Abstract
Abdominal aortic aneurysm (AAA) is a common human disease with a high estimated heritability (0.7); however, only a small number of associated genetic loci have been reported to date. In contrast, over 100 loci have now been reproducibly associated with either blood lipid profile and/or coronary artery disease (CAD) (both risk factors for AAA) in large-scale meta-analyses. This study employed a staged design to investigate whether the loci for these two phenotypes are also associated with AAA. Validated CAD and dyslipidaemia loci underwent screening using the Otago AAA genome-wide association data set. Putative associations underwent staged secondary validation in 10 additional cohorts. A novel association between the SORT1 (1p13.3) locus and AAA was identified. The rs599839 G allele, which has been previously associated with both dyslipidaemia and CAD, reached genome-wide significance in 11 combined independent cohorts (meta-analysis with 7048 AAA cases and 75 976 controls: G allele OR 0.81, 95% CI 0.76–0.85, P = 7.2 × 10−14). Modelling for confounding interactions of concurrent dyslipidaemia, heart disease and other risk factors suggested that this marker is an independent predictor of AAA susceptibility. In conclusion, a genetic marker associated with cardiovascular risk factors, and in particular concurrent vascular disease, appeared to independently contribute to susceptibility for AAA. Given the potential genetic overlap between risk factor and disease phenotypes, the use of well-characterized case–control cohorts allowing for modelling of cardiovascular disease risk confounders will be an important component in the future discovery of genetic markers for conditions such as AAA.
INTRODUCTION
Abdominal aortic aneurysm [AAA (MIM 100070)] is a common condition, particularly in men over the age of 65 years, with a reported prevalence of between 2 and 8% (1). The demographic risk factor profile for AAA is well described and includes smoking, family history of AAA, older age, male sex and concurrent atherosclerotic diseases, particularly coronary artery disease (CAD) (2–4). Whereas historically considered to be a consequence of atherosclerosis, AAA is now considered to be a distinct clinical entity, although one that shares many risk factors with occlusive atherosclerotic vascular disease (2,5). Three genome-wide association studies (GWAS) have been reported for AAA (6–8). To date, only three genetic associations, the ANRIL locus (9p21) (9), DAB2IP (9q33) (8) and LRP1 (12q13) (7), have been reported which reach genome-wide significance (P < 10−8).
The possible association between dyslipidaemia and AAA (4) is somewhat controversial, with both positive (4) and negative (2) associations having been reported. Nevertheless, the inverse association between high-density lipoprotein (HDL) and AAA is widely accepted (10). In addition, examination of human AAA biopsies demonstrates the presence of extensive intimal atherosclerosis (11).
Recently, GWAS have reported numerous loci that are convincingly associated with blood lipid levels (12) and CAD (13). Several loci, such as the PCSK9, SORT1, APOA1 and LDLR, show robust associations with both phenotypes.
This study applied a biological pathway-based screen of a whole-genome AAA case–control data set (14) to determine whether variations in genes previously shown to be associated with either CAD or blood lipids are also associated with AAA. Observations were replicated in multiple international cohorts (final analyses with 7048 AAA cases and 75 976 controls) and independence of the genetic associations determined using logistic regression.
