Abstract
Aiming to identify novel genetic loci for pigmentation and skin cancer, we conducted a series of genome-wide association studies on hair color, eye color, number of sunburns, tanning ability and number of non-melanoma skin cancers (NMSCs) among 10 183 European Americans in the discovery stage and 4504 European Americans in the replication stage (for eye color, 3871 males in the discovery stage and 2496 males in the replication stage). We targeted novel chromosome regions besides the known ones for replication. As a result, we identified a new region downstream of the EDNRB gene on 13q22 associated with hair color and the strongest association was the single-nucleotide polymorphism (SNP) rs975739 (P = 2.4 × 10−14; P = 5.4 × 10−9 in the discovery set and P = 1.2 × 10−6 in the replication set). Using blue, intermediate (including green) and brown eye colors as co-dominant outcomes, we identified the SNP rs3002288 in VASH2 on 1q32.3 associated with brown eye (P = 7.0 × 10−8; P = 5.3 × 10−5 in the discovery set and P = 0.02 in the replication set). Additionally, we identified a significant interaction between the SNPs rs7173419 and rs12913832 in the OCA2 gene region on brown eye color (P-value for interaction = 3.8 × 10−3). As for the number of NMSCs, we identified two independent SNPs on chr6 and one SNP on chromosome 14: rs12203592 in IRF4 (P = 7.2 × 10−14; P = 1.8 × 10−8 in the discovery set and P = 6.7 × 10−7 in the replication set), rs12202284 between IRF4 and EXOC2 (P = 5.0 × 10−8; P = 6.6 × 10−7 in the discovery set and P = 3.0 × 10−3 in the replication set) and rs8015138 upstream of GNG2 (P = 6.6 × 10−8; P = 5.3 × 10−7 in the discovery set and P = 0.01 in the replication set).
INTRODUCTION
Natural hair color, eye color, skin color and sun sensitivity reflect the variation in human basal and transient pigmentation, which has been known to be highly heritable (1,2). More recently, dozens of common genetic variants have been identified to be associated with pigmentary phenotypes from the genome-wide association studies (GWASs) and were reviewed by Gerstenblith et al. (3). These GWASs identified genetic variants in the genes already known to be associated with pigmentation, such as melanocortin 1 receptor (MC1R) (4,5), oculocutaneous albinism II (OCA2) (4), solute carrier family 45, member 2 (SLC45A2) (4), tyrosinase (TYR) (5,6), tyrosinase-related protein 1 (TYRP1) (6), kit ligand (KITLG) (5) and the agouti signaling protein (ASIP) (6), and also identified novel potential pigmentation genes, such as solute carrier family 24, member 4 (SLC24A4) (4,5), two-pore segment channel 2 (TPCN2) (6) and interferon regulatory factor 4 (IRF4) (4). Of interest, some of the genetic determinants are shared across different pigmentation traits, whereas others are distinct, suggesting various mechanisms of pigmentation across different organs (3). Europeans have the majority of variation in pigmentation characteristics compared with other populations, especially in hair and eye colors (5). In this study, we conducted a series of GWASs on hair color, eye colors, tanning ability and the number of sunburns in European Americans, aiming to identify novel genetic loci for pigmentation and to understand the variability of complex pigmentation characteristics in the general population to a greater extent. In addition, given that human pigmentation traits are susceptibility factors for skin cancer (3,7), we further evaluated the newly identified pigmentation loci in association with the risk of melanoma and non-melanoma skin cancer (NMSC). We also conducted a GWAS on the number of NMSCs in this study to identify new loci for the risk of NMSCs (8–10).
RESULTS
The samples used in each GWAS of the present study are summarized in Supplementary Material, Table S1. The quantile–quantile plots based on the meta-analysis did not demonstrate a systematic deviation from the expected distribution, with the overall genomic control inflation factors (λGC) of 0.99–1.04 (Supplementary Material, Fig. S1). The Manhattan plot of each GWAS is presented in Supplementary Material, Figure S2. We focused on the novel chromosome regions suggesting promising associations besides the previously known ones and selected the single-nucleotide polymorphisms (SNPs) with the strongest associations in these regions for replication. A total of 16 SNPs were finally selected for replication, with detailed information presented in Supplementary Material, Table S2. Details of the SNP selection for replication are presented in Materials and Methods.
