Abstract
Background
Beyond estrogen receptor (ER), there are no validated predictors for tamoxifen (TAM) efficacy and toxicity. We utilized a genome-wide cell-based model to comprehensively evaluate genetic variants for their contribution to cellular sensitivity to TAM.
Design
Our discovery model incorporates multidimensional datasets, including genome-wide genotype, gene expression, and endoxifen-induced cellular growth inhibition in the International HapMap lymphoblastoid cell lines (LCLs). Genome-wide findings were further evaluated in NCI60 cancer cell lines. Gene knock-down experiments were performed in four breast cancer cell lines. Genetic variants identified in the cell-based model were examined in 245 Caucasian breast cancer patients who underwent TAM treatment.
Results
We identified seven novel single-nucleotide polymorphisms (SNPs) associated with endoxifen sensitivity through the expression of 10 genes using the genome-wide integrative analysis. All 10 genes identified in LCLs were associated with TAM sensitivity in NCI60 cancer cell lines, including USP7. USP7 knock-down resulted in increasing resistance to TAM in four breast cancer cell lines tested, which is consistent with the finding in LCLs and in the NCI60 cells. Furthermore, we identified SNPs that were associated with TAM-induced toxicities in breast cancer patients, after adjusting for other clinical factors.
Conclusion
Our work demonstrates the utility of a cell-based model in genome-wide identification of pharmacogenomic markers.
Keywords: gene expression, genome-wide association study, HapMap, SNP, tamoxifen
introduction
Tamoxifen (TAM) is one of the most commonly used agents in the treatment and prevention of breast cancer [1]. Despite the broad utility of TAM, it has been reported that up to 35% of patients who receive TAM treatment for breast cancer prevention do not respond to TAM therapy [2]. Moreover, TAM-induced side effects, such as hot flashes and osteoporosis, inflict millions of patients who take this medication.
Considerable effort has been invested in identifying markers that can be used to predict TAM response and toxicity. For example, a seminal paper, demonstrating the relationship between genetic variants in CYP2D6 and the formation of active TAM metabolites [3], suggested the possibility of a useful germline genetic predictor for TAM efficacy and toxicity. Yet, other studies failed to show these genotype–phenotype relationships, thus preventing any firm conclusions as to the clinical utility of this genotype–phenotype pair. Additional work has been done on several other candidate genes, e.g. SULT1A1, ABCC2, and CYP2C19, but none yield robust evidence of an association with TAM's therapeutic effects [4–6]. Thus, the presence of hormone receptors remains, to date, the only validated predictor for TAM treatment efficacy.
Since conflicting results have been obtained from studies focusing on one or several candidate genes, we hypothesized that additional gene/genetic variants may act together to affect TAM sensitivity. Therefore, we set out to examine the human genome, in an unbiased manner, to search for variants that contribute to TAM sensitivity. Utilizing International HapMap cell lines, for which whole-genome genetic (hapmap.ncbi.nlm.nih.gov) and gene expression data [7, 8] are readily available, our goal was to develop a cellular phenotyping assay to quantify TAM sensitivity and to employ a previously developed genome-wide integrative analysis [7] identifying genetic variants associated with TAM sensitivity through their relationship with genome-wide gene expression data. Furthermore, candidate single-nucleotide polymorphisms (SNPs) and genes identified through this approach were prioritized for functional and clinical validation.
patients and methods
cell lines
The 60 unrelated International HapMap CEU (Centre d'Etude du Polymorphisme Humain people from Utah, USA) lymphoblastoid cell lines (LCLs) were purchased from the Coriell Institute for Medical Research (Camden, NJ) and cultured based on Coriell's recommendation. Four breast cancer cell lines (MCF-7, BT-20, BT-549, and ZR-75-1) were purchased from ATCC (www.atcc.org) and maintained following ATCC protocols.
model evaluation—ER expression and function
Western blot was performed to examine the protein expression level for ESR1 in randomly selected LCLs. Real-time polymerase chain reaction was performed for two known estrogen receptor (ER)-regulated genes (IGFBP4 and NMA) [9] in these LCLs after 10−6 M estradiol treatment. Detailed methods can be found in supplementary Materials, available at Annals of Oncology online.