RESULTS
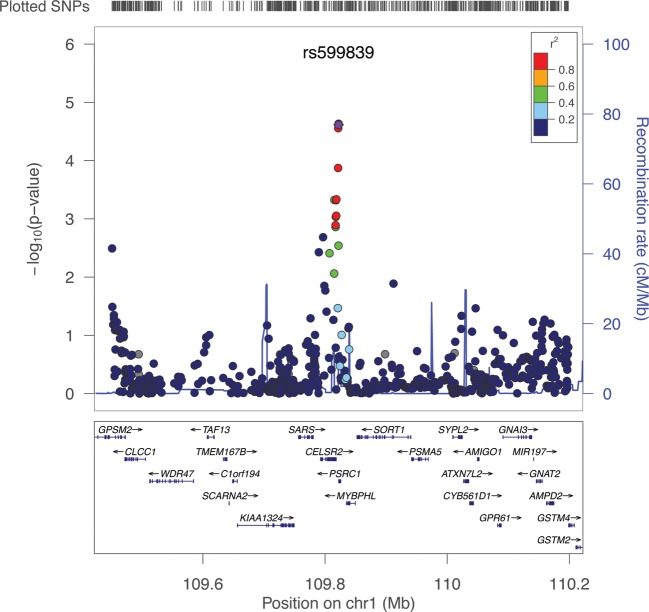
A total of 44 blood lipid- and CAD-associated loci were screened in the New Zealand (NZ) discovery cohort, including four, PCSK9 (1p32.3), SORT1-CELSR2-PSRC1 (1p13.3), APOA1 (11q23.3) and LDLR (19p13.2), which were strongly associated with both phenotypes (Supplementary Material, Table S2). Of these, 15 lead single-nucleotide polymorphisms (SNPs) were associated with AAA with observed P-values < 0.05. Three SNP associations, rs599839 on 1p13.3 (SORT1-CELSR2-PSRC1; Fig. 1), rs4977574 on 9p21 (CDKN2BAS1) and rs4775049 on 15q21–23 (LIPC), had P < 0.1 in the same direction of association as the second NZ cohort (Supplementary Material, Table S3) and were followed up in the WTCCC AAA cohort. Two SNPs remained significant following WTCCC AAA cohort validation. These were 1p13.3 SORT1-CELSR2-PSRC1 rs599839, which represents a novel AAA association, and rs4977574 9p21 CDKN2BAS1, which is in a previously reported AAA locus (9).
Figure 1.
Manhattan plot for the 1p13.3 region in the NZ discovery cohort. The lead SNP in this region was rs599839. Only subjects with call rates >0.98 (MIND >0.98) and common SNPs (MAF >0.05) were included. The plot was generated using LocusZoom.
Replication in additional independent case–control cohorts demonstrated a consistent rs599839 G allele AAA protective association within 6 of the 11 cohorts examined (Table 1). In all of the 11 cohorts, the MAF was lower in cases than in controls, and when all genotyped cohorts were analysed together (7048 AAA cases and 75 976 controls, pHet = 0.68), the meta-P-value was 7.2 × 10−14.
Table 1.
Meta-analysis of the 1p13.3, rs599839 association with AAA
| rs599839 A>G, 1p13.3 | Controls, n | Cases, n | MAF controls | MAF cases | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|
| NZ discovery | 612 | 608 | 0.249 | 0.180 | 0.66 (0.54–0.81) | 3.3 × 10−5 |
| NZ validation | 1766 | 713 | 0.240 | 0.191 | 0.75 (0.64–0.88) | 2.4 × 10−4 |
| WTCCC | 5605 | 1286 | 0.232 | 0.202 | 0.84 (0.75–0.93) | 8.9 × 10−4 |
| Iceland | 61 639 | 1163 | 0.209 | 0.176 | 0.81 (0.72–0.91) | 0.006 |
| The Netherlands | 2792 | 840 | 0.229 | 0.228 | 0.99 (0.45–2.17) | 0.98 |
| Western Australia | 373 | 377 | 0.211 | 0.185 | 0.85 (0.66–1.10) | 0.21 |
| Leeds, UK | 456 | 684 | 0.238 | 0.197 | 0.79 (0.64–0.96) | 0.02 |
| Pennsylvania, USA | 1313 | 773 | 0.222 | 0.193 | 0.84 (0.72–0.99) | 0.03 |
| Mayo eMERGE cohort, USA | 1000 | 230 | 0.226 | 0.209 | 0.89 (0.69, 1.14) | 0.36 |
| Belgium | 267 | 176 | 0.190 | 0.181 | 0.94 (0.65–1.35) | 0.74 |
| Canada | 153 | 198 | 0.260 | 0.214 | 0.77 (0.51–1.16) | 0.21 |
| Combined | 75 976 | 7048 | 0.81 (0.76–0.85) | 7.2 × 10−14 |
The rs599839 G allele was associated with reduced AAA risk in 6 of the 11 cohorts (P-value for heterogeneity 0.68)
A putative functional SNP, rs12740374, associated with SORT1 expression (15), was examined in both NZ cohorts (1325 AAA and 2374 controls) and was in strong linkage disequilibrium (r2 = 0.91, D′ = 0.99, P = 3.3 × 10−16) with rs599839. Whereas rs12740374 was significantly associated with AAA (minor T allele OR 0.80; 95% CI 0.70–0.90; P = 4.1 × 10−4), the rs599839 AAA association was the strongest in the region.
