Abstract
There are a number of related goals of influenza vaccination, including elicitation of protective antibodies and induction of cellular CD4 and CD8 T cell responses. Because CD4 T cell expansion and functionality are influenced by peptide specificity and T cell gene expression can be modified by repeated re-stimulations, it is important to evaluate how frequent influenza vaccinations affect CD4 T cell dependent functions in protective immunity to influenza. Trivalent influenza vaccines (TIV) have production of neutralizing antibodies to HA as their primary goal and main criteria for efficacy. Accordingly, they are not characterized for any other viral components. In the current study, we evaluated whether other influenza virus proteins were present in commercial TIV at levels sufficient for immunogenicity in vivo. Mice that differed with regard to their expressed class II molecules were used in concert with peptide-stimulated cytokine EliSpot assays to comprehensively evaluate the CD4 T cell antigen specificity induced by the TIV. Our studies revealed that NA, NP, M1 and NS1 were present in sufficient quantities in the TIV to prime and boost CD4 T cells. These results suggest that in humans, the broad CD4 T cell repertoire induced by live infection is continually boosted and maintained throughout life by regular vaccination with licensed intramuscular split vaccines. The implications raised by our findings on CD4 T cell functionality in influenza are discussed.
Keywords: vaccine, immmunodominance, CD4 T cell, epitope, immune response
Introduction
Influenza virus is a serious and potentially deadly pathogen in human populations across the globe[1–4]. Seasonal viruses, that have only small genetic variation yearly, have particularly high disease impact on the very young, the elderly and immunocompromised individuals (reviewed in [1, 2, 5, 6]). Pandemic influenza, from newly recombined influenza viruses or adaptation of animal strains to humans, is a risk for all and can have devastating consequences (reviewed in [7–10]). For these reasons, there is continued effort to deploy influenza vaccines to improve their potency and to develop better correlates of immune protection elicited by vaccination (reviewed in [1, 11]).
There are two types of licensed vaccines used in the US [1, 12–15], both typically composed of H1N1, H3N2 and influenza B viral components. The cold-adapted influenza vaccine (“LAIV”) is a 6+2 recombinant that displays HA and NA from the circulating strains and harbors internally expressed virion proteins that carry mutations that prevent replication at 37°C. It is introduced intranasally and has limited replication in vivo, but elicits both cellular T cell responses and antibody responses. The split or subunit vaccine (“TIV”) typically is composed of a 6+2 recombinant membrane HA and NA derived from the circulating strain and internal virion proteins derived from the A/PR8/34 strain to increase yields of the egg-grown stock. The vaccine is prepared from inactivated and detergent solubilized virion particles and is typically introduced intramuscularly. The primary goal of vaccination with TIV is to elicit antibody responses. For this reason, prior to distribution, this vaccine is quantified only for content of HA and vaccine efficacy is primarily evaluated for induction of neutralizing antibody responses and hemagglutination inhibition (reviewed in [14, 16–18]). Although much attention is focused on the B cell response, the cellular response is also critical for protective immunity. CD8 T cells provide direct cytotoxicity toward infected cells in the lung, thus limiting viral replication, while CD4 T cells have many distinct effector functions, including provision of cytokines, help for antibody responses and perhaps direct cytotoxicity (reviewed in [8, 19–22]).
An important question yet to be addressed is whether the antigen specificity of CD4 T cells influences their functional potential. Literature suggests that this may be the case. First, for vaccinia virus, CD4 T cell antigen specificity was linked to B cell specificity for provision of help [23]. If true for influenza, CD4 T cells specific for epitopes within HA may be the most effective in provision of help for production of neutralizing antibodies. Also, T cell function continues to evolve with successive boosts that T cells undergo [24, 25]. This raises the possibility that CD4 T cells specific for conserved epitopes, particularly those abundant in licensed vaccines subject to re-stimulation through yearly vaccination, might have different effector potential than T cells specific for new peptide epitopes from the rapidly evolving HA and NA proteins.
Our previous studies examining the CD4 T cell specificity in the primary response to live intranasal infection with human H1N1 viruses[26–29] revealed that live infection induces an exceptionally broad specificity in CD4 T cells with reactivity to HA, NA, NP, M1 and NS1. Interestingly, this high degree of CD4 T cell diversity persists in memory [30]. Because most humans typically have their first encounter with influenza by infection, we speculate that the influenza specific CD4 memory established in childhood is similarly broad. But it is not clear how memory population is reshaped in humans with repeated infections and vaccination. To shed light on this, in this study evaluated the potential of TIV to elicit or boost CD4 T cells to different influenza viral antigens.
Materials and Methods
Mice
BALB/c, A/JCr and SJL/JCr were purchased from National Cancer Institute-Frederick (Frederick, MD). The DR1 transgenic mice (B10.M/J-TgN-DR1) were obtained from D. Zaller (Merck) through Taconic Laboratories. All mice were maintained in the pathogen-free facility at the University of Rochester according to institutional guidelines.
Ethics Statement
All animal protocols used in this study adhere to the AAALAC, International, the Animal Welfare Act and the PHS Guide and were approved by the University of Rochester Committee on Animal Resources; Animal Welfare Assurance Number A3291-01 on March 4, 2006 (protocol no. 2006-030) and April 10, 2008 (protocol no. 2008-023).
