2004;303(5658):663–666. doi: 10.1126/science.1091639. [DOI] [PubMed] [Google Scholar]
- 3.Hanafusa H, Matsumoto K, Nishida E. Regulation of ERK activity duration by Sprouty contributes to dorsoventral patterning. Nat Cell Biol. 2009;11(1):106–109. doi: 10.1038/ncb1820. [DOI] [PubMed] [Google Scholar]
- 4.Raible F, Brand M. Tight transcriptional control of the ETS domain factors Erm and Pea3 by Fgf signaling during early zebrafish development. Mech Dev. 2001;107(1-2):105–117. doi: 10.1016/s0925-4773(01)00456-7. [DOI] [PubMed] [Google Scholar]
- 5.Gabay L, et al. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development. 1996;122(11):3355–3362. doi: 10.1242/dev.122.11.3355. [DOI] [PubMed] [Google Scholar]
- 6.Qiao F, et al. Derepression by depolymerization; structural insights into the regulation of Yan by Mae. Cell. 2004;118(2):163–173. doi: 10.1016/j.cell.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollenhorst PC. RAS/ERK pathway transcriptional regulation through ETS/AP-1 binding sites. Small GTPases. 2012;3(3):154–158. doi: 10.4161/sgtp.19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farooqui S, et al. Coordinated lumen contraction and expansion during vulval tube morphogenesis in Caenorhabditis elegans. Dev Cell. 2012;23(3):494–506. doi: 10.1016/j.devcel.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Jiménez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: Role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14(2):224–231. [PMC free article] [PubMed] [Google Scholar]
- 11.Cornell RA, Ohlen TV. Vnd/nkx, ind/gsh, and msh/msx: Conserved regulators of dorsoventral neural patterning? Curr Opin Neurobiol. 2000;10(1):63–71. doi: 10.1016/s0959-4388(99)00049-5. [DOI] [PubMed] [Google Scholar]
- 12.Steward R. Relocalization of the dorsal protein from the cytoplasm to the nucleus correlates with its function. Cell. 1989;59(6):1179–1188. doi: 10.1016/0092-8674(89)90773-3. [DOI] [PubMed] [Google Scholar]
- 13.Roth S, Stein D, Nüsslein-Volhard C. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59(6):1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- 14.Rushlow CA, Han KY, Manley JL, Levine M. The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell. 1989;59(6):1165–1177. doi: 10.1016/0092-8674(89)90772-1. [DOI] [PubMed] [Google Scholar]
- 15.Hong JW, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci USA. 2008;105(51):20072–20076. doi: 10.1073/pnas.0806476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rushlow CA, Shvartsman SY. Temporal dynamics, spatial range, and transcriptional interpretation of the Dorsal morphogen gradient. Curr Opin Genet Dev. 2012;22(6):542–546. doi: 10.1016/j.gde.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992;6(8):1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- 18.Mizutani CM, Meyer N, Roelink H, Bier E. Threshold-dependent BMP-mediated repression: A model for a conserved mechanism that patterns the neuroectoderm. PLoS Biol. 2006;4(10):e313. doi: 10.1371/journal.pbio.0040313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson EL, Anderson KV. Decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell. 1992;71(3):451–461. doi: 10.1016/0092-8674(92)90514-d. [DOI] [PubMed] [Google Scholar]
- 20.Shilo BZ. Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp Cell Res. 2003;284(1):140–149. doi: 10.1016/s0014-4827(02)00094-0. [DOI] [PubMed] [Google Scholar]
- 21.Skeath JB. The Drosophila EGF receptor controls the formation and specification of neuroblasts along the dorsal-ventral axis of the Drosophila embryo. Development. 1998;125(17):3301–3312. doi: 10.1242/dev.125.17.3301. [DOI] [PubMed] [Google Scholar]
- 22.Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16(13):1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 23.Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6(9):1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 24.Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277(5329):1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 25.Weiss JB, et al. Dorsoventral patterning in the Drosophila central nervous system: The intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 1998;12(22):3591–3602. doi: 10.1101/gad.12.22.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Ohlen T, Doe CQ. Convergence of dorsal, dpp, and egfr signaling pathways subdivides the drosophila neuroectoderm into three dorsal-ventral columns. Dev Biol. 2000;224(2):362–372. doi: 10.1006/dbio.2000.9789. [DOI] [PubMed] [Google Scholar]
- 27.Yagi Y, Suzuki T, Hayashi S. Interaction between Drosophila EGF receptor and vnd determines three dorsoventral domains of the neuroectoderm. Development. 1998;125(18):3625–3633. doi: 10.1242/dev.125.18.3625. [DOI] [PubMed] [Google Scholar]
- 28.Ajuria L, et al. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development. 2011;138(5):915–924. doi: 10.1242/dev.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cowden J, Levine M. Ventral dominance governs sequential patterns of gene expression across the dorsal-ventral axis of the neuroectoderm in the Drosophila embryo. Dev Biol. 2003;262(2):335–349. doi: 10.1016/s0012-1606(03)00395-6. [DOI] [PubMed] [Google Scholar]