phenotyping of cellular sensitivity to endoxifen
A previously reported cell viability test using AlamarBlue reagent [10] was modified and applied to evaluate LCL sensitivity to 3, 5, 7, and 10 μM endoxifen treatment. The percentage of viable cells at each treatment concentration when compared with control was used as phenotypes in the downstream analysis. For details see supplementary Materials, available at Annals of Oncology online.
identification of genetic predictors using a genome-wide integrative model
A genome-wide approach that integrates genetic variants, mRNA expression, and cellular sensitivity to drug was developed previously [11]. This approach was applied to each of the four cellular sensitivity phenotypes identifying genetic predictors for endoxifen sensitivity.
evaluation of the candidate gene expression in NCI60 cell lines
The expression of 10 genes identified in LCLs and log10(TAM GI50) data from the NCI60 dataset were queried using CellMiner [12]. GI50 refers to the drug concentration required to inhibit cell growth by 50%. Linear regression analysis was performed between each of these gene expression phenotypes and log10(TAM GI50) for all 60 NCI60 samples as well as for a subset of five breast cancer cell lines.
functional evaluation of USP7 in breast cancer cell lines
siRNA knock-down of USP7 gene was performed in four breast cancer cell lines using DharmaFECT transfection kit (Thermo Scientific). Detailed methods can be found in supplementary Materials, available at Annals of Oncology online. Cell viability after knock-down was measured using CellTiter-Glo (Promega). Percentage of cell survival rates was calculated using raw luminescence values between TAM-treated cells and those of control wells.
clinical validation
Two hundred and forty-five Caucasian breast cancer patients who enrolled into a previously reported TAM trial [13] were genotyped for SNPs identified from our cell-based model. We examined the genetic association with three clinical TAM sensitivity phenotypes (average hot flash scores, hip bone loss, and lumbar bone loss) through a multivariate linear regression model, including other clinical factors (e.g. age, menopausal status, prior chemotherapy, and TAM and three metabolites). Details can be found in supplementary Materials, available at Annals of Oncology online.
results
LCL model evaluation
Given the known mechanism of action of TAM, we first set out to evaluate the expression and function of ER in the LCL model. In four randomly selected LCLs, we confirmed the expression of ER alpha protein in all cell lines under different cell culture conditions (supplementary Figure S1, available at Annals of Oncology online). Furthermore, we found significant up-regulation of IGFBP4 and down-regulation of NMA (both are known to be regulated by estrogen) in all four LCLs after 1 μM estradiol treatment (supplementary Figure S1, available at Annals of Oncology online). These results suggested that ER in LCLs is functional, and that LCLs can be used to evaluate TAM sensitivity.
cellular sensitivity to endoxifen
Since the LCL model system does not express high levels of CYP2D6, an enzyme critical to convert TAM to its more active metabolite, endoxifen, we chose to perform phenotyping assay using endoxifen. In addition, TAM/endoxifen competes with hormones or hormone mimicking compounds in the culture media. Therefore, we chose to use phenol red (an estrogen mimicking compound)-free RPMI culture media and 15% charcoal-stripped fetal bovine serum when conducting endoxifen phenotyping assay. When choosing drug concentrations for cellular experiments, a previous study showed that 10 μM of endoxifen induced strong growth inhibition in all breast cancer cell lines evaluated, while 100 μM endoxifen is lethal to all tested cell lines [14]. Therefore, we quantified cellular growth inhibition induced by endoxifen at four increasing concentrations (3, 5, 7, and 10 μM) in the HapMap CEU samples.
We observed a dose-dependent cellular growth inhibition with increasing concentrations of endoxifen treatment in LCLs (supplementary Figure S2, available at Annals of Oncology online). Inter-individual variation was also observed at all four treatment concentrations. The median percent viable cells when compared with no-drug control (with data range) for each endoxifen treatment group are: 86% (66%–103%) at 3 μM; 77% (51%–93%) at 5 μM; 70% (37%–99%) at 7 μM; and 53% (16%–79%) at 10 μM.
identification of genetic predictors using a genome-wide integrative model
As detailed in supplementary Materials, available at Annals of Oncology online, a genome-wide integrative approach was applied to identify the genetic predictors of endoxifen sensitivity. We identified 3527 SNPs associated with at least one endoxifen sensitivity phenotype in the CEU samples (P < 0.0001). Among them, 11 SNPs are associated with all four endoxifen phenotypes. Nine of these 11 are in an intergenic region of chromosome 15 (supplementary Table S1, available at Annals of Oncology online). Furthermore, 20 additional SNPs are associated with at least three phenotypes of endoxifen sensitivity (supplementary Table S1, available at Annals of Oncology online). The pairwise correlation between the phenotypes can be found in supplementary Table S2, available at Annals of Oncology online.