As a result, for the GWAS of natural hair color, the SNP rs975739, which ranked the top besides the known pigmentation SNPs in the discovery set (P = 5.4 × 10−9 in the discovery set), revealed a nominal significance in both of the replication sets (P = 0.05 in the skin cancer set, P = 3.3 × 10−6 in the RF_NHS set and P = 1.2 × 10−6 in the combined replication set, Table 1). This association reached a P-value of 2.4 × 10−14 after combining the discovery set and the replication set. The variant allele (G allele) of this SNP was associated with darker hair color with the regression parameter beta of 0.08 and a standard error (SE) of 0.01. Another tag-SNP rs1146927 in this region, which was in linkage disequilibrium (LD) with rs975739 (r2 = 0.84 in this study), was also replicated in the replication set (P = 2.9 × 10−8 in the discovery set, P = 0.03 in the skin cancer replication set and P = 2.8 × 10−9 in the combined set, Supplementary Material, Table S3). Both of the SNPs (rs975739 and rs1146927) reached the GWAS significance level and revealed a similar association pattern (beta = 0.8 and SE = 0.01 for rs975739; beta = 0.7 and SE = 0.01 for rs1146927), suggesting the same signal represented by them. The association of the other tag-SNP rs58188699 in this region (LD r2 = 0.42 with rs975739 in this study) with natural hair color (P = 7.5 × 10−8 in the discovery set) was not replicated in the replication set (P = 0.27 in the skin cancer replication set, Supplementary Material, Table S3).
Table 1.
Association of the SNP rs975739 downstream of the EDNRB gene with non-red hair colorsa
| Data set | rs975739 T>G |
||||
|---|---|---|---|---|---|
| Sample size | Blond hair color | Beta (SE) | P-valueb | P-value for heterogeneity | |
| Discovery set | |||||
| BC_NHS (female)c | 1741 | 223 | 0.07 (0.02) | 4.8E − 03 | |
| T2D_NHS (female)d | 2377 | 302 | 0.07 (0.02) | 1.6E − 03 | |
| T2D_HPFS (male)e | 1529 | 165 | 0.10 (0.03) | 8.0E − 04 | |
| CHD_NHS (female)f | 783 | 97 | 0.06 (0.04) | 0.09 | |
| CHD_HPFS (male)g | 640 | 87 | 0.08 (0.04) | 0.05 | |
| All (meta-analysis) | 7070 | 874 | 0.07 (0.01) | 5.4E − 09 | 0.92 |
| Replication set | |||||
| Skin cancer (female and male)h | 1639 | 183 | 0.05 (0.03) | 0.05 | |
| RF_NHS (female)i | 2516 | 314 | 0.10 (0.02) | 3.3E − 06 | |
| All (meta-analysis) | 4155 | 497 | 0.08 (0.02) | 1.2E − 06 | 0.16 |
| Combined set (meta-analysis) | 11 225 | 1371 | 0.08 (0.01) | 2.4E − 14 | 0.74 |
aNon-red hair colors were coded as: 1, blond; 2, light brown; 3, dark brown; 4, black; beta > 0 means association with darker hair color.
bAdjusted for the top four principal components of genetic variance in the discovery sets and adjusted for gender in the skin cancer replication set.
cPostmenopausal invasive breast cancer case–control study nested within the NHS.
dType 2 diabetes case–control study nested within the NHS.
eType 2 diabetes case–control study nested within the HPFS.
fCoronary heart disease case–control study nested within the NHS.
gCoronary heart disease case–control study nested within the HPFS.
hSkin cancer case–control study nested within the NHS and HPFS.
iRenal function study nested within the NHS.