The 1p13.3 locus has been shown to have a strong effect on circulating low-density lipoprotein (LDL)-cholesterol (12,13,15,16), as well as CAD and myocardial infarction (13,16). We therefore tested the possible confounding effect of rs599839 SNP effect on LDL cholesterol, after adjusting for statin use. Carriers of the AAA protective rs599839 G allele had significantly lower LDL in both the NZ (P = 0.015) and Leeds (P = 0.012) cohorts. These G allele effects were of consistent magnitude and direction (−4.48 mg/dl) with that previously reported by Teslovich et al. (12). Nevertheless, when LDL-cholesterol was added to the multivariate model, along with other known AAA demographic risk factors including a history of CAD and HDL-cholesterol, the rs599839 G allele AAA association remained significant (Table 2).
Table 2.
Results from multivariate analyses for AAA rs599839 association
| Risk factor | NZ, 1262 AAA/1202 controls |
Leeds, UK, 532 AAA/432 controls |
||
|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |
| G allele | 0.75 (0.64–0.88) | 2 × 10−4 | 0.78 (0.63–0.98) | 0.031 |
| with age | 0.69 (0.57–0.81) | 2 × 10−5 | 0.80 (0.64–1.01) | 0.058 |
| with sex | 0.75 (0.64–0.88) | 2 × 10−4 | 0.79 (0.63–0.98) | 0.033 |
| with history of CAD | 0.76 (0.64–0.89) | 5 × 10−4 | 0.78 (0.62–0.97) | 0.029 |
| with smoking | 0.70 (0.59–0.84) | 4 × 10−5 | 0.78 (0.62–0.97) | 0.031 |
| with DM | 0.74 (0.63–0.87) | 2 × 10−4 | 0.78 (0.63–0.98) | 0.031 |
| with dyslipidaemia | 0.75 (0.64–0.88) | 2 × 10−4 | 0.77 (0.61–0.96) | 0.019 |
| with HDL-cholesterol | 0.74 (0.62–0.87) | 2 × 10−4 | 0.78 (0.63–0.98) | 0.029 |
| with LDL-cholesterola | 0.76 (0.65–0.90) | 5 × 10−4 | 0.77 (0.62–0.96) | 0.022 |
| Combined modelb | 0.68 (0.55–0.83) | 1.2 × 10−4 | 0.79 (0.62–1.01) | 0.057 |
| Meta-analysis | 0.72 (0.62–0.85), P = 5.6 × 10−5 | |||
The results were adjusted for cardiovascular risk factors in the combined NZ discovery and replication studies and the Leeds cohort, UK. CAD was defined as a history of previous myocardial infarction or angiographically proved coronary stenosis. Smoking history was based on pack-years (1 pack/day for 1 year equals 1 pack-year). Dyslipidaemia was defined as either self-reported history or treatment (specifically) for hyperlipidaemia. Diabetes mellitus (DM) includes both type 1 and type 2. The adjusted odds ratios were generated in separate analyses using logistic regression comparing genotypic effect with each risk factor.
aLDL-cholesterol corrected for statin therapy.
bDyslipidaemia excluded from combined model to avoid multicollinearity with LDL and HDL measures. Inclusion of dyslipidaemia, instead of HDL and LDL, did not significantly alter the combined model result.