Peptides
17-mer peptides overlapping by 11 amino of the HA and NA the H1N1 virus A/New Caledonia/20/99, the HA and NA from the H3N2 virus A/New York/384/2005, the NS1 sequence from the A/New York/444/2001, and the NP, M1 and PB1 from the H1N1 virus A/New York/348/2003, were obtained from BEI Resources, ATCC.
Immunizations
Mice were immunized subcutaneously in the hind-footpads with 50μl of the Influenza Virus Vaccine, Fluzone, 2006–2007 formula or 2007–2008 formula (Sanofi Pasteur, obtained from BEI Resources, ATCC) emulsified in Complete Freund’s Adjuvant (CFA)(Sigma-Aldrich, St. Louis, MO). Influenza vaccine was dialyzed and concentrated, and, each mouse was immunized with 22.5μg HA per footpad. At ten to thirteen days post immunization, serum was collected for ELISAs, and the spleen and lymph nodes were excised, pooled and used as sources of CD4 T cells for ELISPOT analyses. Cells were depleted of red blood cells (RBC) using ACK Lysis Buffer (0.15M NH4Cl, 1mM KHCO3, and 0.1 mM Na2-EDTA in H2O, pH7.2), followed by depletion of B cells, CD8 cells and macrophages by negative selection using MACS depletion (Miltenyi Biotech, Gladbach, Germany), according to the manufacturer instructions.
ELISPOT Assays
ELISPOT assays were performed as described [27–29]}. Briefly, 96-well filter plates (Millipore, Billerica, MA) were coated rat anti-mouse IL-2 or IFN-g (clone JES6-1A12 and clone AN-18, respectively, BD Biosciences, San Jose, CA), washed and blocked. CD4 T cells were co-cultured with APC and with the indicated peptide or peptide pool at a final concentration of 10μM or 2μM, respectively, for 18–20 hours at 37°C and 5% CO2. Plates were washed and incubated with biotinylated rat anti-mouse IL-2 or IFN-g (clone JES6-5H4, clone XMG1.2, BD Biosciences), and developed, dried and quantified with an Immunospot reader series 2A, using Immunospot software, version 3.2.
Antibody ELISA Assays
Mouse sera were collected from individual mice and HA and NP-specific antibodies were determined by ELISA assay. Plates (Costar) were coated with 200ng of purified NP protein, cloned from A/New Caledonia/20/99 or HA protein from, A/New Caledonia/20/99, purchased from Protein Sciences (Meriden, CT). Wells were rinsed, incubated with blocking buffer (3% BSA in PBS) and then diluted serum samples (in 0.5% BSA-PBS) were added to the plates and incubated for 2–3 hours at room temperature. The wells were washed, incubated sequentially with 100μl/well alkaline phosphatase conjugated goat anti-mouse IgG secondary antibody (SouthernBiotech) and, one p-nitrophenyl phosphate substrate. After washing, absorbance at 405nm was read.
Vaccine Boost post Influenza Infection
Mice were infected as we have previously described with A/New Caledonia/20/99 [27]. At day 10 (peak responses) or 60 (memory), mice were sacrificed and spleen and mediastinal lymph nodes were used as sources of CD4 T cells for ELISPOT analyses. For intramuscular boost mice were immunized with Influenza vaccine 2006–2007 formula at a total dose of 45μg total HA given in two injections, each of 22.5μg (immunization volume 50μl in PBS), into the tibialis anterior muscle of each leg. Seven days post immunization, lymphoid tissues were used as sources of CD4 T cells for ELISPOT analyses, as described above.
Results
We first evaluated the distribution of influenza protein-specific patterns within CD4 T cells elicited in the primary response to trivalent influenza vaccines. Patterns in immunogenicity among different influenza proteins will presumably reflect the abundance of the proteins in virions and the final protein products of the vaccine after commercial purification, as well as differences in uptake by antigen presenting cells and yield of antigenic peptides liberated. It is not possible to study the unbiased primary CD4 T cell response to vaccines in adult subjects therefore we used mice never exposed to influenza. Because different alleles of class II dramatically bias the specificity of the CD4 T cells to many antigens including influenza virus [28], different mouse strains were studied, each of which express unrelated MHC class II proteins, allowing “sampling” of many different influenza-derived peptides by diverse MHC class II proteins. We anticipated this approach would collectively reveal the CD4 T cell response potential of humans that have many distinct class II molecules.
