Abstract
Accurate duplication of DNA prior to cell division is essential to suppress mutagenesis and tumour development. The high fidelity of eukaryotic DNA replication is due to a combination of accurate incorporation of nucleotides into the nascent DNA strand by DNA polymerases, the recognition and removal of mispaired nucleotides (proofreading) by the exonuclease activity of DNA polymerases δ and ɛ, and post-replication surveillance and repair of newly synthesized DNA by the mismatch repair (MMR) apparatus. While the contribution of defective MMR to neoplasia is well recognized, evidence that faulty DNA polymerase activity is important in cancer development has been limited. We have recently shown that germline POLE and POLD1 exonuclease domain mutations (EDMs) predispose to colorectal cancer (CRC) and, in the latter case, to endometrial cancer (EC). Somatic POLE mutations also occur in 5–10% of sporadic CRCs and underlie a hypermutator, microsatellite-stable molecular phenotype. We hypothesized that sporadic ECs might also acquire somatic POLE and/or POLD1 mutations. Here, we have found that missense POLE EDMs with good evidence of pathogenic effects are present in 7% of a set of 173 endometrial cancers, although POLD1 EDMs are uncommon. The POLE mutations localized to highly conserved residues and were strongly predicted to affect proofreading. Consistent with this, POLE-mutant tumours were hypermutated, with a high frequency of base substitutions, and an especially large relative excess of G:C>T:A transversions. All POLE EDM tumours were microsatellite stable, suggesting that defects in either DNA proofreading or MMR provide alternative mechanisms to achieve genomic instability and tumourigenesis.
INTRODUCTION
Faithful replication of DNA is essential to maintain genomic stability and to prevent mutagenesis and tumour development. The remarkable accuracy of eukaryotic DNA replication—associated with an error frequency of ∼10−9–10−10 nucleotides replicated—results from a combination of the high fidelity of DNA polymerases and post-replication surveillance by the mismatch repair apparatus (1). The principal replicases at the eukaryotic replication fork are DNA polymerases ɛ and δ, of which the major components are encoded by POLE and POLD1, respectively, in humans. POLɛ synthesizes the leading strand, while POLδ synthesizes Okazaki fragments of the lagging strand (2–5). Both the enzymes display high accuracy of dNTP incorporation into the nascent DNA strand by their polymerase domains, with error rates of 10−4–10−5. In addition, both POLE and POLD1 contain a 3′–5′ exonuclease (proofreading) domain, which increases replication fidelity ∼100-fold by the recognition and excision of mispaired bases. Yeast and mice with mutations in the exonuclease domain of Polɛ or Polδ, or their homologues, show an increase in the rate of spontaneous mutations (6–10). A more complex range of phenotypes, including both hyper- and hypomutation, can result from genetic variants in the polymerase domain (11).
Humans with germline mutations in mismatch repair genes MSH2, MLH1, MSH6 and PMS2 are predisposed to a variety of cancer types as part of Lynch syndrome, which is typified by microsatellite instability (MSI) and a high risk of colorectal and endometrial carcinoma (CRC and EC) (12). Other individuals, with bi-allelic germline mutations in the base excision repair gene MUTYH, develop multiple colorectal adenomas and cancer (13,14). In both these cases, DNA polymerases, especially Polδ, are involved in the repair process. We have recently shown that germline mutations in the exonuclease domains of POLE and POLD1 predispose to CRC, and, in the case of the POLD1 variant, EC (15). In addition, by analysis of recently published datasets (16) we have shown that somatic POLE exonuclease domain mutations (EDMs) characterize a group of hypermutated, microsatellite stable (MSS) colorectal tumours (15–17). In the light of these findings, we hypothesized that sporadic ECs might also acquire somatic POLE and/or POLD1 mutations and have investigated the frequency and potential significance of somatic and germline mutations in the exonuclease domains of POLE and POLD1 in EC.
RESULTS
POLE exonuclease domain mutations are common in endometrial cancer
Screening of the exonuclease domains of POLE (residues 268–471) and POLD1 (residues 304–517) in 173 endometrial cancers (Table 1) identified 14 non-synonymous variants, 13 in POLE and 1 in POLD1. The change in POLD1 (p.Arg311Cys, rs201010746) was also present in a single individual from the 2012 release of 1000 genomes project (minor allele frequency 0.28%, N = 181) and one of the variants found in POLE (p.Arg446Gln, rs151273553) was also identified in the NHLBI GO Exome Sequencing Project at a frequency of 0.00054% (N = 13 006). All other variants were not present in public databases of unselected or cancer-free individuals (Table 2, Supplementary Material, Fig. S1). All of the mutations identified were missense changes. Analysis of paired constitutional DNA demonstrated that 12 changes were somatic, while the variant in POLD1 and one in POLE were present in the germline (Table 2). The overall frequency of somatic EDMs in EC was thus 6.9%. Mutations were found in tumours of endometrioid (11/154, 7.1%), serous (2/8, 25%) and mixed histologies (Table 1). There was a significant association between EDM and high tumour grade (Table 1). However, no difference in tumour stage or age of disease onset between EDM and non-EDM cancers was evident.
Table 1.