These novel hair-color-related SNPs identified in this study are located in the region downstream of the endothelin receptor type B (EDNRB) gene. We additionally evaluated the association of all the SNPs within 200 kb surrounding the gene region with hair color. The regional association plot in the discovery set is presented in Figure 1. After adjustment for the SNP rs975739, the associations of 1611 SNPs in this region were substantially attenuated, and none of them remained regionally significant (0.05/1611 = 3.1 × 10−5, the smallest P = 1.5 × 10−4 after adjustment, Fig. 1). We also constructed the haplotypes based on the three tag-SNPs (rs975739, rs1146927 and rs58188699). When we used the most prevalent haplotype (the combination of the common allele of rs58188699 with the rare alleles of both rs975739 and rs1146927, 38.7% of the population) as a reference, the haplotypes with the common alleles of both rs975739 and rs1146927, together with either the rare allele (37.8% of the population) or the common allele (18.9% of the population) of rs58188699, were marginally associated with lighter hair color (P = 0.08, beta = 0.07, SE = 0.04 with the rare allele of rs58188699, and P = 0.07, beta = 0.05, SE = 0.03 with the common allele of rs58188699), suggesting that these associations were originally from the same signal as rs975739 and rs1146927. Additionally, all of the 117 SNPs with P < 1 × 10−7 in the discovery stage were in LD (r2 > 0.6).
Figure 1.
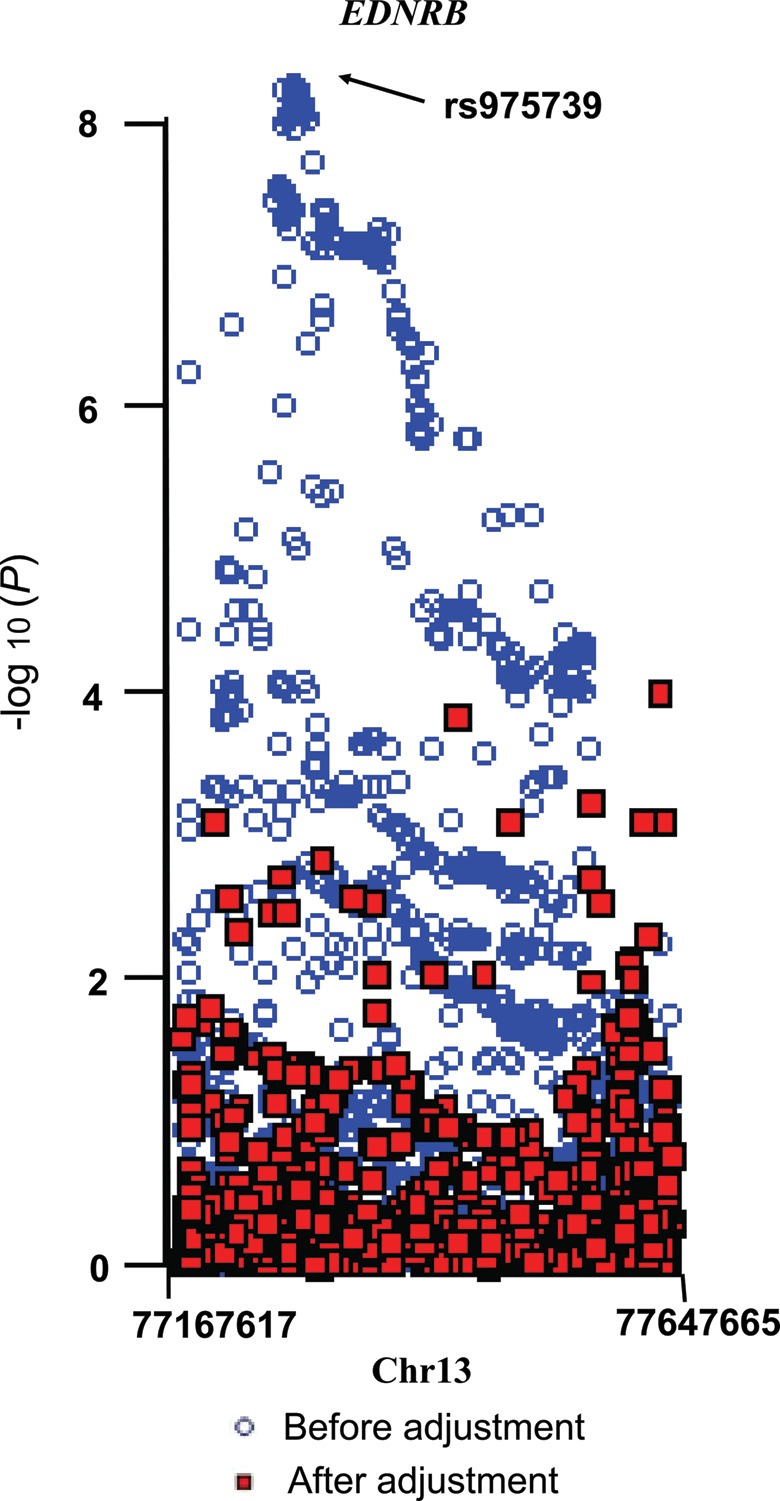
Regional plot of the EDNRB gene. Manhattan plots for the GWAS of hair color in the region within 200 kb surrounding EDNRB on chromosome 13, before and after adjustment by the SNP rs975739.