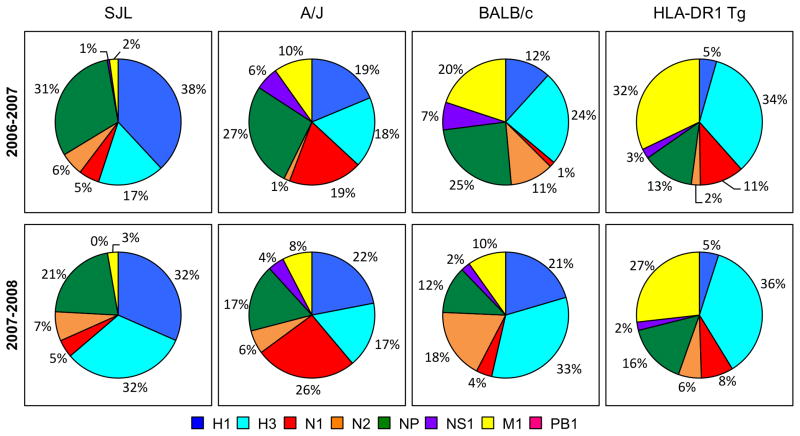
Two different yearly vaccines (2006 and 2007) were examined that had related but distinct H1N1, H3N2 and Influenza B viruses, to avoid any “batch-specific” effects in protein composition. Mice expressing distinct MHC alleles, including SJL (I-As), A/J (I-Ak, I-Ek), BALB/c (I-Ad, I-Ed) and DR1 transgenic mice. The vaccines were emulsified in CFA in order to reveal the potential immunogenicity of the components in the vaccine. Naïve mice do not make a detectable response to vaccine in saline but do once immunological memory is established (see Figure 6). CD4 T cells from the draining lymph node were screened for reactivity towards different viral proteins by using pools of synthetic peptides representing the entire viral protein sequence of each protein tested (H1, H3, N1, N2, NP, NS1, M1 and PB1). The number of reactive CD4 T cells to each protein was quantified by cytokine EliSpot assays. Shown in Figure 1 are the results of these analyses. As expected, based on their expression of distinct class II molecules, each strain displayed differing patterns of CD4 T cell reactivity. The A/J strain displayed the broadest specificity, with high reactivity towards H1, H3, N1 and NP and lower reactivity toward the NS1 and M1. SJL mice had dominant specificities for H1, H3 and NP. BALB/c and DR1 transgenic mice displayed more modest reactivity to the vaccine epitopes, but both were broad in their specificity, with CD4 T cell responses distributed among many different viral proteins. BALB/c displayed fairly even reactivity across H1, H3, N2, NP, M1 while the DR1-Tg mice showed more dominant reactivity toward H3, NP and M1. The relative distribution of epitopes among all the viral proteins can be presented as a fractional response to each viral protein shown in Figure 2, which illustrates the breadth in protein specificity of CD4 T cells elicited by the split vaccines. Collectively, these results demonstrate that although these split vaccines’ main purpose is to elicit neutralizing anti-HA antibodies, they in fact elicit CD4 T cells of broad antigen specificity including most viral proteins tested. The ability to detect this priming depends on the MHC molecules expressed in the host.
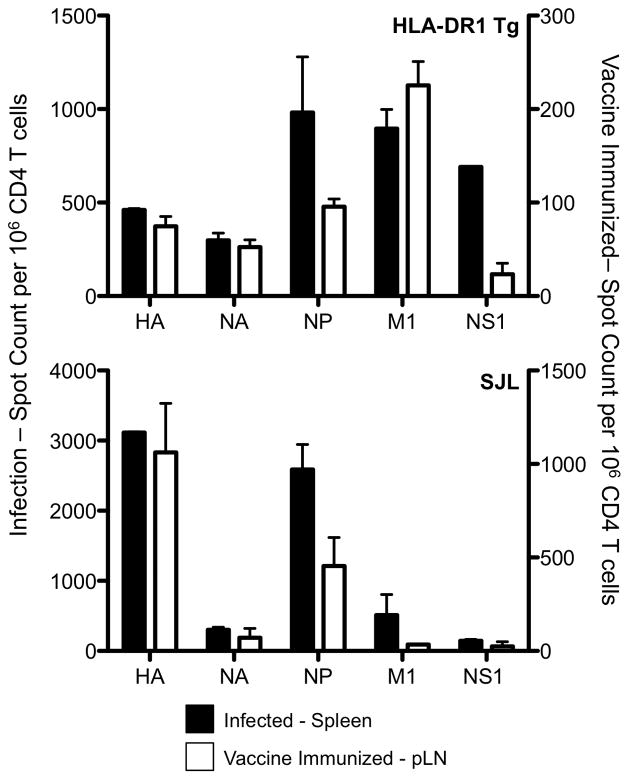
Figure 6. The antigen specificity CD4 T cells boosted with TIV.

DR1 Tg and SJL mice primed 4–8 weeks earlier by intranasal infection with live with A/New Caledonia/20/99 H1N1 virus were vaccinated intramuscularly in the thigh with TIV in saline. The number of IL-2 (left (DR1 Tg) and middle (SJL) panels) or IFN-γ (right panel, SJL) producing CD4 T cells purified from the inguinal lymph nodes (draining site) were enumerated in ELISPOT assays. CD4 T cells from HLA-DR1 mice predominantly produce IL-2 over IFN-γ (ref. 27). Reactivity in the mice infected alone is shown in black bars at the memory time point before boost, and reactivity in CD4 T cells from mice infected and then boosted is shown in white bars. Shown is the average of two independent experiments of 4–6 pooled mice and the error bars represent the range of the response.
Figure 1. Distribution of influenza protein specific patterns elicited in the primary response to trivalent influenza vaccine immunization.