Patient and tumour characteristics
| All cases | Non-EDM cases | EDM cases | P | |
|---|---|---|---|---|
| Number | 173 | 159 | 14 | |
| Median age (range) | 59 (24–73) | 59 (24–73) | 57 (52–66) | NS |
| Histology | ||||
| Atypical hyperplasia | 2 | 2 | 0 | P = 0.17 (endo versus other) |
| MMT | 4 | 4 | 0 | |
| Endometrioid | 154 | 143 | 11 | |
| Serous | 8 | 6 | 2 | |
| Mixed endo/serous | 2 | 1 | 1 | |
| Mixed endo/clear cell | 1 | 1 | 0 | |
| Mucinous | 1 | 1 | 0 | |
| Not known | 1 | 1 | 0 | |
| Grade | ||||
| 1 | 64 | 61 | 3 | P = 0.001 (1 versus 2 versus 3) |
| 2 | 59 | 58 | 1 | |
| 3 | 45 | 35 | 10 | |
| Not known | 5 | 5 | 0 | |
| Stage | ||||
| 1 | 114 | 106 | 8 | P = 0.52 (1 versus others) |
| 2 | 18 | 17 | 1 | |
| 3 | 15 | 14 | 1 | |
| 4 | 8 | 6 | 2 | |
| Not known | 18 | 16 | 2 | |
| MSI | ||||
| MSI− | 144 | 130 | 13 | P = 0.22 (MSI− versus MSI+) |
| MSI+ | 24 | 24 | 0 | |
| Not known | 5 | 5 | 1 | |
Association statistics were calculated using Fisher's exact or χ2 tests for categorical variables and by t test for continuous variables.
Table 2.
Incidence, site and predicted consequence of tumour POLE and POLD1 exonuclease domain mutations
| Nucleotide change | Amino acid change | No. | Origin | Site | SIFT score | PolyPhen-2 score | PhastCons score | MutationTaster score | A-GVGD prediction | MSI |
|---|---|---|---|---|---|---|---|---|---|---|
| POLE | ||||||||||
| c.824A>T | p.Asp275Val | 1 | Somatic | Active site, Exo I motif | 0.000 | 1.000 | 1.00 | 4.15 | Class 65 | ND |
| c.857C>G | p.Pro286Arg | 6 | Somatic | Flanking Exo I motif | 0.000 | 1.000 | 1.00 | 2.81 | Class 65 | MSS |
| c.890C>T | p.Ser297Phe | 2 | Somatic | DNA binding interface | 0.000 | 1.000 | 1.00 | 4.23 | Class 65 | MSS |
| c.1231G>C | p.Val411Leu | 2 | Somatic | DNA binding interface | 0.000 | 1.000 | 1.00 | 0.87 | Class 25 | MSS |
| c.1337G>A | p.Arg446Gln | 1 | Germline | Solvent exposed | 0.200 | 0.994 | 1.00 | 1.17 | Class 0 | MSS |
| c.1366G>C | p.Ala456Pro | 1 | Somatic | Exo III motif | 0.000 | 1.000 | 1.00 | 0.74 | Class 0 | MSS |
| POLD1 | ||||||||||
| c.931C>T | p.Arg311Cys | 1 | Germline | Flanking Exo I motif | 0.000 | 0.999 | 1.00 | 4.91 | Class 65 | MSI |
Exonuclease domain missense mutations affect highly conserved residues and are predicted to affect function
All somatic POLE EDMs affected highly or universally conserved residues and were predicted to affect protein function (Fig. 1A, Table 2). The most common change (p.Pro286Arg) was found in six tumours. Sequence alignment revealed that proline at this site is absolutely conserved in all POLɛ orthologues (Fig. 1B), and also in the related exonucleases POLδ and the bacteriophage T4 polymerase (not shown). Residue 286 is immediately adjacent to the Exo I motif required for exonuclease function (residues 271–285) (18). Structural modelling demonstrated that this residue sits on the edge of the DNA binding pocket of POLE adjacent to the exonuclease active site, and close to the nascent single-stranded DNA (∼2.4 Å, a typical distance for polar interactions or hydrogen bonds) (Fig. 1C, highlighted in inset). Substitution of arginine at this residue will result in substantial perturbation of the DNA-binding pocket, and is thus predicted to have significant effects on protein function (Table 2). Replacement of proline by leucine at the equivalent residue in T4 polymerase (p.Pro123Leu) results in a strong mutator phenotype (19). The POLE p.Pro286Arg mutation has been reported in a CRC previously (17) and a p.Pro286His substitution has been found in another CRC (16). In addition, we have reported a probably pathogenic germline mutation at the homologous site, residue 327, in POLD1 (15).
Figure 1.
POLE exonuclease mutations in endometrial cancer affect conserved residues required for proofreading. (A) Localization of POLE mutations within the exonuclease domain (residues 268–471). Conserved exo motifs I–V are highlighted in pink, and the exo I motif active sites at codons 275 and 277 are shown in red. Mutations in endometrioid and serous tumours are indicated by open and closed circles, respectively. (B) Sequence alignment demonstrates that Asp275 and Pro286 are invariant in POLE orthologues. (C) Structural assessment of POLE EDMs using a composite of the conserved yeast Pol δ structure (PDB 3IAY) and the ssDNA component of the T4 polymerase complex (PDB 1NOY). Mutated residues are highlighted. With the exception of Arg446, which is solvent exposed and located away from the active site and polymerase domain, all changes are likely to distort the active site architecture. Pro286 flanks the exo I motif with its side chain 2.6 Å from the nascent DNA strand, while the side chain of active site residue Asp275 lies 3.7 Å from the DNA backbone (C, inset). Substitution of either residue will result in steric and electrostatic perturbation of exonuclease function.