Subsequently, when evaluating the potential function of the 3527 SNPs, we found nine SNPs are associated with 16 gene expression traits (P < 3.8 × 10−6) in LCLs. Finally, we conducted linear regression between each of the 16 target genes and each of the 4 phenotypes of endoxifen sensitivity. Ten of the 16 genes were found to be correlated to at least one endoxifen sensitivity phenotype (P < 0.05), which consequently further narrowed down the SNP list from 9 to 7. The final findings are summarized in Table 1.
Table 1.
Summary of genome-wide integrative analysis of endoxifen sensitivity in 60 CEU LCLs
| SNP information |
SNP–phenotype association (P-value) |
SNP–target gene association (P-value) | Target gene–phenotype association (P-value) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Chromosome (position) | 3 μM | 5 μM | 7 μM | 10 μM | 3 μM | 5 μM | 7 μM | 10 μM | |
| rs17394957 | Chr. 1 (159 580 061) | 0.003 | 4 × 10−5 | 0.002 | 0.024 | GLTSCR2 (3 × 10−6) | 0.386 | 0.014 | 0.171 | 0.048 |
| rs13064915 | Chr. 3 (138 603 933) | 0.010 | 0.003 | 5 × 10−5 | 0.013 | ILVBL (1 × 10−6) | 0.055 | 0.003 | 0.004 | 0.007 |
| SNRPB (3 × 10−6) | 0.098 | 0.025 | 0.096 | 0.168 | ||||||
| rs478437 | Chr. 7 (144 424 767) | 0.016 | 0.007 | 0.040 | 6 × 10−5 | USP7 (9 × 10−7) | 0.014 | 0.013 | 0.027 | 0.007 |
| rs8068198 | Chr. 17 (68 597 219) | 0.007 | 1 × 10−5 | 0.001 | 0.0045 | SYT17 (3 × 10−6) | 0.226 | 0.026 | 0.104 | 0.048 |
| rs310786 | Chr. 12 (75 960 279) | 0.445 | 0.069 | 0.012 | 5 × 10−5 | TMPRSS3 (1 × 10−6) | 0.243 | 0.045 | 0.009 | 0.006 |
| rs1534882 | Chr. 22 (35 659 491) | 0.093 | 0.003 | 1 × 10−5 | 5 × 10−5 | KLF7 (3 × 10−6) | 0.654 | 0.001 | 0.002 | 0.001 |
| TES (1 × 10−6) | 0.717 | 0.294 | 0.085 | 0.328 | ||||||
| GPR55 (2 × 10−6) | 0.896 | 0.253 | 0.066 | 0.123 | ||||||
| rs738149 | Chr. 22 (35 660 195) | 0.096 | 0.002 | 7 × 10−6 | 3 × 10−5 | SMARCA2 (2 × 10−6) | 0.683 | 0.078 | 0.023 | 0.021 |
| GPR55 (3 × 10−6) | 0.896 | 0.253 | 0.066 | 0.123 | ||||||
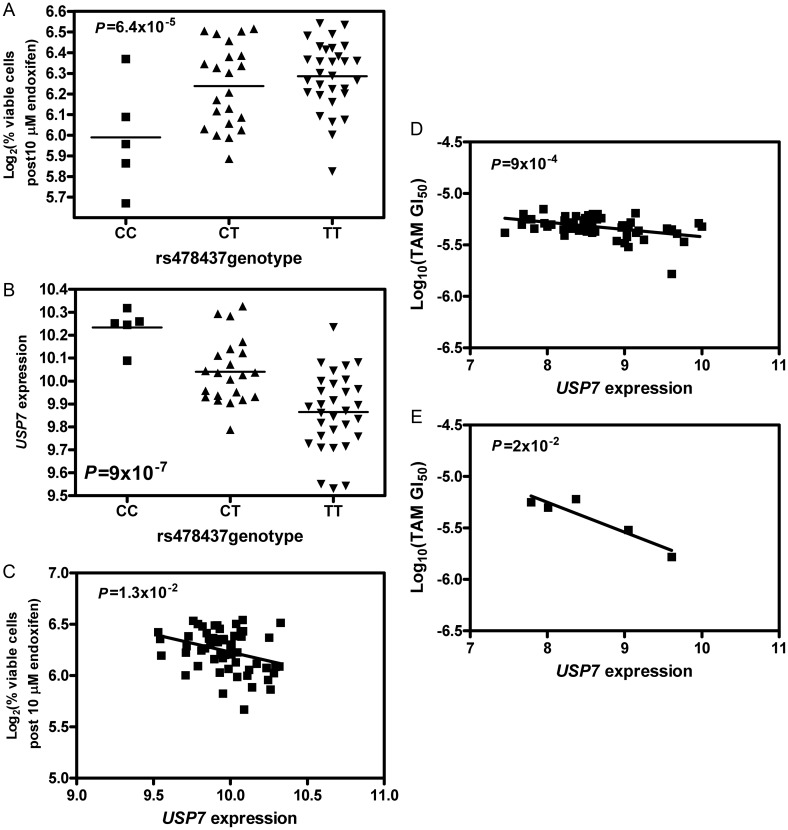