The SNP rs975739 was not associated with eye colors (P = 0.78 for brown eye color, P = 0.33 for intermediate eye color, P = 0.46 for blue eye color), tanning ability (P = 0.72) or the number of sunburns (P = 0.67). Similar results were detected for the other two SNPs (rs1146927 and rs58188699). We also evaluated the association between the two significant SNPs and red hair color phenotype, and no significant association was found (P = 0.95 for rs975739 and P = 0.90 for rs1146927).
We also replicated the association between the SNP rs12421680 in the neurotrimin (NTM) gene on chromosome 11 and the number of sunburns (P = 5.3 × 10−5 in the discovery set and P = 0.02 in the replication set, Table 2). After combining the discovery and replication sets, P-value for the association of this SNP reached 5.6 × 10−6. The variant allele (A allele) was associated with an increased number of sunburns (beta = 0.41, SE = 0.09). Additionally, based on the transcript expression profiling data of 79 HapMap CEU cell lines (NCBI GEO database, accession GSE7792) (11), we found that the AA genotype was associated with a lower expression of NTM than the GG/GA genotypes (P < 0.001, Supplementary Material, Fig. S3). There was no association between this SNP and eye colors (P = 0.54 for brown eye color, P = 0.69 for intermediate eye color, P = 0.80 for blue eye color), natural hair color (P = 0.55) or tanning ability (P = 0.49). Because the imputation quality of this SNP was not particularly high in four subsets in the discovery stage (imputation r2: 0.61–0.62 for T2D_NHS, CHD_NHS, T2D_HPFS and CHD_HPFS), we re-genotyped this SNP in the subset of T2D_HPFS using TaqMan assay to validate the result. The association of this SNP with the number of sunburns was comparable by using the re-genotyped data (beta = 0.82, SE = 0.30, P = 0.01) and the imputed data (beta = 0.88, SE = 0.38, P = 0.02).
Table 2.
Association of the SNP rs12421680 in the NTM gene with the number of sunburnsa
| Data set | rs12421680 G>A |
||||
|---|---|---|---|---|---|
| Sample size | Sunburns 10+ | Beta (SE) | P-valuei | P-value for heterogeneity | |
| Discovery set | |||||
| BC_NHS (female)b | 2153 | 852 | 0.37 (0.19) | 0.05 | |
| T2D_NHS (female)c | 2915 | 1430 | 0.55 (0.22) | 0.01 | |
| T2D_HPFS (male)d | 2170 | 1075 | 0.88 (0.38) | 0.02 | |
| CHD_NHS (female)e | 1056 | 668 | 0.41 (0.35) | 0.24 | |
| CHD_HPFS (male)f | 989 | 631 | 0.67 (0.56) | 0.23 | |
| All (meta-analysis) | 9283 | 4656 | 0.49 (0.12) | 5.3E − 05 | 0.79 |
| Replication set | |||||
| Skin cancer (female and male)g | 1655 | 558 | 0.44 (0.23) | 0.05 | |
| RF_NHS (female)h | 2581 | 891 | 0.24 (0.17) | 0.16 | |
| All (meta-analysis) | 4236 | 1449 | 0.31 (0.14) | 0.02 | 0.49 |
| Combined set (meta-analysis) | 13 519 | 6105 | 0.41 (0.09) | 5.6E − 06 | 0.32 |
aThe number of sunburns was coded as a continuous variable; beta > 0 means association with increased number of sunburns.
bPostmenopausal invasive breast cancer case–control study nested within the NHS.
cType 2 diabetes case–control study nested within the NHS.
dType 2 diabetes case–control study nested within the HPFS.
eCoronary heart disease case–control study nested within the NHS.
fCoronary heart disease case–control study nested within the HPFS.
gSkin cancer case–control study nested within the NHS and HPFS.
hRenal function study nested within the NHS.
iAdjusted for age and the top four principal components of genetic variance in the discovery sets and adjusted for age and gender in the skin cancer replication set.