The POLE p.Asp275Val change present in one tumour is highly likely to be functional. Asp275 and Glu277, which are invariate in Polɛ, Polδ and T4 polymerases (Fig. 1B and C), are POLɛ proofreading active site residues. Substitution of the equivalent residues in yeast Polɛ—Asp290 and Glu292—for alanine preserves polymerase activity while decreasing proofreading ability 100-fold, resulting in a 22-fold increase in the rate of spontaneous mutation (10). Mice with the corresponding amino acid changes also display increased mutagenesis and develop tumours (9).
A serine to phenylalanine substitution at codon 297 (p.Ser297Phe) was present in two cancers. Whilst strongly predicted to affect protein function (Table 2), this mutation has not previously been reported in the germline or soma. Amino acid 297 is absolutely conserved in POLɛ orthologues (Supplementary Material, Fig. S2). Whilst it is outside the Exo motifs, structural mapping (Fig. 1C) demonstrates that the equivalent amino acid in Saccharomyces cerevisiae Pol3p, Ala343, interacts with the active site aspartic acid at codon 321 (equivalent to human POLE D275). Thus, the p.Ser297Phe change is likely to affect exonuclease function by altering the active site conformation. Also detected in two tumours was a valine to leucine substitution at codon 411 (p.Val411Leu). Although it does not directly interact with DNA, this residue is conserved in POLɛ orthologues (Supplementary Material, Fig. S2) and somatic mutation of this site has previously been reported in CRC (16). Another somatic change in POLE was p.Ala456Pro, which lies within the Exo III motif. Alanine is conserved at this site from humans to drosophila, though the equivalent residue in S. cerevisiae and Schizosaccharomyces pombe is serine (Supplementary Material, Fig. S2). The p.Ala456Pro change is variably predicted to affect function (Table 2), but mutation of the equivalent flanking Exo III residues in yeast Pol2p (Arg511, Arg512 or Tyr516) results in defective proofreading and a 20- to 80-fold increase in spontaneous mutation (20,21). Structural analysis suggests that the change at codon 456 will directly alter DNA binding (Fig. 1C).
Assessment of germline POLE and POLD1 variants
In one EC patient, we found the germline POLE variant p.Arg446Gln (rs151273553). This residue is conserved from humans to S. cerevisiae, but not S. pombe (Supplementary Material, Fig. S2). A somatic p.Arg446Gln alteration has previously been found in a lung tumour (22). It is predicted to affect function (Table 2), but structural analysis demonstrated that this residue is solvent exposed and distant from both exonuclease and polymerase active sites (Fig. 1C). The carrier of this variant had no family or personal history of either endometrial or colorectal tumours and p.Arg446Gln must currently be classified as of uncertain significance. The germline change in POLD1 was a rare, but previously reported, variant (p.Arg311Cys, rs201010746). This invariant residue flanks the Exo I motif and is strongly predicted to affect protein function (Table 2). The equivalent residue has not been mutated in model organisms, and we have not found it to date in other patients with CRC or EC. It too must currently be classified as a variant of uncertain significance. The two ECs from the germline variant carriers were both grade 3, stage IV endometrioid tumours.
We examined whether the somatic p.Pro286Arg and p.Val411Arg variants might also be present in the germline of patients with EC. However, screening of 444 additional EC cases failed to detect either change.
‘Second hits’ at POLE in endometrial cancers
Mice with germline defects in Polɛ proofreading only display increased mutagenesis and tumour development in the homozygous state (9). Although we found no tumour with two somatic EDMs, it remains possible that ‘second hits’ at POLE occur through mutations elsewhere in the gene. Although screening the entire POLE gene for mutations was impractical in the FFPE samples available, we investigated whether loss of heterozygosity (LOH) at POLE had occurred within tumours by examination of flanking microsatellite markers. In the 10 cases for which markers were informative, no tumour LOH was detected. Thus, in contrast to the mouse, our findings are consistent with an effect of POLE EDMs in the heterozygous state, although our data do not exclude all types of second hit, such as promoter methylation or POLE mutations outside the EDM.
POLE EDM cancers are microsatellite-stable
We and others have previously shown that POLE EDM CRCs are hypermutant, but MSS (15,16), although a single case of a POLE EDM MSI-high CRC, has been reported (23). The overall frequency of MSI in our ECs was 18.5%, and as anticipated, MSI was only present in endometrioid tumours. However, all POLE EDM cancers were microsatellite stable. The single POLD1 EDM mutant cancer was MSI-high, contrasting with our previous findings of microsatellite stable CRCs in germline POLD1 EDM carriers, but consistent with the limited data available from the TCGA study of sporadic CRC (15,16).