Despite the apparent independence of the rs599839 AAA association, an additional analysis was performed to determine whether the CAD or lipid risk alleles simply represented a proxy for AAA association. CAD (13) or lipid (12) risk allele lead trait effects were correlated with their respective AAA odds ratios using Pearson's correlation coefficients. Suggestive AAA SNP effects (OR ≤ 0.9 or >1.1) significantly correlated with CAD SNP effects (n = 13, r = 0.78, P < 0.001, 12 of 13 SNPs with consistent directions of effect). Lipid SNP effects showed a weaker suggestive correlation (n = 10, r = 0.62, P = 0.06, 8 of 10 SNPs with consistent directions of effect). Because of this suggestive association, an expanded lipid SNP set was formed by including all HDL or LDL SNPs with P < 10−4 from the meta-analysis data sets published online by Teslovich et al. (12). When aligned to the NZ AAA discovery data set, this corresponded to 4960 HDL (specific) SNPs and 4229 LDL (specific) SNPs. The lead AAA-associated SNP in each lipid locus, along with any other (LD-filtered) AAA associated (P < 0.05) SNPs, was selected. This resulted in 120 HDL and 96 LDL SNPs. An additional 18 SNPs were associated with both HDL and LDL and were not included in lipid subgroup analysis. No significant correlations between AAA odds ratios and the lipid effect sizes were observed in any of these (expanded) groups (HDL: r = 0.11, P = 0.24, 50 of 120 consistent direction of effects; LDL: r = 0.04, P = 0.68, 53 of 96 consistent direction of effects). These correlations did not change significantly when only SNPs with suggestive (OR ≤ 0.9 or >1.1) AAA associations were included.
Aortic tissue gene and protein expression
The 1p13.3 rs599839 SNP is in the vicinity of the CELSR2, PSRC1 and SORT1 genes. Aortic RNA expression for SORT1, CELSR2 and PSRC1 was assessed using RT–PCR. Only SORT1 showed robust expression in AAA tissues (Supplementary Material, Fig. S1).
SORT1 encodes the sortilin-1 protein, the levels of which were assessed by western blot analysis to confirm the presence of protein within control and AAA abdominal aortic tissue. Sortilin-1 protein (both cytoplasmic and extracellular domains) was detected in all aortic samples and was found to have significantly higher protein levels in AAA compared with controls (Supplementary Material, Fig. S2). AAA sortilin-1 tissue protein levels did not correlate with aneurysm size (r = 0.19, P = 0.11, Spearman's correlation), nor was there any association between either rs599839 or rs12740374 genotype and tissue sortilin-1 levels (not shown).
Immunohistochemical analysis confirmed cytoplasmic staining for sortilin-1 within aortic tissues. In non-aneurysmal controls, staining was generally weak and when present appeared restricted to cells in the thickened (Supplementary Material, Fig. S3) or atherosclerotic intima. In AAA tissue, staining was observed throughout the atherosclerotic intima as well as the residual media and inflamed adventitia. In particular, T-cell-rich lymphoid aggregates in the AAA adventitia appeared strongly positive for sortilin-1 (Supplementary Material, Fig. S4).
DISCUSSION
Evidence suggests that functionally related genes can collectively influence disease susceptibility (17). Biological pathway-based analysis of whole-genome data sets has therefore been suggested as an approach to facilitate more powerful analysis of genome-wide data sets (14). We applied this strategy on two levels, first by testing lead SNPs that have been convincingly associated with two AAA risk factors, namely blood lipid profile and concurrent CAD. Second, by examining a larger number of SNPs in the vicinity of these loci to identify any other (LD-filtered) SNPs that also appear to be associated with AAA. Using this approach, we were able to identify a novel association between the SNP rs599839 and AAA.
The genetic variant rs599839 (or SNPs in strong linkage disequilibrium) has been shown to have highly significant associations with both circulating lipoprotein profile (12,16) and CAD risk (13). This SNP is in the region 1p13.3, and includes the genes CELSR2, PSRC1 and SORT1. Evidence suggests that the lipoprotein association is primarily driven by altered liver SORT1 expression (15,18). In this study, rs599839 was consistently associated with AAA at a genome-wide level of significance. In addition, we also observed strong SORT1 (but not CELSR2 or PSRC1) gene and sortilin-1 protein expression within abdominal aortic tissues. This is consistent with the high SORT1 gene expression levels previously reported within ascending thoracic aortic tissue by Folkersen et al. (18). Although we observed significantly higher SORT1 protein levels in AAA tissue compared with controls, there did not appear to be an association between aortic wall sortilin-1 protein and rs599839 genotype. Again, this observation appears consistent with Folkersen et al.'s observations of the rs599839 SNP having a genotype effect on SORT1 gene expression in human liver, but not in mammary artery or ascending aortic tissues (18). Although this may tend to suggest that the SORT1 genetic association with vascular diseases (such as CAD and AAA) acts primarily by altering liver-related functions, such as lipid metabolism, it should be noted that the rs599839 AAA effect persisted despite modelling for lipoprotein profile and other cardiovascular risk factors. It should also be noted that we examined relatively small numbers of end-stage AAA biopsies available from patients undergoing open surgery. Thus, it is possible that rs599839 could influence aortic SORT1 expression prior to or during the early stages of AAA formation and we would have failed to detect this association during our analyses.