As for the GWASs on eye color, we replicated the association of the SNP rs3002288 in vasohibin 2 (VASH2) with brown eye color (P = 7.0 × 10−8 in the combined set; P = 4.3 × 10−5 in the discovery set and P = 3.3 × 10−5 in the replication set), as well as the association of the SNP rs12520016 upstream of DNA polymerase sigma (POLS) with intermediate (including green) eye color (P = 9.1 × 10−7 in the combined set; P = 4.7 × 10−5 in the discovery set and P = 0.03 in the replication set, Table 3). The variant allele of rs3002288 (A allele) showed an inverse association with brown eye color (beta = −0.22, SE = 0.04), and the variant allele of rs12520016 (G allele) showed a positive association with intermediate eye color (beta = 0.42, SE = 0.08). The association between the SNP rs3002288 and brown eye color was approaching the GWAS significance level of 10−8. Neither of the two SNPs revealed an association with natural hair color (P = 0.37 for rs3002288 and P = 0.58 for rs12520016), tanning ability (P = 0.93 for rs3002288 and P = 0.71 for rs12520016) or the number of sunburns (P = 0.57 for rs3002288 and P = 0.91 for rs12520016).
Table 3.
Association of the SNP rs3002288 in the VASH2 gene with brown eye color and the SNP rs12520016 upstream of POLS gene with intermediate eye colora
| Data set | Sample size | rs3002288 G>A |
rs12520016 T>G |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Brown eye color | Beta (SE) | P-valuef | P-value for heterogeneity | Intermediate eye color | Beta (SE) | P-valuef | P-value for heterogeneity | ||
| Discovery set | |||||||||
| T2D_HPFS (male)b | 2292 | 717 | −0.25 (0.07) | 4.0E − 04 | 761 | 0.52 (0.14) | 3.4E − 04 | ||
| CHD_HPFS (male)c | 1064 | 316 | −0.12 (0.11) | 0.24 | 348 | 0.58 (0.19) | 2.8E − 03 | ||
| KS_HPFS (male)d | 515 | 165 | −0.50 (0.15) | 5.1E − 04 | 146 | 0.15 (0.31) | 0.62 | ||
| All (meta-analysis) | 3871 | 1033 | −0.25 (0.06) | 4.3E − 06 | 0.11 | 1109 | 0.49 (0.11) | 4.7E − 06 | 0.48 |
| Replication set | |||||||||
| PrCa_HPFS (male)e | 2496 | 760 | −0.18 (0.06) | 3.3E − 03 | 805 | 0.29 (0.14) | 0.03 | ||
| Combined set (meta-analysis) | 6367 | 1793 | −0.22 (0.04) | 7.0E − 08 | 0.38 | 1914 | 0.42 (0.08) | 9.1E − 07 | 0.25 |
aEye colors were coded as co-dominant outcomes (brown versus non-brown eye color and green versus non-green eye color).
bType 2 diabetes case–control study nested within the HPFS.
cCoronary heart disease case–control study nested within the HPFS.
dKidney stone case–control study nested within the HPFS.
eProstate cancer case–control study nested within the HPFS.
fAdjusted for the top four principal components of genetic variance in the discovery sets.