POLE EDM endometrial tumours are hypermutated
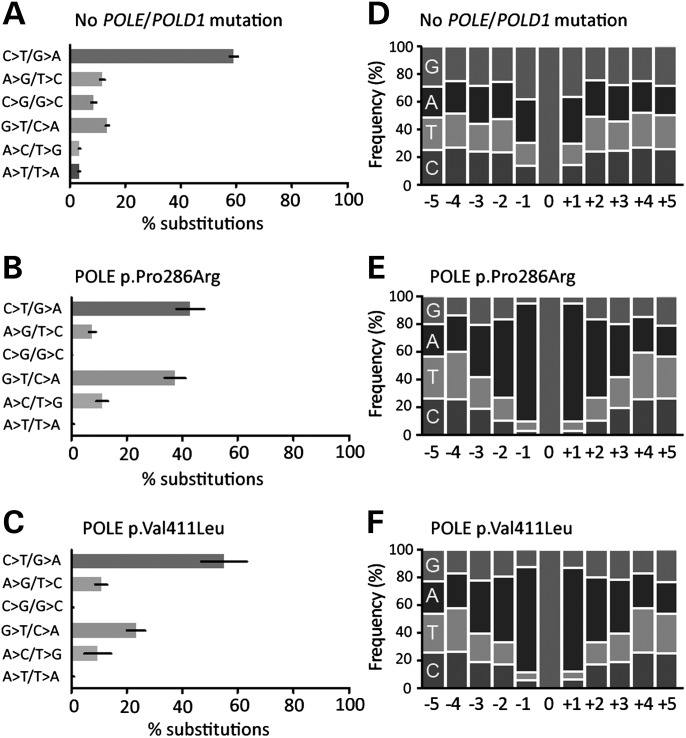
We next compared the overall somatic mutation status of a subset of POLE EDM ECs with a set of non-EDM, MSS ECs matched for tumour grade, using an extended Ion Torrent panel comprising 75 cancer genes. The EDM cancers demonstrated an ∼6 fold increase in mutations in the genes analysed (48.5 versus 8 mutations/tumour, P = 0.029, Mann–Whitney U test). Notably, and consistent with our previous observations in CRC, while the frequency of all types of mutations was increased in POLE EDM cancers, there was a relative excess of the proportion of transversions (47.3 versus 34.4% of total mutations) due largely to an increase in G:C>T:A substitutions (26.2 versus 15.5% of all mutations) (Fig. 2A–C versus D–F). As proofreading accuracy may depend partly on the DNA sequence flanking mispaired bases, we examined the sequence context of the G:C>T:A transversions in POLE EDM tumours. Although comparison with tumours lacking POLE EDM was precluded by a lack of mutations in this group, p.Pro286Arg cancers demonstrated an apparent enrichment of A:T base pairs flanking the mutated G:C bases, and more specifically for adenine 5′ and 3′ to the substituted guanine (Fig. 2G). This was most notable at the −1 and +1 positions relative to the transversion, with the excess diminishing further away from the substitution.
Figure 2.
Hypermutation of POLE EDM endometrial cancers is associated with a relative excess of G:C>T:A transversions associated with bias of flanking A:T base pairs. The frequency of each type of base change is shown as a proportion of the total number of substitutions detected by Ion Torrent sequencing. Tumours without DNA polymerase EDM (A–C) display a preponderance of C:G>T:A transitions, while POLE EDM tumours with Pro286Arg changes (D–F) are characterized by an relative increase in the proportion of G:C>T:A transversions. (G) Examination of the sequence context of G:C>T:A transversions in POLE p.Pro286Arg EDM cancers revealed an apparent over-representation of A:T base pairs 5′ and 3′ to the mutated base. The low frequency of G:C>T:A changes precluded analysis of flanking sequence in the POLE EDM non-mutants. For clarity in (G), all nucleotides are shown relative to the mutated guanine.
Examination of known EC drivers (see Methods) showed that mutations in PTEN, PIK3CA, and FBXW7 were present in both EDM and non-EDM cancers (Supplementary Material, Table S1), though our limited sample set precluded firm conclusions on mutation spectra.
Analysis of TCGA endometrial cancer exome sequencing data
Though unpublished, whole exome somatic mutation data from The Cancer Genome Atlas (TCGA, available at cancergenome.nih.gov) analysis of EC have recently been made available to the research community, and we therefore sought to validate our findings in their cohort of 248 patients. Missense mutations in the exonuclease domain of POLE were detected in 21 tumours (8.5%) and of POLD1 in 1 tumour (0.4%), highly concordant with our findings of 7.5 and 0.6% respectively. The commonest POLE change in the TCGA data was also p.Pro286Arg, detected in eight tumours (3.2%), with the p.Val411Leu substitution present in five cases (2.0%), and the p.Ser297Phe and p.Ala456Pro variants present in one tumour each.