Gene ontology analysis (AmiGO version 1.8, release date September 2012) indicates that SORT1 may be involved in a wide range of processes, including not only lipoprotein metabolism but also insulin-responsive glucose transport, G-protein-coupled muscle differentiation, nerve growth factor receptor signalling and apoptosis. The role of SORT1 in normal and pathobiological processes within the vascular wall remains unclear; however, this study suggests that SORT1 may not only have systemic effects, for example by altering circulating lipoprotein profiles, but also have the potential to have actions directly within the blood vessel wall. The immunohistochemical localization of sortilin-1 protein within adventitial inflammatory cell aggregates may suggest a role in this key pathological aspect of aneurysm biology. Indeed, the rs599839 G allele has been associated with increased SORT1 mRNA expression within lymphocytes, indicating that the genotype effects of this SNP may extend beyond that of liver tissues (19).
Pathway loci selection for this study was driven by the availability of high-quality meta-analysis data on two phenotypes implicated in the pathogenesis of AAA, specifically concurrent CAD (13) and blood lipid profile (12). The evidence from several large AAA screening studies has strongly suggested a positive correlation between the risk of AAA and concurrent atherosclerotic occlusive diseases, especially CAD (2–4). However, these studies offer contradictory associations with regard to AAA and dyslipidaemia (blood lipid profile) (2,4). Golledge et al. (20), examining subjects from the Australian Health in Men Study, reported that HDL-cholesterol, but not LDL or triglycerides, was an important risk factor for (small) AAA. In addition, in middle-aged men, aortic atherosclerotic plaque, but not blood lipid profile, has been correlated with abdominal aortic size (21). Similarly, carotid plaque and CAD, but not total cholesterol, were shown to be associated with aortic size in the Tromsø Study (2). These observations appeared to match the strong correlation and consistency of effect for the CAD SNPs tested. In contrast, a weaker correlation and consistency of effect was observed for the lipid loci SNPs. Taken together these data suggest that vessel wall atherosclerotic burden may be a more significant risk factor for AAA than circulating lipid profile.
In conclusion, this study identified a genetic variant rs599839, which has been previously associated with SORT1 expression, circulating LDL-cholesterol profile and risk of CAD, to be consistently associated with AAA risk. This association appears to be independent of other known cardiovascular risk factors, including concurrent vascular disease and dyslipidaemia.
MATERIALS AND METHODS
Ethics statement
All samples used in this study were collected under the approval of the appropriate local ethics committee, and written informed consent was obtained for all individuals.
Discovery cohort
The NZ AAA discovery cohort consisted of 612 AAA patients and 608 elderly controls screened to exclude AAA (Supplementary Material, Table S1). All discovery participants were genotyped using the Affymetrix SNP6 GeneChip array and had call rates >95% (mean 99.2%). Imputation was then conducted using IMPUTE 2.2 run on the BCISNPmax database platform (version 3.5, BCI Platforms, Espoo, Finland). The reference haplotypes were based on the 1000 Genomes June 2011 release. Imputed calls were filtered by quality score (>0.9) to restrict to higher quality imputed SNPs. The genomic inflation factor (λ) was 1.06 (MAF > 0.05, 5.4 million SNPs).
Selection of SNPs for analyses
A biological pathway-based screen of the NZ AAA discovery data set was used to identify possible blood lipids or CAD SNP associations with AAA. Validated loci associated with blood lipids [27 loci (12) with P < 10−20] and CAD [20 loci (13) with P < 10−8] were identified from the published literature. A PLINK association analysis was performed using a panel of either the lead CAD or blood lipid-associated SNPs within each locus. In addition, an analysis was conducted consisting of all SNPs with an MAF >0.05 in the gene locus regions (including those within the 200 kbp flanking regions). PLINK set-based tests were performed to identify the best SNP associations within the blood lipid and CAD loci (Supplementary Material, Table S2).