SJL, A/J, BALB/c and HLA-DR1 Tg mice were immunized subcutaneously in the hind footpads with influenza vaccine, 2006–2007 formula in the top row and 2007–2008 formula in the bottom row, emulsified in CFA. Ten days post immunization the number of IL-2 producing CD4 T cells reactive to the pools of peptides from the influenza proteins (2μM of each peptide) were enumerated in ELISPOT assays.
Figure 2. The relative distribution of influenza protein specific patterns elicited in the primary response to trivalent influenza vaccine immunization represented as the fractional response to each protein.
As indicated above the panels, SJL, A/J, BALB/c and HLA-DR1 Tg mice were immunized subcutaneously in the hind footpads with influenza vaccine, 2006–2007 formula (top panels) and 2007–2008 formula (bottom panels), emulsified in CFA. Ten days post immunization the number of IL-2 producing CD4 T cells reactive to the pools of peptides from the influenza proteins (2μM final concentration of each peptide) were enumerated in ELISPOT assays and the fractional response for each protein was calculated by dividing the total response for each protein by the sum of the responses to all of the proteins. The individual proteins tested are colored coded as indicated beneath the Figure.
Because CD4 T cell responses to NP were prominent, we evaluated if NP-specific antibodies were elicited by the split vaccine and how these compared to the HA-specific antibodies. ELISA assays (Figure 3) revealed that all mice mounted readily detectable IgG antibody responses to HA and NP after vaccination, confirming that both are present in sufficient quantity and immunogenicity to elicit CD4 T cell and B cell responses.
Figure 3. All four strains of mice have detectable IgG antibody responses to HA and NP after vaccination.
BALB/c (closed circle), A/J (closed square), SJL (open square) and HLA-DR1 Tg (open circle) mice were immunized subcutaneously with 2006–2007 formula trivalent influenza vaccine, serum was collected 13 days later and tested for HA and NP specific IgG antibodies. Shown is the average for three individual mice and the error bars represent the range of the response.
Based on the broad specificity of CD4 T cells elicited by TIV, inclusive of many viral proteins it was of interest to compare the repertoire of CD4 T cells elicited by infection and vaccination (Figure 4). From infected SJL and DR1 Tg mice, robust reactivity was elicited toward all viral proteins tested, depending on the MHC class II allele. In contrast, as discussed previously, the major specificities elicited by the vaccine were specific for HA, NP and M1. In contrast, as discussed previously, the major specificities elicited by the vaccine were specific for HA, NP and M1. The selective CD4 specificities towards NS1 from infection compared to vaccination are not surprising because NS1 is not expressed at high levels within virions [32]. NS1 can be detected at low levels in some commercial poultry vaccines in immunogenic form, presumably as a result of carryover from the allantoic fluid [33, 34]. We have confirmed that the commercial human TIV vaccines studied here contain NS1 protein detectable by Western blotting (not shown) and this appears to be present at levels sufficient to be immunogenic, depending on the class II molecules expressed. The apparent deficit in M1-specific reactivity in SJL mice immunized with the vaccine may be due the mismatch between the seasonal A/New Caledonia sequences used for infection which the peptides matched, while the vaccine is derived from A/PR/8 recombinant to increase yields in eggs, as well as the overall magnitude of the response. In response to live infection, The SJL strain recognized only 3 peptides from M1 ([28]), of which 2 from the vaccine backbone have an amino acid substitution with the tested peptides. In constrast, HLA-DR1 mice infected with influenza recognize 11 M1 peptides, of which only 4 have substitutions with the vaccine derived sequences and their response magnitude is approximately 7 times as great as SJL ([34]). We conclude from this that most of the viral proteins expressed in virions are contained in the vaccines and can elicit CD4 T cell responses similar to that elicited from live infection.
Figure 4. The total response of CD4 T cells to live influenza infection compared to vaccination.
HLA-DR1 Tg (top panel) and SJL (bottom panel) mice were infected intranasally with A/New Caledonia/20/99 H1N1 virus or were immunized with trivalent influenza vaccine formula 2006–2007 emulsified in CFA. Ten days post inoculation the number of IL-2 producing CD4 T cells, from the draining popliteal lymph nodes for vaccinated animals (white bars) and the spleen for infected animals (black bars), were enumerated in ELISPOT assays. For the HLA-DR1 Tg mice, where the response is very broad, total pools of influenza peptides were used in restimulation; while for the SJL mice CD4 T cells were co-cultured with the known influenza single peptides, whose responses were summed and shown here as the total response to each influenza protein. The left axis is used to enumerate the number of IL-2 producing CD4 T cells during influenza infection and the right axis is used to enumerate the number of IL-2 producing CD4 T cells after vaccination. Shown is the average of two independent experiments and the error bars represent the range of the responses.