Of interest, we found that the SNP rs7173419 located in the intron region of the OCA2 gene revealed a significant association with brown eye color (T allele, beta = 0.33, SE = 0.06; P = 1.3 × 10−7 in the discovery set), whereas this association was inverted (T allele, beta = −0.48, SE = 0.08; P = 1.5 × 10−8 in the discovery set) after adjustment by the top SNP in this region (rs12913832, A allele, beta = 2.82, SE = 0.10; P = 1.2 × 10−177 in the discovery stage of brown eye color GWAS). The SNP rs12913832 is located upstream of OCA2 (in the intron of HERC2) and known as a pigmentation SNP by controlling the expression of OCA2 (12,13). We replicated this finding in the replication set (rs7173419, T allele; beta = 0.56, SE = 0.07, P = 4.1 × 10−15 before adjustment by rs12913832; and beta = −0.21, SE = 0.10, P = 0.04 after adjustment by rs12913832). We further conducted a stratification analysis of the SNP rs12913832 by the genotypes of rs7173419 among the pooled population of the discovery set and the replication set. As shown in Table 4, the effect of the SNP rs12913832 on brown eye color decreased with more counts of the variant allele (T allele) of rs7173419, and the interaction of these two SNPs was significant (P = 3.8 × 10−3).
Table 4.
Interaction between the SNPs rs7173419 and rs12913832 on brown eye colora and melanoma risk
| rs7173419 | Eye color |
rs12913832 G>Ab |
P-value for interaction | |||
|---|---|---|---|---|---|---|
| Sample size | Brown eye color, % (SD) | Beta | SD | P-valuec | ||
| CC | 3755 | 26.3 (0.7) | 3.92 | 0.13 | 6.9E − 215 | 3.8E − 03 |
| CT | 2178 | 36.1 (1.0) | 2.54 | 0.12 | 6.9E − 103 | |
| TT | 337 | 45.4 (2.7) | 2.18 | 0.26 | 6.6E − 17 | |
| rs7173419 | Melanoma risk | rs12913832 G>Ab | P-value for interaction | |||
| Cases | Controls | OR | 95% CI | P-valued | ||
| CC | 1134 | 610 | 0.64 | 0.53–0.77 | 3.6E − 06 | 0.06 |
| CT | 592 | 363 | 0.76 | 0.61–0.94 | 0.01 | |
| TT | 78 | 53 | 1.05 | 0.63–1.73 | 0.86 | |
aEye colors were coded as brown versus non-brown eye color.
brs12913832 was coded in an additive model, with G as wild-type allele and A as variant allele.
cAdjusted for the top four principal components.
dAdjusted for age, gender and the top two principal components.
As for the number of NMSCs, we identified two independent SNPs on chromosome 6 (LD r2 = 0.12): rs12203592 in IRF4 with a P-value of 7.2 × 10−14 in the combined set (T allele, beta = 0.21, SE = 0.03; P = 1.8 × 10−8 in the discovery stage and P = 6.7 × 10−7 in the replication stage) and rs12202284 between IRF4 and exocyst complex component 2 (EXOC2) with a P-value of 5.0 × 10−8 in the combined set (A allele, beta = 0.15, SE = 0.03; P = 6.6 × 10−7 in the discovery stage and P = 3.0 × 10−3 in the replication stage; Table 5). After mutual adjustment by each other, the association of each SNP with the number of NMSCs remained significant (P = 6.0 × 10−4 for rs12203592 and P = 5.8 × 10−3 for rs12202284), suggesting independent effects of these two SNPs. We previously reported an association of the SNP rs12203592 with hair color (4) and tanning ability (14). In this study, we additionally found a significant association of this SNP with the number of sunburns (T allele, beta = 1.13, SE = 0.24; P = 2.2 × 10−6; Table 6). We also identified a novel SNP rs8015138 upstream of guanine nucleotide-binding protein, gamma 2 (GNG2), on chromosome 14 associated with the number of NMSCs with a P-value of 6.6 × 10−8 in the combined set (C allele, beta = −0.11, SE = 0.02; P = 5.3 × 10−7 in the discovery stage and P = 0.01 in the replication stage; Table 5).
Table 5.