In order to determine the relative effect of lipid fractions, an additional analysis was conducted whereby SNPs associated (P < 10−4) with either HDL or LDL were identified from the meta-analysis conducted by Teslovich et al. (12) (www.sph.umich.edu/csg/abecasis/public). These lists were LD-filtered (r2 > 0.5), and the correlation between either all SNP or those with suggestive (OR ≤ 0.9 or >1.1) AAA associations and the respective HDL and LDL traits determined using Pearson's correlation coefficients.
Replication sets
A second NZ cohort was used to validate discovery cohort observations. This cohort included 713 AAA patients and 1766 screened controls (Supplementary Material, Table S1).
NZ observations were further validated in an additional nine independent AAA case–control cohorts. These were from the Wellcome Trust Case Control Consortium (WTCCC2, 1286 cases/5605 controls), deCODE Genetics (Icelandic, 1163/61 639; Dutch, 840/2792), Western Australia (Health in Men Study, 377/373), Leeds, UK (684/456), Geisinger Clinic case–control sample, Pennsylvania, USA (773/1313) (6,8), the Mayo Clinic eMERGE data set (230/1000), Belgium (176/267) and Canada (198/153) (8,9). A more detailed description of each cohort is provided in the Supplementary Material.
The rs599839 SNP was re-genotyped in NZ (discovery and validation cohorts), Western Australian, Leeds, Mayo Clinic and Geisinger Clinic cohorts using the TaqMan allelic discrimination method (using a pre-designed assay available from Life Technologies). SNPs for the other cohorts were from imputed data sets as previously reported (7). The SNP rs12740374, a putative functional SNP (15) in high LD with rs599839, was also re-genotyped in both NZ samples cohorts using the TaqMan methodology.
A meta-analysis of the cohort odds ratios was conducted using a Maentel–Haenzel method.
The known effect of rs599839 on LDL-cholesterol (12) was modelled in both the Otago (NZ) and Leeds (UK) cohorts. Statin use reduced LDL by an average factor of 0.68 compared with untreated individuals. This difference was the same in both case and control groups. All statin-treated LDL-levels were multiplied by the reciprocal of 0.68 and these values used for linear regression. Similar rs599839 effects on LDL were obtained when both uncorrected LDL and statin use were included in a linear regression model.
Confirmation of aortic gene and protein expression
Quantitative PCR probes for SORT1 (NM_002959), CELSR2 (NM_001408), PSRC1 (NM_032636), ACTB (NM_001101) and GAPDH (NM_002046, SA Biosciences, Germany) were used to determine gene expression in RNA extracted from 11 AAA samples. Total RNA was extracted using Trizol (Life Technologies, NY, USA) and RNeasy kits (Qiagen, Germany) according to the manufacturers' instructions, and was followed by DNase treatment. RNA quantity and purity were confirmed by a NanoDrop (Thermo Scientific, DE, USA). Target gene probe intensities were standardized against ACTB or GAPDH probe intensities, with similar results being obtained regardless of which reference gene was used.
Infra-renal aortic tissue samples from 20 elderly non-aneurysmal post-mortem controls [74% male, mean age 63 years (range 24–83)] and 98 AAA [78% male, mean age 73 years (range 51–87)] patients were collected. Solubilized protein was extracted from samples using Trizol according to the manufacturer's instructions. Two separate polyclonal antibodies against human SORT1, one targeting the cytoplasmic domain (LS-C94912, Lifespan Biosciences, Inc., Seattle, WA, USA) and the other against the extracellular region (S0697, Sigma-Aldrich, St Louis, MI, USA), were used to detect protein levels by western blot analysis. Recombinant human Sortilin (Q99523, R&D Systems, Minneapolis, MN, USA) was used as a positive control. Protein levels were standardized against ACTB (SC-130301, Santa Cruz Biotechnology, CA, USA).
Immunohistochemical analysis was performed on both aneurysmal (n = 4) and non-aneurysmal aortic (n = 4) formalin-fixed paraffin-embedded tissue using the SORT1 cytoplasmic domain (LS-C94912) antibody. Smooth muscle (A2547 Sigma, St Louis, MO, USA), T-cells and B-cells (ab5690 and ab24081, respectively, Abcam, Cambridge, UK) were also immunostained to provide cellular localization of SORT1.