To begin to model the immune response to vaccination in humans, the ability of the vaccine to recall CD4 T cells elicited from viral infection was studied. SJL mice were first exposed to an H1N1 influenza virus via infection, the route common in most human populations. After infection, mice were rested for 60 days as immunological memory became established. Figure 5 shows the specificity of memory (D60) after priming by infection compared to the specificity at the peak of the response. We then examined the consequences of boosting the previously infected mice. Here, we tested an intramuscular injection of vaccine in saline alone in order to mimic the use of the licensed vaccine in humans and included both SJL and HLA-DR1 mice (Fig 6). Previous studies by our laboratory (not shown) had indicated very poor priming of mice using this type of vaccination strategy in naïve mice. However, we speculated that after priming by infection and establishment of a diverse memory population, it might be possible to recall CD4 T cells with the intramuscular injection. Because of the low yield of lymphocytes from mice primed with vaccine in saline alone, only selected peptide epitopes were tested, chosen based on the magnitude of the response they elicit in the primary response to live influenza (see Figure 5) and the protein they were derived from, as we wished to sample epitopes from different viral proteins. Our hypothesis that CD4 T cell responses would be detectable in hosts that had immunological memory toward influenza was confirmed in the experiment shown in Figure 6. HA, M1 and NP specific memory is detectable at baseline within many draining lymph nodes after virus priming and rest (Figure 6 filled bars) and this reactivity is boosted dramatically in all the lymph nodes tested after intramuscular vaccination in the thigh (Figure 6, open bars). Collectively, these results indicate that in the typical human host that has established immunological memory to influenza induced by live virus or attenuated vaccines, vaccination with inactivated split vaccines will likely boost reactivity of CD4 T cells to many viral proteins.
Figure 5. The CD4 T cell response at peak and memory time points after infection.

SJL mice were infected with A/New Caledonia/20/99 H1N1 virus and then rested for 8 weeks allowing for the memory compartment to become established. At peak (D10, white bars) and memory (D60, black bars) time points post infection the number of IL-2 producing T cells from the spleen were enumerated in ELISPOT assays. CD4 T cells were co-cultured with syngeneic splenocytes and the known influenza single peptides (10μM) for 18 hrs. Shown is the average of two independent experiments and the error bars represent the range of the responses.
Discussion
There are a number of related goals of influenza vaccination. The original and still-dominant goal is to elicit neutralizing antibodies that will protect from future infections, a process that requires B cells and helper CD4 T cells. Another goal is to elicit a cellular response by CD8 and CD4 T cells that provide effector function in the lung (reviewed in [21, 35, 40]). There is also great interest in vaccines that will provide heterosubtypic immunity against novel or pandemic influenza. The general strategy to achieve this latter goal is to focus immune responses to T cell or B cell epitopes that are shared among even distantly related viruses. Candidates include M2e [41, 42], or “headless” HA [43, 44]. For T cells, cross-reactive epitopes are enriched within internal viral proteins not subject to antibody-mediated selection, such as M1, NP and the polymerase proteins. However, because of the possible requirement for linked recognition of CD4 and B cell epitopes, CD4 T cell help for HA-derived epitopes might be most useful for B cell responses to HA.
Understanding links between CD4 T cell specificity and function in influenza-specific responses is clearly needed for both vaccine design and vaccine evaluation. Although infection with live or attenuated viruses has been shown to elicit T cells specific for both internal and cell surface associated viral proteins [29, 45, 46], the protein and specificity of CD4 T cells elicited from vaccination with split vaccines has never been evaluated. In fact, viral protein composition is not reported by manufacturers, and the production procedures themselves are proprietary and not described in detail by the manufacturers (reviewed in [16, 47]). In the unbiased study performed here, we have found that licensed human TIV elicits robust CD4 T cell responses to M1, NS1 and NP in addition to HA and NA. Responses to these proteins are boosted in hosts previously been exposed through live infection. In addition to HA, TIV also induces IgG antibody responses toward NP, the only other protein tested here. Yearly TIV vaccination will boost and maintain T cell diversity toward internal and highly conserved viral proteins, much as the cold adapted vaccine will. B cells specific for these proteins will also be primed and established and available for future responses.
Based on these findings, several important issues need to be resolved in order to understand if this complex mixture of immunogens has positive or negative consequences toward vaccine efficacy. The first is the issue of competition. Are B cell and CD4 T cell responses to HA compromised due to the co-introduced and immunogenic NP and M1 proteins? There is ample evidence for both the benefits and the potential detrimental effects of competitive responses for T cells [48–57] and the balance between these events is not yet known but should be evaluated. If increased diversity diminishes quantity or quality of protective antibody, then it might be useful to consider selective enrichment of HA and NA prior to final preparation of the vaccine. The second issue to consider is whether CD4 T cell specificity for help is linked to B cell specificity. If B cells specific for HA internalize only HA via their immunoglobulin receptor, via uptake of membrane fragments containing this membrane-associated antigen, they will only display HA-derived peptides and therefore can only recruit help by CD4 T cells specific for HA. Helper cells specific for other viral proteins such as NP, that are also boosted during vaccination with TIV may occupy immunological “space” that does not contribute to the neutralizing antibody response. Finally, it is useful to consider is whether repeated boosting changes T cell effector function in a way that is relevant for protective immunity to influenza. There is some evidence that CD8 T cells continue to “evolve” in gene expression patterns upon each re-stimulation [24, 25]. If these changes also occur in CD4 T cells then repeated stimulation through TIV vaccination might lead to CD4 T cells specific for conserved proteins having a distinct phenotype compared to those specific for newly encountered epitopes. While prime-boost strategies of vaccination are generally thought to be efficacious for pathogen-specific responses (reviewed in [58–62]), the consequences of repeated cycles of re-stimulation over many years have not been explored. There is evidence that regulatory T cells and IL-10 producing cells can become enriched with repeated vaccination [63, 64], and thus frequent boosting of CD4 T cells specific for highly conserved internal influenza proteins may be detrimental for protective immunity or T cell helper function. Clearly, these issues need to be evaluated in future studies that seek to optimize both the short term and long term effects of influenza vaccination.