Association of the SNP rs12203592 in the IRF4 gene, rs12202284 between IRF4 and EXOC2 and rs8015138 upstream of GNG2 with the number of NMSCsa
| Data set | Sample size | NMSCs, 5+ | rs12203592 C>T |
rs12202284 C>A |
rs8015138 A>C |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (SE) | P-valueh | P-value for heterogeneity | Beta (SE) | P-valueh | P-value for heterogeneity | Beta (SE) | P-valueh | P-value for heterogeneity | |||
| Discovery set | |||||||||||
| BC_NHS (female)b | 1620 | 45 | 0.20 (0.07) | 0.0037 | 0.21 (0.08) | 0.005 | −0.09 (0.05) | 0.081 | |||
| T2D_NHS (female)c | 2427 | 65 | 0.21 (0.08) | 0.011 | 0.14 (0.07) | 0.039 | −0.12 (0.04) | 0.005 | |||
| T2D_HPFS (male)d | 1308 | 83 | 0.24 (0.15) | 0.11 | 0.17 (0.12) | 0.16 | −0.07 (0.08) | 0.41 | |||
| CHD_NHS (female)e | 760 | 23 | 0.21 (0.14) | 0.13 | 0.36 (0.11) | 0.0015 | −0.23 (0.08) | 0.002 | |||
| CHD_HPFS (male)f | 632 | 39 | 0.47 (0.25) | 0.056 | 0.49 (0.21) | 0.017 | −0.32 (0.13) | 0.01 | |||
| All (meta-analysis) | 6747 | 255 | 0.22 (0.03) | 1.8E − 8 | 0.89 | 0.21 (0.04) | 6.6E − 7 | 0.42 | −0.14 (0.03) | 5.3E − 7 | 0.26 |
| Replication set | |||||||||||
| Skin cancer (female and male)g | 2078 | 118 | 0.20 (0.04) | 6.7E − 7 | 0.10 (0.03) | 0.003 | −0.08 (0.03) | 0.01 | |||
| Combined set (meta-analysis) | 8825 | 373 | 0.21 (0.03) | 7.2E − 14 | 0.50 | 0.15 (0.03) | 5.0E − 8 | 0.05 | −0.11 (0.02) | 6.6E − 8 | 0.11 |
aThe number of NMSCs was coded as: 1-0, 2-1, 3-2-4, 4-5+; beta > 0 means association with increased number of NMSCs.
bPostmenopausal invasive breast cancer case–control study nested within the NHS.
cType 2 diabetes case–control study nested within the NHS.
dType 2 diabetes case–control study nested within the HPFS.
eCoronary heart disease case–control study nested within the NHS.
fCoronary heart disease case–control study nested within the HPFS.
gSkin cancer case–control study nested within the NHS and HPFS.
hAdjusted for the top four principal components of genetic variance in the discovery sets and adjusted for gender in the skin cancer replication set.
Table 6.
Summary of pigmentary SNPs identified from the previous and the present GWASs and their associations with pigmentation traits, melanoma risk and NMSCs in this studya
| SNP | Gene | Chr. | Location | Ref. | A1 | Hair color |
Tanning ability |
Number of sunburns |
Blue eye color |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P-value | Beta | SE | P-value | Beta | SE | P-value | Beta | SE | P-value | |||||||||
| rs16891982b | SLC45A2 | 5 | 33 987 450 | G (major) | C (minor) | – | – | – | – | – | – | – | – | – | −0.42 | 0.15 | 5.4E − 03 | |||
| rs12203592 | IRF4 | 6 | 341 321 | C | T | 0.35 | 0.03 | 1.0E − 28 | −0.35 | 0.03 | 3.3E − 23 | 1.13 | 0.24 | 2.2E − 06 | 0.08 | 0.18 | 0.66 | |||