Of the final seven SNPs, we identified that an SNP, rs478437, is associated with all four phenotypes of endoxifen sensitivity with P < 0.05 (Table 1). A representation of genotype and phenotype (% cell growth inhibition after 10 μM endoxifen treatment) association is shown in Figure 1A (P = 6.4 × 10−5). The CC genotype is associated with higher cellular sensitivity. This same SNP is also associated with USP7 expression (P = 9 × 10−7, Figure 1B). USP7 expression is correlated with all four endoxifen sensitivity phenotypes (P < 0.05, Table 1), with lower expression correlated with more resistance to endoxifen (Figure 1C) in LCLs.
Figure 1.
Relationships between rs478437, USP7, and endoxifen sensitivity. (A) Association between rs478437 and percent viable cells after 10 μM endoxifen treatment in LCLs; (B) association between rs478437 and USP7 expression in LCLs; (C) association between USP7 expression and percent viable cells after 10 μM endoxifen treatment in LCLs; and (D) association between USP7 expression and log10(TAM GI50) in NCI60 cell lines. USP7 expression was characterized by Affymetrix Genechip HG-133. (E) Association between USP7 expression and log10(TAM GI50) in five breast cancer cell lines of NCI60. USP7 expression was characterized by Affymetrix Genechip HG-133.
evaluation of the candidate gene expression in NCI60 cell lines
Utilizing data obtained from the NCI60 cancer lines, including levels of gene expression and growth inhibition after TAM treatment, we found that all 10 target genes identified in CEU LCLs are correlated with TAM-induced growth inhibition in NCI60 cancer cell lines (P < 0.05). Figure 1D illustrates the correlation between the expression of USP7 and log10(TAM GI50) (P = 0.0009) in all 60 cancer cell lines. This is in agreement with the observation in LCLs that lower USP7 expression was correlated with more resistance to TAM. Furthermore, the expression for 6 of the 10 target genes (USP7, SYT17, GLTSCR2, SPRR1A, GPR55, and KLF7) are also correlated with TAM-induced growth inhibition in five NCI60 breast cancer cell lines (P < 0.05). Figure 1E illustrates the significant correlation between the USP7 expression and log10(TAM GI50) (P = 0.02) in these breast cancer cell lines.