- 30.Stathopoulos A, Levine M. Localized repressors delineate the neurogenic ectoderm in the early Drosophila embryo. Dev Biol. 2005;280(2):482–493. doi: 10.1016/j.ydbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Grimm O, et al. Torso RTK controls Capicua degradation by changing its subcellular localization. Development. 2012;139(21):3962–3968. doi: 10.1242/dev.084327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- 33.Lecuit T, Samanta R, Wieschaus E. slam encodes a developmental regulator of polarized membrane growth during cleavage of the Drosophila embryo. Dev Cell. 2002;2(4):425–436. doi: 10.1016/s1534-5807(02)00141-7. [DOI] [PubMed] [Google Scholar]
- 34.Dubuis JO, Samanta R, Gregor T. Accurate measurements of dynamics and reproducibility in small genetic networks. Mol Syst Biol. 2013;9:639. doi: 10.1038/msb.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia M, Stathopoulos A. Lateral gene expression in Drosophila early embryos is supported by Grainyhead-mediated activation and tiers of dorsally-localized repression. PLoS ONE. 2011;6(12):e29172. doi: 10.1371/journal.pone.0029172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Astigarraga S, et al. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26(3):668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanodia JS, et al. A computational statistics approach for estimating the spatial range of morphogen gradients. Development. 2011;138(22):4867–4874. doi: 10.1242/dev.071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiménez G, Shvartsman SY, Paroush Z. The Capicua repressor—a general sensor of RTK signaling in development and disease. J Cell Sci. 2012;125(pt 6):1383–1391. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dissanayake K, et al. ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicúa. Biochem J. 2011;433(3):515–525. doi: 10.1042/BJ20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raz E, Shilo BZ. Establishment of ventral cell fates in the Drosophila embryonic ectoderm requires DER, the EGF receptor homolog. Genes Dev. 1993;7(10):1937–1948. doi: 10.1101/gad.7.10.1937. [DOI] [PubMed] [Google Scholar]
- 41.Sundaram MV. RTK/Ras/MAPK signaling. WormBook. 2006;Feb 11:1–19. doi: 10.1895/wormbook.1.80.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang SH, Sharrocks AD, Whitmarsh AJ. MAP kinase signalling cascades and transcriptional regulation. Gene. 2013;513(1):1–13. doi: 10.1016/j.gene.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto T, et al. Continuous ERK activation downregulates antiproliferative genes throughout G1 phase to allow cell-cycle progression. Curr Biol. 2006;16(12):1171–1182. doi: 10.1016/j.cub.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 44.Albeck JG, Mills GB, Brugge JS. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol Cell. 2013;49(2):249–261. doi: 10.1016/j.molcel.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarcic G, et al. EGR1 and the ERK-ERF axis drive mammary cell migration in response to EGF. FASEB J. 2012;26(4):1582–1592. doi: 10.1096/fj.11-194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitehurst A, Cobb MH, White MA. Stimulus-coupled spatial restriction of extracellular signal-regulated kinase 1/2 activity contributes to the specificity of signal-response pathways. Mol Cell Biol. 2004;24(23):10145–10150. doi: 10.1128/MCB.24.23.10145-10150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4(8):556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 48.Schilling M, et al. Theoretical and experimental analysis links isoform-specific ERK signalling to cell fate decisions. Mol Syst Biol. 2009;5:334. doi: 10.1038/msb.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Kriegsheim A, et al. Cell fate decisions are specified by the dynamic ERK interactome. Nat Cell Biol. 2009;11(12):1458–1464. doi: 10.1038/ncb1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bettegowda C, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fryer JD, et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334(6056):690–693. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghiglione C, Perrimon N, Perkins LA. Quantitative variations in the level of MAPK activity control patterning of the embryonic termini in Drosophila. Dev Biol. 1999;205(1):181–193. doi: 10.1006/dbio.1998.9102. [DOI] [PubMed] [Google Scholar]
- 53.Liang HL, et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456(7220):400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanodia JS, et al. Dynamics of the Dorsal morphogen gradient. Proc Natl Acad Sci USA. 2009;106(51):21707–21712. doi: 10.1073/pnas.0912395106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tootle TL, Lee PS, Rebay I. CRM1-mediated nuclear export and regulated activity of the Receptor Tyrosine Kinase antagonist YAN require specific interactions with MAE. Development. 2003;130(5):845–857. doi: 10.1242/dev.00312. [DOI] [PubMed] [Google Scholar]
- 56.Kim Y, et al. Gene regulation by MAPK substrate competition. Dev Cell. 2011;20(6):880–887. doi: 10.1016/j.devcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung K, et al. A microfluidic array for large-scale ordering and orientation of embryos. Nat Methods. 2011;8(2):171–176. doi: 10.1038/nmeth.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coppey M, Boettiger AN, Berezhkovskii AM, Shvartsman SY. Nuclear trapping shapes the terminal gradient in the Drosophila embryo. Curr Biol. 2008;18(12):915–919. doi: 10.1016/j.cub.2008.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanodia JS, et al. Pattern formation by graded and uniform signals in the early Drosophila embryo. Biophys J. 2012;102(3):427–433. doi: 10.1016/j.bpj.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]