TCGA EC POLE mutations not found in our own tumour panel included two cases of a p.Leu424Val change. We have recently demonstrated this mutation to predispose to CRC when present in the germline (15). In addition, the TCGA cancers included one case each of p.Ala428Thr, p.Met444Lys, p.Gln453Arg and p.Ala465Val substitutions. Although a germline serine to asparagine substitution at codon 478 in POLδ, the equivalent residue to POLɛ codon 428 predisposes to both CRC and EC (15), the POLE p.Ala428Thr change is likely to be tolerated, as several POLE orthologues contain threonine at this site (Supplementary Material, Fig S2). The other 3 POLE changes have not previously been reported, but mostly localize to conserved residues within, or close to the Exo motifs required for exonuclease activity and are likely to affect protein function (Supplementary Material, Fig. S2, Supplementary Material, Table S2). The only POLD1 EDM detected, p.Val392Met, has previously been reported in a kidney tumour (TCGA-B0-5080-01A-01D-1501-10) and is predicted to alter protein activity (Supplementary Material, Table S2). Eleven tumours lacking POLE/POLD1 exonuclease domain mutations had mutations at other sites in POLE and POLD1 (five and six cases respectively), though no particular pattern of non-EDM mutations was evident, none of the changes were recurrent and the predicted effects on protein function were variable (not shown).
With the exception of the tumour containing the p.Ala428Thr change, POLE EDM cancers from the TCGA cohort displayed striking levels of hypermutation, with a mean of 6811 exonic base substitutions per tumour (range: 227–14 695, 95% CI: 4691–8932) compared to 174 (range 22–2014, 95% CI 139–209) in cancers lacking POLE/POLD1 mutations. The single POLD1 EDM mutant cancer harboured a more modest 1637 changes, while tumours with non-exonuclease domain POLE/POLD1 mutations had means of 636 and 606 mutations/tumour, respectively. Further analysis of POLE EDM tumours revealed a variation in both the number and type of somatic mutations depending on the specific EDM variant (Fig 3, Supplementary Material, Table S3). Tumours with P286R changes were characterized by high levels of hypermutation (mean 7355 mutations/tumour) and a marked increase in the proportion of G:C>T:A transversions compared with tumours lacking POLE/POLD1 mutations (37.4 versus 13.5%) (Fig. 3A andB). Other POLE mutations associated with a preponderance of G:C>T:A substitutions were the p.Leu424Val, p.Met444Lys, p.Ser297Phe, p.Ala456Pro and, to a lesser extent, p.Val411Leu variants (Fig. 3C). Like the changes at codons 286, 297 and 456 previously discussed, both Met444 and Leu424 also localize within or close to the POLE Exo motifs within 10.2 Å of the nascent DNA strand, and the reported mutations are thus likely to affect proofreading activity. Interestingly, the tumours with POLE p.Gln453Arg and pAla465Val and POLD1 p.Val392Met mutations demonstrated a more modest level of mutation without an increase in the proportion of G:C>T:A substitutions, possibly related to the location of these residues away from the Exo motifs.
Figure 3.
Whole exome data from the TCGA EC study confirm disproportionate increase in G:C>T:A transversions in hypermutant POLE EDM cancers with variation by the EDM mutation type. The frequency of each type of base change is shown as a proportion of all substitutions for (A) cancers without POLE/POLD1 mutations (n = 215), (B) POLE EDM tumours with p.Pro286Arg (n = 8) and (C) p.Val411Leu (n = 5) changes. Examination of sequence context of G:C>T:A transversions by POLE EDM status (D–F) confirmed enrichment of A:T base pairs 5′ and 3′ to mutated G:C base pairs, greatest in the adjacent residue. For clarity in (D–F) all nucleotides are shown relative to the mutated guanine. Error bars in (A–C) indicate 95% confidence intervals.
We next sought to confirm the apparent nucleotide bias flanking G:C>T:A transversions in POLE EDM tumours in the TCGA set. Compared with cancers lacking POLE mutations, there was a notable over-representation of A:T base pairs flanking the mutated G:C bases in cancers with p.Pro286Arg, and to a lesser extent p.Val411Leu (Fig. 3D–F), suggesting that this sequence context renders recognition of mispaired C:Gs particularly defective in cancers with deficient proofreading.
Although 10 POLE EDM cancers harboured ‘second hits’ outside the POLE proofreading domain, only two contained mutations within the polymerase domain predicted to have functional consequences. One tumour with a p.Pro286Arg EDM change also contained a c.2445C>A substitution resulting in a p.Phe815Leu change, and the single p.Ala456Pro mutated tumour, also contained a c.3311T>C change causing a p.Thr1104Met alteration. The variant at codon 815 lies close to the polymerase active site and might contribute to the high number of mutations present in this cancer (11 726 substitutions), while codon 1104 lies outside the active sites and is of uncertain significance. We cannot, of course, determine whether these mutations are in cis or in trans with the EDM change.
As MSI status was not available for the TCGA ECs and could not easily be deduced from the type and number of insertion–deletion mutations, we analysed the mutation status of the mismatch repair genes and MLH1 promoter methylation status. We detected no validated pathogenic mutations in MLH1, MSH2, MSH6 or PMS2 in POLE EDM tumours. MLH1 methylation was detected in three POLE EDM cancers, though none of these harboured either Pro286Arg or Val411Leu changes (Supplementary Material, Table S4). Analysis of EC driver mutation status demonstrated that mutations in FBXW7 (33.3 versus 7.9%, P = 0.0021, Fisher's exact test) and biallelic PTEN mutations (38.1 versus 12.1%, P = 0.0043) were more common in POLE EDM tumours than in those lacking POLE/POLD1 changes (Supplementary Material, Table S4). The type of driver mutation also differed, with fewer PIK3CA mutations at hotspot codons 542, 545, 546 and 1047 in EDM tumours (7.1 versus 40.1%, P = 0.0174) and more at position 88 (28.6 versus 4.6%, P = 0.009). POLE EDM tumours also demonstrated an increased frequency of PTEN missense mutations at codon 130 (62.5 versus 25.6%, P = 0.0035).