functional validation of USP7 in TAM sensitivity among breast cancer cell lines
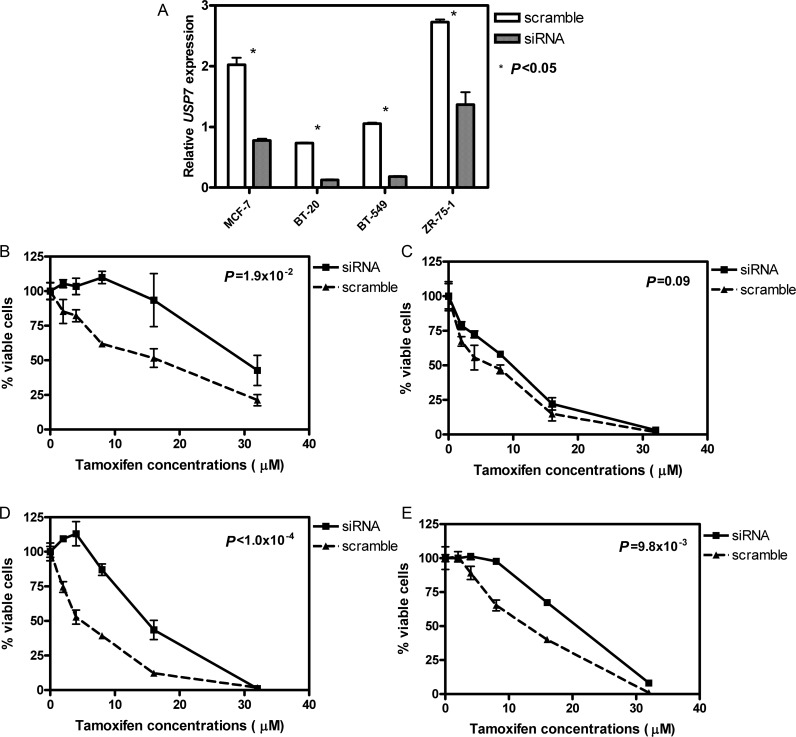
To validate the role of USP7 in TAM therapy, we knocked down this gene in four breast cancer cell lines independently and examined TAM sensitivity in the USP7 knock-down cell lines in comparison with that in the cell lines treated with scramble siRNA. The knock-down experiments were successfully performed in all four cell lines (Figure 2A) with significantly decreased USP7 levels compared with the scramble controls (P < 0.05). In addition, the lasting knock-down effect was observed at 5, 24, and 48 h post transfection. Knocking down USP7 also resulted in significant increases in TAM resistance in all four breast cancer cell lines [Figure 2B–E, with the two-way analysis of variance (ANOVA) P-values: 0.019, 0.090, <0.0001, and 0.0098, respectively]. Interestingly, we found the smallest change in TAM sensitivity after siRNA experiment in BT-20, which has the lowest USP7 expression levels prior to knock-down among the four cell lines tested (Figure 2A).
Figure 2.
Changes of TAM sensitivity in breast cancer cell lines with knock-down of USP7 by siRNA. (A) Decrease in relative USP7 mRNA levels in four breast cancer cell lines after the treatment of USP7 siRNA. (B) TAM sensitivity in MCF-7 cell line. (C) TAM sensitivity in BT-20 cell line. (D) TAM sensitivity in BT-549 cell line. (E) TAM sensitivity in ZR-75-1 cell line. P-value in each graph was calculated based on two-way ANOVA analyses.
clinical validation among breast cancer patients
To examine the role of genetic variants identified in our cell-based model in clinical samples, we evaluated 245 American Caucasian patients who had undergone TAM treatment. First, we tested the correlation between the phenotype of interest (the average score of hot flashes and hip/lumbar bone loss) and potential covariates. We found that prior chemotherapy is correlated with average hot flash scores (P = 0.057), while menopausal status and patient age are correlated with hip bone loss (P = 0.058 and 0.032, respectively). In addition, we found that age, menopausal status, and prior chemotherapy treatment are significantly correlated with lumbar bone loss (P = 0.0002, 0.001, and 0.0005, respectively). Therefore, these factors were incorporated, along with SNP genotype, into the corresponding subsequent multivariate regression model as independent variables, while each TAM sensitivity phenotype was treated as the dependent variable.
We found that rs310786 is significantly associated with lumbar bone loss after taking into consideration the age, menopausal status, and prior chemotherapy treatment (P = 0.044). Using rank as phenotype, we found that the SNP–lumbar bone loss association remains significant (P = 0.043). This finding is consistent with the results from LCLs that the CC genotype of rs310786 is associated with higher cellular sensitivity to endoxifen (supplementary Figure S3A, available at Annals of Oncology online), while patients who are carriers of CC genotype demonstrated more lumbar bone loss (supplementary Figure S3B, available at Annals of Oncology online).
discussions
Using an integrative genomic approach, we identified seven SNPs that nominally associated with cellular sensitivity to endoxifen through their effects on 10 gene expression. Since our genome-wide discovery was performed in LCLs derived from apparently healthy individuals, we evaluated these findings in cancer cells and confirmed the role of all TAM sensitivity-related genes identified in LCLs in a set of 60 cancer cell lines from the NCI60 dataset. More importantly, 6 of the 10 genes of interest were also found to relate to TAM sensitivity in five breast cancer cell lines that are a part of NCI60.