Highlights.
Mouse models of influenza vaccination were used to evaluate CD4 T cell specificity.
Licensed split influenza vaccines elicit CD4 T cells of diverse specificity.
HA, NA, M1, NP and NS1 reactive CD4 T cells were elicited after primary vaccination.
Vaccination continually boosts and maintains an extremely diverse CD4 repertoire.
Acknowledgments
This work was supported by grants HHSN266200700008C and R01AI51542 to A. J. Sant from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med. 2010 Nov 18;363(21):2036–44. doi: 10.1056/NEJMra1002842. [DOI] [PubMed] [Google Scholar]
- 2.Monto AS. Seasonal influenza and vaccination coverage. Vaccine. 2010 Sep 7;28(Suppl 4):D33–44. doi: 10.1016/j.vaccine.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Nair H, Brooks WA, Katz M, Roca A, Berkley JA, Madhi SA, et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. 2011 Dec 3;378(9807):1917–30. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- 4.Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007 Jun 28;25(27):5086–96. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 5.Kumar D, Blumberg EA, Danziger-Isakov L, Kotton CN, Halasa NB, Ison MG, et al. Influenza vaccination in the organ transplant recipient: review and summary recommendations. Am J Transplant. 2011 Oct;11(10):2020–30. doi: 10.1111/j.1600-6143.2011.03753.x. [DOI] [PubMed] [Google Scholar]
- 6.McElhaney JE. Influenza vaccine responses in older adults. Ageing Res Rev. 2011 Jul;10(3):379–88. doi: 10.1016/j.arr.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuyama S, Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol. 2011 Aug;23(4):481–6. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valkenburg SA, Rutigliano JA, Ellebedy AH, Doherty PC, Thomas PG, Kedzierska K. Immunity to seasonal and pandemic influenza A viruses. Microbes Infect. 2011 May;13(5):489–501. doi: 10.1016/j.micinf.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katz JM, Veguilla V, Belser JA, Maines TR, Van Hoeven N, Pappas C, et al. The public health impact of avian influenza viruses. Poult Sci. 2009 Apr;88(4):872–9. doi: 10.3382/ps.2008-00465. [DOI] [PubMed] [Google Scholar]
- 10.Yen HL, Webster RG. Pandemic influenza as a current threat. Curr Top Microbiol Immunol. 2009;333:3–24. doi: 10.1007/978-3-540-92165-3_1. [DOI] [PubMed] [Google Scholar]
- 11.Subbarao K, Murphy BR, Fauci AS. Development of effective vaccines against pandemic influenza. Immunity. 2006 Jan;24(1):5–9. doi: 10.1016/j.immuni.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis. 2012 Jan;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 13.Carter NJ, Curran MP. Live attenuated influenza vaccine (FluMist(R); Fluenz): a review of its use in the prevention of seasonal influenza in children and adults. Drugs. 2011 Aug 20;71(12):1591–622. doi: 10.2165/11206860-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 14.Minor PD. Vaccines against seasonal and pandemic influenza and the implications of changes in substrates for virus production. Clin Infect Dis. 2010 Feb 15;50(4):560–5. doi: 10.1086/650171. [DOI] [PubMed] [Google Scholar]
- 15.Schultz-Cherry S, Jones JC. Influenza vaccines: the good, the bad, and the eggs. Adv Virus Res. 2010;77:63–84. doi: 10.1016/B978-0-12-385034-8.00003-X. [DOI] [PubMed] [Google Scholar]
- 16.Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003 May 1;21(16):1776–9. doi: 10.1016/s0264-410x(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 17.Wood JM, Major D, Heath A, Newman RW, Hoschler K, Stephenson I, et al. Reproducibility of serology assays for pandemic influenza H1N1: collaborative study to evaluate a candidate WHO International Standard. Vaccine. 2012 Jan 5;30(2):210–7. doi: 10.1016/j.vaccine.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 18.Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti Infect Ther. 2011 Jun;9(6):669–83. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 19.Whitmire JK. Induction and function of virus-specific CD4+ T cell responses. Virology. 2011 Mar 15;411(2):216–28. doi: 10.1016/j.virol.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas PG, Keating R, Hulse-Post DJ, Doherty PC. Cell-mediated protection in influenza infection. Emerg Infect Dis. 2006 Jan;12(1):48–54. doi: 10.3201/eid1201.051237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinstry KK, Strutt TM, Swain SL. Hallmarks of CD4 T cell immunity against influenza. J Intern Med. 2011 May;269(5):507–18. doi: 10.1111/j.1365-2796.2011.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strutt TM, McKinstry KK, Swain SL. Functionally diverse subsets in CD4 T cell responses against influenza. J Clin Immunol. 2009 Mar;29(2):145–50. doi: 10.1007/s10875-008-9266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sette A, Moutaftsi M, Moyron-Quiroz J, McCausland MM, Davies DH, Johnston RJ, et al. Selective CD4+ T cell help for antibody responses to a large viral pathogen: deterministic linkage of specificities. Immunity. 2008 Jun;28(6):847–58. doi: 10.1016/j.immuni.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirth TC, Xue HH, Rai D, Sabel JT, Bair T, Harty JT, et al. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 2010 Jul 23;33(1):128–40. doi: 10.1016/j.immuni.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masopust D, Ha SJ, Vezys V, Ahmed R. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J Immunol. 2006 Jul 15;177(2):831–9. doi: 10.4049/jimmunol.177.2.831. [DOI] [PubMed] [Google Scholar]