WEB RESOURCES
The URLs for data presented herein are as follows:
IMPUTE 2.2, http://mathgen.stats.ox.ac.uk/impute/impute_v2
LocusZoom, http://csg.sph.umich.edu/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org/
AmiGo (the Gene Ontology project), http://amigo.geneontology.org/cgi-bin/amigo/go.cgi
Blood Lipid Meta-analysis data (12) www.sph.umich.edu/csg/abecasis/public
SUPPLEMENTARY MATERIAL
FUNDING
Funding for the New Zealand project was provided by the Health Research Council of New Zealand (08/075). The WTCCC project was funded by the Wellcome Trust under awards 076113, 085475 and 084695. The collection of samples and genotyping at Geisinger Clinic was supported by a grant from the Pennsylvania Commonwealth Universal Research Enhancement Program (to D.J.C.); and a Grant-In-Aid from the American Heart Association (to D.J.C.). The Geisinger MyCode® Project was funded in part by a grant from the Ben Franklin Technology Development Fund of Pennsylvania. The deCODE study was funded in part through grants from the European Community's Seventh Framework Programme (FP7/2007–2013), FAD project grant agreement; HEALTH-F2–2008-200647 and ENGAGE project, grant agreement HEALTH-F4-2007- 201413. The Netherland cohort was funded by the Netherlands Heart Foundation (2009T001). LEADS (Leeds Aneurysm Development Study) is supported by the Garfield Weston Trust for Medical Research into Diseases of the Heart. The Health In Men Study (Western Australia) was funded by National Health and Medical Research Project Grant 303232. J.G. holds a Practitioner Fellowship from the National Health and Medical Research Council, Australia (1019921) and a Senior Clinical Research Fellowship from the Queensland Government. The Mayo eMERGE study was supported by the grant U01-HG04599 from the National Human Genome Research Institute. The Belgian and Canadian sample collections were funded in part by the National Heart, Lung, and Blood Institute of the National Institutes of Health (HL045996 and HL06410 to H.K. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would also like to thank all the study participants, without whom this study would not have been possible. The LEADS (Leeds Aneurysm Development Study) authors would like to acknowledge the work of the research nurses who have been key to the successes of the project.
Conflict of Interest statement. deCODE genetics is a biotechnology company, and authors employed by deCODE own stock or stock options in the company.
REFERENCES
- 1.Sakalihasan N., Limet R., Defawe O.D. Abdominal aortic aneurysm. Lancet. 2005;365:1577–1589. doi: 10.1016/S0140-6736(05)66459-8. [DOI] [PubMed] [Google Scholar]
- 2.Johnsen S.H., Forsdahl S.H., Singh K., Jacobsen B.K. Atherosclerosis in abdominal aortic aneurysms: a causal event or a process running in parallel? The Tromsø Study. Arterioscler. Thromb. Vasc. Biol. 2010;30:1263–1268. doi: 10.1161/ATVBAHA.110.203588. [DOI] [PubMed] [Google Scholar]
- 3.Kent K.C., Zwolak R.M., Egorova N.N., Riles T.S., Manganaro A., Moskowitz A.J., Gelijns A.C., Greco G. Analysis of risk factors for abdominal aortic aneurysm in a cohort of more than 3 million individuals. J. Vasc. Surg. 2010;52:539–548. doi: 10.1016/j.jvs.2010.05.090. [DOI] [PubMed] [Google Scholar]
- 4.Lederle F.A., Johnson G.R., Wilson S.E., Chute E.P., Hye R.J., Makaroun M.S., Barone G.W., Bandyk D., Moneta G.L., Makhoul R.G. The aneurysm detection and management study screening program: validation cohort and final results. Aneurysm Detection and Management Veterans Affairs Cooperative Study Investigators. Arch. Intern. Med. 2000;160:1425–1430. doi: 10.1001/archinte.160.10.1425. [DOI] [PubMed] [Google Scholar]
- 5.Golledge J., Norman P.E. Atherosclerosis and abdominal aortic aneurysm: cause, response, or common risk factors? Arterioscler. Thromb. Vasc. Biol. 2010;30:1075–1077. doi: 10.1161/ATVBAHA.110.206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmore J.R., Obmann M.A., Kuivaniemi H., Tromp G., Gerhard G.S., Franklin D.P., Boddy A.M., Carey D.J. Identification of a genetic variant associated with abdominal aortic aneurysms on chromosome 3p12.3 by genome wide association. J. Vasc. Surg. 2009;49:1525–1531. doi: 10.1016/j.jvs.2009.01.041. [DOI] [PubMed] [Google Scholar]