DISCUSSION
We have demonstrated that somatic POLE exonuclease domain mutations occur in ∼7% of sporadic ECs and are associated with a MSI-negative hypermutator phenotype that is characterized by an excess of substitution mutations, in particular a relative excess of G:C>T:A substitutions. The majority of POLE EDMs affect invariant or highly conserved residues, and using a variety of methods, including characterization of the mutator phenotypes resultant from the equivalent changes in model organisms and structural assessment, we have provided strong evidence that most of the changes we report are pathogenic. In addition, analysis of POLE mutation frequency, type and correlates using recent TCGA data reveals excellent concordance with our results. In addition to being hypermutant, POLE EDM tumours are more frequently high grade, and display a characteristic pattern of driver mutations that differ from tumours lacking POLE/POLD1 changes. In contrast to the relatively high frequency of POLE mutations, somatic POLD1 mutation appears to be uncommon in EC. The reasons for this disparity are, at present, unclear.
Though preliminary examination of the frequency and type of substitution mutations associated with specific POLE EDMs reveals noteworthy variation. In general, the severity of hypermutation and relative excess of G:C>T:A transversions appears to correlate with the degree of perturbation of the residues within, and close to the Exo motifs required for exonuclease activity. For example, p.Val411Leu is predicted to have a mildly disruptive effect and tumours with this mutation have a relatively subtle excess of G:C>T:A changes compared with p.Pro286Arg mutants which have stronger predicted effects and a greater mutational bias. A similar difference between mutations at these sites is also seen in CRC (unpublished data). However, these must be regarded as preliminary observations and further mechanistic insights into these findings await detailed structural and functional studies. Similarly, the reason for the over-representation of A:T base pairs immediately 5′ and 3′ to mutated G:Cs is, at present, unclear.
Concordant with the majority of previous data in CRC (15,16), all POLE EDM tumours in our cohort are microsatellite stable, suggesting that defective proofreading provides an alternative mechanism to achieve genomic instability and tumourigenesis. It is noteworthy that POLE exonuclease domain mutations are found in both endometrioid and serous tumours, in contrast to MSI, which is typically restricted to endometrioid cancers (24). Although, three POLE EDM cancers in the TCGA had MLH1 promoter methylation, the correlation between this and MSI in EC is imperfect (25,26) and further evaluation of the apparent exclusivity between POLE EDM and MSI status in our data awaits confirmation. Interestingly, the single POLD1 EDM tumour in our panel was MSI-positive, as has been demonstrated previously for sporadic CRCs with POLD1 exonuclease mutations (15). Whether this is related to the apparent attenuation of hypermutation in cancers with POLD1 EDMs relative to those with POLE EDMs is uncertain, though the combination of highly deficient proofreading and defective mismatch repair is lethal in both yeast and mice (9,21,27).
In summary, somatic mutations of the POLE proofreading exonuclease domain are relatively common in EC and appear to define a hypermutated, MSI-negative group of tumours. CRCs with hypermutation due to mismatch repair deficiency comprise a distinct tumour subtype characterized by high tumour grade and favourable prognosis (28,29), though the corresponding data in EC are limited. It will be of interest to determine whether POLE EDMs in EC and CRC are of similar prognostic and therapeutic significance, and to extend the study of POLE and POLD1 to other tumour types.
MATERIALS AND METHODS
Cases and samples
Peripheral blood and formalin-fixed paraffin embedded archival tumour samples were obtained from 173 patients enrolled in the UK National Study of Endometrial Cancer Genetics (NSECG) between 2008 and 2012, without selection for age, histology or any other clinico-pathological feature. The recruitment criteria to the study were any woman of European ethnic origins who presented with primary endometrial carcinoma at ≤70 years of age. Lynch syndrome was excluded wherever possible on the basis of testing already performed in NHS diagnostic laboratories. Full clinico-pathological information was obtained from each case. Most cases (n = 154) were endometrioid (Type I) tumors, while 10 patients had serous (Type II) or mixed endometrioid/serous histology and the remainder less common subtypes (Table 1). EC tumour blocks were obtained. Haematoxylin and eosin-stained slides were reviewed and DNA extracted from consecutive sections containing >75% cancer cells by standard methods. All work was performed according to local ethical guidelines (approval 04/Q0803/148).
Germline genotyping and mutation screening
Bi-directional Sanger sequencing was used to screen the regions encoding the exonuclease domains of POLE (codons 268–471) and POLD1 (codons 304–517) for somatic mutations (details of PCR primers and reaction conditions available from the authors). Variants were identified both automatically and manually in the Mutation Surveyor program (Soft Genetics). All somatic mutations were confirmed in an independent PCR reaction, followed by sequencing and the origin of each mutation detected was tested using constitutional DNA. In-house Kaspar competitive allele specific PCR (LGC Genomics) assays were used to screen for POLE variants at codons 286 and 411 in germline DNA extracted from blood samples from 444 EC patients (primer sequences available on request). Two positive controls were included on each plate and outliers were confirmed by sequencing.