- 26.Sant AJ, Chaves FA, Krafcik FR, Lazarski CA, Menges P, Richards K, et al. Immunodominance in CD4 T-cell responses: implications for immune responses to influenza virus and for vaccine design. Expert Rev Vaccines. 2007 Jun;6(3):357–68. doi: 10.1586/14760584.6.3.357. [DOI] [PubMed] [Google Scholar]
- 27.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol. 2007 Jul;81(14):7608–19. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nayak JL, Richards KA, Chaves FA, Sant AJ. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol. 2010 Apr;23(2):169–80. doi: 10.1089/vim.2009.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards KA, Chaves FA, Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol. 2009 Jul;83(13):6566–77. doi: 10.1128/JVI.00302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards KA, Chaves FA, Sant AJ. The memory phase of the CD4 T-cell response to influenza virus infection maintains its diverse antigen specificity. Immunology. 2011 Jun;133(2):246–56. doi: 10.1111/j.1365-2567.2011.03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krug RM, Etkind PR. Cytoplasmic and nuclear virus-specific proteins in influenza virus-infected MDCK cells. Virology. 1973 Nov;56(1):334–48. doi: 10.1016/0042-6822(73)90310-3. [DOI] [PubMed] [Google Scholar]
- 32.Suarez DL. Overview of avian influenza DIVA test strategies. Biologicals. 2005 Dec;33(4):221–6. doi: 10.1016/j.biologicals.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 33.Tumpey TM, Alvarez R, Swayne DE, Suarez DL. Diagnostic approach for differentiating infected from vaccinated poultry on the basis of antibodies to NS1, the nonstructural protein of influenza A virus. J Clin Microbiol. 2005 Feb;43(2):676–83. doi: 10.1128/JCM.43.2.676-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaves FA, Lee AH, Nayak JL, Richards KA, Sant AJ. The utility and limitations of current Web-available algorithms to predict peptides recognized by CD4 T cells in response to pathogen infection. J Immunol. 2012 May 1;188(9):4235–48. doi: 10.4049/jimmunol.1103640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Combadiere B, Siberil S, Duffy D. Keeping the memory of influenza viruses. Pathol Biol (Paris) 2010 Apr;58(2):e79–86. doi: 10.1016/j.patbio.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Waffarn EE, Baumgarth N. Protective B cell responses to flu--no fluke! J Immunol. 2011 Apr 1;186(7):3823–9. doi: 10.4049/jimmunol.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMurry JA, Johansson BE, De Groot AS. A call to cellular & humoral arms: enlisting cognate T cell help to develop broad-spectrum vaccines against influenza A. Hum Vaccin. 2008 Mar-Apr;4(2):148–57. doi: 10.4161/hv.4.2.5169. [DOI] [PubMed] [Google Scholar]
- 38.Kaur K, Sullivan M, Wilson PC. Targeting B cell responses in universal influenza vaccine design. Trends Immunol. 2011 Nov;32(11):524–31. doi: 10.1016/j.it.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Epstein SL, Price GE. Cross-protective immunity to influenza A viruses. Expert Rev Vaccines. 2010 Nov;9(11):1325–41. doi: 10.1586/erv.10.123. [DOI] [PubMed] [Google Scholar]
- 40.Hillaire ML, Osterhaus AD, Rimmelzwaan GF. Induction of virus-specific cytotoxic T lymphocytes as a basis for the development of broadly protective influenza vaccines. J Biomed Biotechnol. 2011;2011:939860. doi: 10.1155/2011/939860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiers W, De Filette M, El Bakkouri K, Schepens B, Roose K, Schotsaert M, et al. M2e-based universal influenza A vaccine. Vaccine. 2009 Oct 23;27(45):6280–3. doi: 10.1016/j.vaccine.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Ebrahimi SM, Tebianian M. Influenza A viruses: why focusing on M2e-based universal vaccines. Virus Genes. 2011 Feb;42(1):1–8. doi: 10.1007/s11262-010-0547-7. [DOI] [PubMed] [Google Scholar]
- 43.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009 Mar;16(3):265–73. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010 Apr;1(1) doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol. 2010 Nov 1;185(9):4998–5002. doi: 10.4049/jimmunol.1001395. [DOI] [PubMed] [Google Scholar]