Although none of these discoveries from genome-wide analyses has been previously reported to relate to TAM, many of them are known to play critical roles in hormone biosynthesis and tumor suppression, such as USP7 [15], TMPRSS3 [16], TES [17], and SMARCA2 [18]. USP7, an ubiquitin-specific protease, is known to cleave ubiquitin from its substrates, such as p53 and PTEN, and therefore stabilizes these proteins [15]. Higher levels of USP7 expression are expected to result in higher levels of p53 and PTEN in the cells. Indeed, we observed a positive correlation on the mRNA expression levels between USP7 and PTEN in LCLs. Both p53 and PTEN are well-known human tumor suppressors and have been found to inhibit cell proliferation and cell growth [19, 20]. Since we observed that increased expression of USP7 leads to increased sensitivity to TAM in both LCLs and cancer cell lines, we hypothesized that higher levels of USP7 would lead to higher PTEN expression and higher cell growth inhibition. This hypothesis was supported in the LCL model, where the expression of PTEN was also correlated with cellular sensitivity to endoxifen at 7 and 10 μM treatment concentrations (P = 0.045 and 0.023, respectively). But the expression of p53 is not. These findings suggested a potential genetic regulatory network from an SNP (rs478437) to USP7 to PTEN, which eventually influences TAM sensitivity. The validity of this network remains to be further evaluated; however, the USP7 knock-down experiments conducted in four breast cancer cell lines further support the role of USP7 in TAM sensitivity in breast cancer.
In addition to evaluating the mechanism of action for the observed genotype–phenotype association, we also evaluated the genetic variants identified in LCLs in clinical patient samples. The major clinical problem for TAM is the lack of response. In our clinical study, only a very small percentage of patients had the disease recurrent event. Therefore, we do not have power to evaluate clinical TAM efficacy. However, various studies have reported that the increased incidences of hot flashes induced by TAM correlate with improved treatment efficacy [21]. Therefore, we chose to evaluate the genetic effect on TAM-induced toxicities, namely hot flashes and bone loss. We found that an SNP regulating TMPRSS3, namely rs310786, is related to TAM-induced lumbar bone loss, after controlling for relevant clinical factors. The CC genotype is related to both increased cellular sensitivity to endoxifen and more lumbar bone loss induced by TAM.
Our study focused on SNPs regulating gene expression. Therefore, we might have missed other important genetic variants that associate with endoxifen sensitivity, but are not correlated with gene expression at P = 3.8 × 10−6, such as the 11 SNPs related to all four endoxifen sensitivity phenotypes. They might contribute to endoxifen sensitivity through miRNA or other epigenetic factors (e.g. DNA methylation and histone remodeling). In an extended analysis in the clinical samples of SNPs that were identified in LCLs to be associated with at least three endoxifen sensitivity phenotypes, we found six of them are also associated with TAM side effects (P < 0.05) after corresponding covariates were controlled for; for example, rs10983932, rs10984098, and rs4959825 to hot flash; rs10983920 and rs9862879 to both hot flash and hip bone loss; rs4959825 to both hot flash and lumbar bone loss. These discoveries warrant further validation.
To date, only a few genetic predictors have been reported to play a role in TAM sensitivity, most notably CYP2D6 [22]. We did not observe them in our genome-wide integrative studies. Given the lack of major cytochrome P450 enzymes expression in the LCLs, this is not surprising. In fact, the genetic variants reported in this study may be taken as complimentary to those identified in CYP2D6 or other P450 enzymes. A recent genome-wide association study among Japanese breast cancer patients who underwent TAM therapy reported nine chromosomal loci significantly associated with recurrence-free survival [23]. None of these loci were among the SNPs found to be associated with cellular sensitivity to endoxifen in LCLs at P = 0.0001. This discrepancy could be explained by the following: (i) different phenotypes were evaluated (recurrence-free survival versus cellular sensitivity to TAM); (ii) different ethnic groups were the focus (Japanese versus Europe Caucasian).