Microsatellite instability (MSI) analysis
MSI was determined using a panel of three markers (BAT25, BAT26 and D2S123). Tumours were classified as MSI-high if they had two or more unstable markers and MSI-negative/MSS if all markers were stable. The status of samples in which the initial panel was inconclusive was clarified using two further markers (D5S346, BAT40).
Loss of heterozygosity (LOH) analysis
LOH was assessed by genotyping microsatellites mapping close to POLE (D12S1723, D12S1628, D12S357, D12S1638) or POLD1 (D19S867, D19S904, D19S907). Microsatellites were analysed in the Gene Marker (Soft Genetics) program (PCR conditions available from the authors). LOH was scored if the intensity of any allele was reduced by ≥50% relative to the other allele after taking account of the relative allelic intensities in paired constitutional DNA. In the case of discordance between microsatellites, precedence was given to the one closest to POLE or POLD1.
Somatic mutation spectra and driver mutation screening
Four POLE EDM ECs and four ECs lacking EDMs were screened for exonic variants in a custom panel of 75 cancer-related genes using the Ion Torrent Ampliseq method (details available on request). We searched specifically for mutations in known EC driver genes (defined as recurrent missense changes recorded in COSMIC or premature truncations in the case of tumour suppressors) and examined the overall mutation number and spectrum. Mutations were called using the Torrent variant caller plugin v3.4.49252 (Life Technologies). We applied a cut-off of 10% supporting reads for mutation calls and excluded variants flanked by homopolymers of three or more bases following optimization experiments. In addition, bam files were inspected manually in order to remove likely artefacts and to detect any mutations in known EC driver genes–especially insertions and deletions–that were not called by the Ion Torrent software.
Structural analysis
Human mutations in both POLE and POLD1 were visualized in PyMOL (http://pymol.org) on the catalytic subunit of the yeast DNA polymerase δ [PDB ID: 3IAY (30)] with the ssDNA component of the T4 polymerase complex [PDBID: 1NOY (31)] modelled into the exonuclease active site.
Somatic variant and methylation analysis of The Cancer Genome Atlas (TCGA) Uterine Corpus Endometrioid Carcinoma [UCEC] dataset
Somatic variant calls and beta scores generated from IlluminaHumanMethylation27/450 arrays for 248 ECs were downloaded from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/tcgaCancerDetails.jsp?diseaseType=UCEC&diseaseName=Uterine%20Corpus%20Endometrioid%20Carcinoma). refGene annotations were generated using ANNOVAR (32). DNA sequence flanking mutations was downloaded using Galaxy (33). Two probes (cg00893636, cg13846866) mapping to the MLH1 promoter CpG island were identified. Tumours were called as methylated if the beta score was >0.2 based on a step-change in beta value at this point in the dataset as a whole. The two probes were highly correlated (R2 0.98).
Data analysis and statistics
Data were analysed in Excel and graphs plotted using Prism5 (Graphpad software). Comparison of categorical variables was made by Fisher's exact test and continuous variables using the t test, or Mann–Whitney U-test. All P values were 2-sided, and statistical significance was accepted at P < 0.05.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by funding from Cancer Research UK (C6199/A10417), the European Union Seventh Framework Programme (FP7/207- 2013) grant 258236 collaborative project SYSCOL, and the Oxford NIHR Comprehensive Biomedical Research Centre. We also acknowledge core funding to the Wellcome Trust Centre for Human Genetics from the Wellcome Trust (090532/Z/09/Z). DNC was partly funded a Development Fund Award from The Oxford Cancer Research Centre. The results published here are in part based on data generated by The Cancer Genome Atlas project funded by the NCI and NIH (cancergenome.nih.gov). Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the many individuals who participated in the NSECG Project and the numerous institutions and their staff who have supported recruitment.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. doi:10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 2.Pursell Z.F., Isoz I., Lundström E-B., Johansson E., Kunkel T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. doi:10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M.J., Kunkel T.A. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. doi:10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larrea A.A., Lujan S.A., Nick McElhinny S.A., Mieczkowski P.A., Resnick M.A., Gordenin D.A., Kunkel T.A. Genome-wide model for the normal eukaryotic DNA replication fork. Proc. Natl Acad. Sci. USA. 2010;107:17674–17679. doi: 10.1073/pnas.1010178107. doi:10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyabe I., Kunkel T.A., Carr A.M. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. doi:10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon M., Giot L., Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldsby R.E., Lawrence N.A., Hays L.E., Olmsted E.A., Chen X., Singh M., Preston B.D. Defective DNA polymerase delta proofreading causes cancer susceptibility in mice. Nat. Med. 2001;7:638–639. doi: 10.1038/88963. doi:10.1038/88963. [DOI] [PubMed] [Google Scholar]