- 46.Alam S, Sant AJ. Infection with seasonal influenza virus elicits CD4 T cells specific for genetically conserved epitopes that can be rapidly mobilized for protective immunity to pandemic H1N1 influenza virus. J Virol. 2011 Dec;85(24):13310–21. doi: 10.1128/JVI.05728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthews JT. Egg-based production of influenza vaccine: 30 years of commerical experience. Bridge. 2006;36:17–24. [Google Scholar]
- 48.Srinivasan A, Foley J, McSorley SJ. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated Salmonella causes interclonal competition. J Immunol. 2004 Jun 1;172(11):6884–93. doi: 10.4049/jimmunol.172.11.6884. [DOI] [PubMed] [Google Scholar]
- 49.Stockinger B, Barthlott T, Kassiotis G. The concept of space and competition in immune regulation. Immunology. 2004 Mar;111(3):241–7. doi: 10.1111/j.1365-2567.2004.01831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson BE, Moran TM, Bona CA, Kilbourne ED. Immunologic response to influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. III. Reduced generation of neuraminidase-specific helper T cells in hemagglutinin-primed mice. Journal of Immunology. 1987;139(6):2015–9. [PubMed] [Google Scholar]
- 51.Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, et al. T cells compete for access to antigen-bearing antigen-presenting cells. J Exp Med. 2000;192(8):1105–13. doi: 10.1084/jem.192.8.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith AL, Wikstrom ME, Fazekas de St Groth B. Visualizing T cell competition for peptide/MHC complexes: a specific mechanism to minimize the effect of precursor frequency. Immunity. 2000;13(6):783–94. doi: 10.1016/s1074-7613(00)00076-5. [DOI] [PubMed] [Google Scholar]
- 53.Kastenmuller W, Gasteiger G, Gronau JH, Baier R, Ljapoci R, Busch DH, et al. Cross-competition of CD8+ T cells shapes the immunodominance hierarchy during boost vaccination. J Exp Med. 2007 Sep 3;204(9):2187–98. doi: 10.1084/jem.20070489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarkar S, Teichgraber V, Kalia V, Polley A, Masopust D, Harrington LE, et al. Strength of stimulus and clonal competition impact the rate of memory CD8 T cell differentiation. J Immunol. 2007 Nov 15;179(10):6704–14. doi: 10.4049/jimmunol.179.10.6704. [DOI] [PubMed] [Google Scholar]
- 55.Di Genova G, Savelyeva N, Suchacki A, Thirdborough SM, Stevenson FK. Bystander stimulation of activated CD4+ T cells of unrelated specificity following a booster vaccination with tetanus toxoid. Eur J Immunol. 2010 Apr;40(4):976–85. doi: 10.1002/eji.200940017. [DOI] [PubMed] [Google Scholar]
- 56.Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, et al. T cells compete for access to antigen-bearing antigen-presenting cells. J Exp Med. 2000 Oct 16;192(8):1105–13. doi: 10.1084/jem.192.8.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willis RA, Kappler JW, Marrack PC. CD8 T cell competition for dendritic cells in vivo is an early event in activation. Proc Natl Acad Sci U S A. 2006 Aug 8;103(32):12063–8. doi: 10.1073/pnas.0605130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paris RM, Kim JH, Robb ML, Michael NL. Prime-boost immunization with poxvirus or adenovirus vectors as a strategy to develop a protective vaccine for HIV-1. Expert Rev Vaccines. 2010 Sep;9(9):1055–69. doi: 10.1586/erv.10.106. [DOI] [PubMed] [Google Scholar]
- 59.Nicol MP, Grobler LA. MVA-85A, a novel candidate booster vaccine for the prevention of tuberculosis in children and adults. Curr Opin Mol Ther. 2010 Feb;12(1):124–34. [PubMed] [Google Scholar]
- 60.Hill AV, Reyes-Sandoval A, O’Hara G, Ewer K, Lawrie A, Goodman A, et al. Prime-boost vectored malaria vaccines: progress and prospects. Hum Vaccin. 2010 Jan;6(1):78–83. doi: 10.4161/hv.6.1.10116. [DOI] [PubMed] [Google Scholar]
- 61.Lu S. Heterologous prime-boost vaccination. Curr Opin Immunol. 2009 Jun;21(3):346–51. doi: 10.1016/j.coi.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Newman MJ. Heterologous prime-boost vaccination strategies for HIV-1: augmenting cellular immune responses. Curr Opin Investig Drugs. 2002 Mar;3(3):374–8. [PubMed] [Google Scholar]
- 63.Rech AJ, Vonderheide RH. Clinical use of anti-CD25 antibody daclizumab to enhance immune responses to tumor antigen vaccination by targeting regulatory T cells. Ann N Y Acad Sci. 2009 Sep;1174:99–106. doi: 10.1111/j.1749-6632.2009.04939.x. [DOI] [PubMed] [Google Scholar]
- 64.Ha SJ, West EE, Araki K, Smith KA, Ahmed R. Manipulating both the inhibitory and stimulatory immune system towards the success of therapeutic vaccination against chronic viral infections. Immunol Rev. 2008 Jun;223:317–33. doi: 10.1111/j.1600-065X.2008.00638.x. [DOI] [PubMed] [Google Scholar]