- 7.Bown M.J., Jones G.T., Harrison S.C., Wright B.J., Bumpstead S., Baas A.F., Gretarsdottir S., Badger S.A., Bradley D.T., Burnand K., et al. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am. J. Hum. Genet. 2011;89:619–627. doi: 10.1016/j.ajhg.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gretarsdottir S., Baas A.F., Thorleifsson G., Holm H., den Heijer M., de Vries J.P., Kranendonk S.E., Zeebregts C.J., van Sterkenburg S.M., Geelkerken R.H., et al. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat. Genet. 2010;42:692–697. doi: 10.1038/ng.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helgadottir A., Thorleifsson G., Magnusson K.P., Gretarsdottir S., Steinthorsdottir V., Manolescu A., Jones G.T., Rinkel G.J., Blankensteijn J.D., Ronkainen A., et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat. Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 10.Golledge J., Tsao P.S., Dalman R.L., Norman P.E. Circulating markers of abdominal aortic aneurysm presence and progression. Circulation. 2008;118:2382–2392. doi: 10.1161/CIRCULATIONAHA.108.802074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones G.T. The histopathology of abdominal aortic aneurysm. In: Grundmann R.T., editor. Diagnosis, Screening and Treatment of Abdominal, Thoracoabdominal and Thoracic Aortic Aneurysms. InTech, Rijeka, Croatia; 2011. DOI:10.5772/746. [Google Scholar]
- 12.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K., Li M., Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 15.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M., et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleber M.E., Renner W., Grammer T.B., Linsel-Nitschke P., Boehm B.O., Winkelmann B.R., Bugert P., Hoffmann M.M., Marz W. Association of the single nucleotide polymorphism rs599839 in the vicinity of the sortilin 1 gene with LDL and triglyceride metabolism, coronary heart disease and myocardial infarction. The Ludwigshafen Risk and Cardiovascular Health Study. Atherosclerosis. 2010;209:492–497. doi: 10.1016/j.atherosclerosis.2009.09.068. [DOI] [PubMed] [Google Scholar]
- 17.Ramanan V.K., Shen L., Moore J.H., Saykin A.J. Pathway analysis of genomic data: concepts, methods, and prospects for future development. Trends Genet. 2012;28:323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folkersen L., van't Hooft F., Chernogubova E., Agardh H.E., Hansson G.K., Hedin U., Liska J., Syvanen A.C., Paulsson-Berne G., Franco-Cereceda A., et al. Association of genetic risk variants with expression of proximal genes identifies novel susceptibility genes for cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3:365–373. doi: 10.1161/CIRCGENETICS.110.948935. [DOI] [PubMed] [Google Scholar]
- 19.Linsel-Nitschke P., Heeren J., Aherrahrou Z., Bruse P., Gieger C., Illig T., Prokisch H., Heim K., Doering A., Peters A., et al. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 2010;208:183–189. doi: 10.1016/j.atherosclerosis.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 20.Golledge J., van Bockxmeer F., Jamrozik K., McCann M., Norman P.E. Association between serum lipoproteins and abdominal aortic aneurysm. Am. J. Cardiol. 2010;105:1480–1484. doi: 10.1016/j.amjcard.2009.12.076. [DOI] [PubMed] [Google Scholar]
- 21.Paivansalo M.J., Merikanto J., Jerkkola T., Savolainen M.J., Rantala A.O., Kauma H., Lilja M., Reunanen Y.A., Kesaniemi A., Suramo I. Effect of hypertension and risk factors on diameters of abdominal aorta and common iliac and femoral arteries in middle-aged hypertensive and control subjects: a cross-sectional systematic study with duplex ultrasound. Atherosclerosis. 2000;153:99–106. doi: 10.1016/s0021-9150(00)00374-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.