- 8.Goldsby R.E. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc. Natl Acad. Sci. USA. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. doi:10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albertson T.M., Ogawa M., Bugni J.M., Hays L.E., Chen Y., Wang Y., Treuting P.M., Heddle J.A., Goldsby R.E., Preston B.D. DNA polymerase and proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl Acad. Sci. USA. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. doi:10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison A., Bell J.B., Kunkel T.A., Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′-5′ exonuclease activity. Proc. Natl Acad. Sci. USA. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. doi:10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesan R.N., Hsu J.J., Lawrence N.A., Preston B.D., Loeb L.A. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase delta. J. Biol. Chem. 2006;281:4486–4494. doi: 10.1074/jbc.M510245200. doi:10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- 12.Lynch H.T., de la Chapelle A. Hereditary colorectal cancer. N. Engl. J. Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. doi:10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 13.Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R., et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. doi: 10.1038/ng828. doi:10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 14.Sieber O.M., Lipton L., Crabtree M., Heinimann K., Fidalgo P., Phillips R.K., Bisgaard M.L., Orntoft T.F., Aaltonen L.A., Hodgson S.V., et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. doi:10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 15.Palles C., Cazier J-B., Howarth K.M., Domingo E., Jones A.M., Broderick P., Kemp Z., Spain S.L., Almeida E.G., Salguero I., et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45:136–144. doi: 10.1038/ng.2503. doi:10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. doi:10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seshagiri S., Stawiski E.W., Durinck S., Modrusan Z., Storm E.E., Conboy C.B., Chadhuri S., Guan Y., Janakiraman V., Jaiswal B.S., et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. doi:10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shevelev I.V., Hübscher U. The 3′–5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. doi:10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 19.Reha-Krantz L.J. Amino acid changes coded by bacteriophage T4 DNA polymerase mutator mutants. Relating structure to function. J. Mol. Biol. 1988;202:711–724. doi: 10.1016/0022-2836(88)90552-9. doi:10.1016/0022-2836(88)90552-9. [DOI] [PubMed] [Google Scholar]
- 20.Jin Y.H., Obert R., Burgers P.M.J., Kunkel T.A., Resnick M.A., Gordenin D.A. The 3′->5′ exonuclease of DNA polymerase can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl Acad. Sci. USA. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. doi:10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herr A.J., Ogawa M., Lawrence N.A., Williams L.N., Eggington J.M., Singh M., Smith R.A., Preston B.D. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011;7:e1002282. doi: 10.1371/journal.pgen.1002282. doi:10.1371/journal.pgen.1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. doi:10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida R., Miyashita K., Inoue M., Shimamoto A., Yan Z., Egashira A., Oki E., Kakeji Y., Oda S., Maehara Y. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur. J. Hum. Genet. 2010;19:320–325. doi: 10.1038/ejhg.2010.216. doi:10.1038/ejhg.2010.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeramian A., Moreno-Bueno G., Dolcet X., Catasus L., Abal M., Colas E., Reventos J., Palacios J., Prat J., Matias-Guiu X. Endometrial carcinoma: molecular alterations involved in tumor development and progression. Oncogene. 2013;32:403–413. doi: 10.1038/onc.2012.76. doi:10.1038/onc.2012.76. [DOI] [PubMed] [Google Scholar]
- 25.Kanaya T., Kyo S., Maida Y., Yatabe N., Tanaka M., Nakamura M., Inoue M. Frequent hypermethylation of MLH1 promoter in normal endometrium of patients with endometrial cancers. Oncogene. 2003;22:2352–2360. doi: 10.1038/sj.onc.1206365. doi:10.1038/sj.onc.1206365. [DOI] [PubMed] [Google Scholar]
- 26.Simpkins S.B., Bocker T., Swisher E.M., Mutch D.G., Gersell D.J., Kovatich A.J., Palazzo J.P., Fishel R., Goodfellow P.J. MLH1 promoter methylation and gene silencing is the primary cause of microsatellite instability in sporadic endometrial cancers. Hum. Mol. Genet. 1999;8:661–666. doi: 10.1093/hmg/8.4.661. doi:10.1093/hmg/8.4.661. [DOI] [PubMed] [Google Scholar]
- 27.Tran H.T., Gordenin D.A., Resnick M.A. The 3′>5′ exonucleases of DNA polymerases delta and epsilon and the 5′>3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth A.D., Tejpar S., Delorenzi M., Yan P., Fiocca R., Klingbiel D., Dietrich D., Biesmans B., Bodoky G., Barone C., et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J. Clin. Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. doi:10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 29.Bertagnolli M.M., Redston M., Compton C.C., Niedzwiecki D., Mayer R.J., Goldberg R.M., Colacchio T.A., Saltz L.B., Warren R.S. Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III colon cancer—a study of CALGB 9581 and 89803. J. Clin. Oncol. 2011;29:3153–3162. doi: 10.1200/JCO.2010.33.0092. doi:10.1200/JCO.2010.33.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swan M.K., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. doi:10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Yu P., Lin T.C., Konigsberg W.H., Steitz T.A. Crystal structures of an NH 2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochem. 1996;35:8110–8119. doi: 10.1021/bi960178r. doi:10.1021/bi960178r. [DOI] [PubMed] [Google Scholar]
- 32.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. doi:10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goecks J., Nekrutenko A., Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. doi:10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.