In conclusion, our work demonstrates the utility of the cell-based model in the genome-wide identification of pharmacogenomic markers for TAM. The SNPs and their corresponding target genes identified by our study, once further validated, may be used to predict TAM sensitivity in breast cancer patients.
funding
This study was supported by the University of Chicago Cancer Center support grant (#P30 CA14599) and National Institutes of Health/National Institute of General Medical Science (Pharmacogenomics of Anticancer Agents grant U01GM61393). Patient data were collected by the Consortium on Breast Cancer Pharmacogenomics (COBRA) investigators. RSH also received support from National Institute of General Medical Science K08 (GM089941), the National Cancer Institute R21 (CA139278), the University of Chicago Breast Cancer SPORE Career Development Award (CA125183), and the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1RR024999).
disclosure
The authors have declared no conflicts of interest. The authors did not use editorial support for preparation of the manuscript.
Supplementary Material
references
- 1.Jordan VC. Tamoxifen treatment for breast cancer: concept to gold standard. Oncology (Williston Park) 1997;11:7–13. [PubMed] [Google Scholar]
- 2.Clarke R, Leonessa F, Welch JN, et al. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol Rev. 2001;53:25–71. [PubMed] [Google Scholar]
- 3.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 4.Moyer AM, Suman VJ, Weinshilboum RM, et al. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics. 2011;12:1535–1543. doi: 10.2217/pgs.11.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Schaik RH, Kok M, Sweep FC, et al. The CYP2C19*2 genotype predicts tamoxifen treatment outcome in advanced breast cancer patients. Pharmacogenomics. 2011;12:1137–1146. doi: 10.2217/pgs.11.54. [DOI] [PubMed] [Google Scholar]
- 6.Kiyotani K, Mushiroda T, Imamura CK, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CY, Strom A, Vega VB, et al. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang RS, Kistner EO, Bleibel WK, et al. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genome-wide approach. Am J Hum Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shankavaram U, Varma S, Kane D, et al. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Y, Hayes DF, Li L, et al. Estrogen receptor genotypes influence hot flash prevalence and composite score before and after tamoxifen therapy. J Clin Oncol. 2008;26:5849–5854. doi: 10.1200/JCO.2008.16.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmad A, Ali SM, Ahmad MU, et al. Orally administered endoxifen is a new therapeutic agent for breast cancer. Breast Cancer Res Treat. 2010;122:579–584. doi: 10.1007/s10549-009-0704-7. [DOI] [PubMed] [Google Scholar]
- 15.Colland F, Formstecher E, Jacq X, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 16.Fasquelle L, Scott HS, Lenoir M, et al. Tmprss3, a transmembrane serine protease deficient in human DFNB8/10 deafness, is critical for cochlear hair cell survival at the onset of hearing. J Biol Chem. 2011;286:17383–17397. doi: 10.1074/jbc.M110.190652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coutts AS, MacKenzie E, Griffith E, et al. TES is a novel focal adhesion protein with a role in cell spreading. J Cell Sci. 2003;116:897–906. doi: 10.1242/jcs.00278. [DOI] [PubMed] [Google Scholar]
- 18.Marshall TW, Link KA, Petre-Draviam CE, et al. Differential requirement of SWI/SNF for androgen receptor activity. J Biol Chem. 2003;278:30605–30613. doi: 10.1074/jbc.M304582200. [DOI] [PubMed] [Google Scholar]
- 19.Razak ZR, Varkonyi RJ, Kulp-McEliece M, et al. p53 differentially inhibits cell growth depending on the mechanism of telomere maintenance. Mol Cell Biol. 2004;24:5967–5977. doi: 10.1128/MCB.24.13.5967-5977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu EC, Tarnawski AS. PTEN regulatory functions in tumor suppression and cell biology. Med Sci Monit. 2004;10:RA235–RA241. [PubMed] [Google Scholar]
- 21.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 22.Brauch HB, Schroth W, Ingle JN, et al. CYP2D6 and tamoxifen: awaiting the denouement. J Clin Oncol. 2011;29:4589–4590. doi: 10.1200/JCO.2011.38.8611. author reply 4590–4581. [DOI] [PubMed] [Google Scholar]
- 23.Kiyotani K, Mushiroda T, Tsunoda T, et al. A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Hum Mol Genet. 2012;21:1665–1672. doi: 10.1093/hmg/ddr597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.