| rs1408799 | TYRP1 | 9 | 12 662 097 | C | T | 0.04 | 0.01 | 5.6E − 03 | 0.04 | 0.01 | 4.3E − 04 | −0.17 | 0.10 | 0.09 | −0.27 | 0.05 | 4.9E − 07 | |||
| rs35264875c | TPCN2 | 11 | 68 602 975 | T (minor) | A (major) | 0.06 | 0.02 | 3.8E − 04 | 0.02 | 0.02 | 0.21 | 0.14 | 0.13 | 0.26 | – | – | – | |||
| rs3829241 | TPCN2 | 11 | 68 611 939 | G | A | −0.04 | 0.01 | 2.1E − 03 | −0.02 | 0.01 | 0.13 | 0.09 | 0.10 | 0.37 | −0.03 | 0.05 | 0.54 | |||
| rs1126809c | TYR | 11 | 88 657 609 | G | A | −0.03 | 0.01 | 0.04 | −0.12 | 0.01 | 5.0E − 21 | 0.60 | 0.11 | 1.8E − 08 | – | – | – | |||
| rs1042602b | TYR | 11 | 88 551 344 | C | A | – | – | – | – | – | – | – | – | – | −0.08 | 0.05 | 0.14 | |||
| rs12821256 | KITLG | 12 | 87 852 466 | C | T | 0.18 | 0.02 | 6.8E − 19 | 0.04 | 0.02 | 0.04 | −0.21 | 0.16 | 0.17 | −0.18 | 0.08 | 0.03 | |||
| rs12896399 | SLC24A4 | 14 | 91 843 416 | G | T | −0.16 | 0.01 | 1.5E − 36 | −0.02 | 0.01 | 0.07 | 0.08 | 0.10 | 0.43 | 0.33 | 0.05 | 4.1E − 11 | |||
| rs12913832 | HERC2 | 15 | 26 039 213 | G | A | 0.39 | 0.01 | 1.4E − 167 | 0.14 | 0.01 | 1.4E − 22 | −0.48 | 0.11 | 2.1E − 05 | −2.84 | 0.11 | 1.2E − 158 | |||
| rs1805007 | MC1R | 16 | 88 513 618 | C | T | −0.16 | 0.03 | 2.9E − 09 | −0.39 | 0.02 | 1.1E − 65 | 1.66 | 0.18 | 1.5E − 19 | 0.01 | 0.10 | 0.91 | |||
| rs1805008c | MC1R | 16 | 88 513 645 | C | T | −0.09 | 0.02 | 7.2E − 05 | −0.16 | 0.02 | 1.3E − 13 | 0.71 | 0.18 | 4.8E − 05 | – | – | – | |||
| rs4911414 | ASIP Haplotype | 20 | 32 193 105 | G | T | −0.05 | 0.01 | 1.3E − 04 | −0.07 | 0.01 | 3.8E − 09 | 0.26 | 0.10 | 0.01 | 0.05 | 0.05 | 0.32 | |||
| rs1015362 | ASIP Haplotype | 20 | 32 202 273 | C | T | 0.01 | 0.01 | 0.50 | 0.02 | 0.01 | 0.20 | −0.12 | 0.11 | 0.26 | 0.00 | 0.05 | 1.00 | |||
| rs975739d | EDNRB | 13 | 77 279 147 | G | T | −0.07 | 0.01 | 5.4E − 09 | 0.00 | 0.01 | 0.72 | −0.04 | 0.10 | 0.67 | 0.04 | 0.05 | 0.46 | |||
| rs12421680d | NTM | 11 | 130 856 178 | G | A | −0.01 | 0.02 | 0.55 | −0.01 | 0.02 | 0.49 | 0.49 | 0.12 | 5.3E − 05 | 0.01 | 0.06 | 0.80 | |||
| rs3002288d | VASH2 | 1 | 211 193 188 | G | A | −0.01 | 0.01 | 0.37 | 0.00 | 0.01 | 0.93 | −0.06 | 0.10 | 0.57 | 0.11 | 0.05 | 0.04 | |||
| rs12520016d | POLS | 5 | 6 820 312 | G | T | −0.02 | 0.03 | 0.58 | 0.01 | 0.03 | 0.71 | 0.03 | 0.21 | 0.91 | 0.36 | 0.12 | 2.1E − 03 | |||
| Intermediate/green eye color | Brown eye color | Melanoma risk | Mole count | BCC risk | SCC risk | Number of NMSCs | ||||||||||||||
| Beta | SE | P-value | Beta | SE | P-value | Beta | SE | P-value | Beta | SE | P-value | Beta | SE | P-value | Beta | SE | P-value | Beta | SE | P-value |
| 0.26 | 0.13 | 0.05 | 0.11 | 0.13 | 0.41 | −0.52 | 0.19 | 0.01 | – | – | – | −0.12 | 0.15 | 0.42 | −0.49 | 0.34 | 0.15 | – | – | – |
| 0.12 | 0.19 | 0.54 | −0.21 | 0.20 | 0.28 | 0.16 | 0.07 | 0.02 | 0.05 | 0.03 | 0.05 | 0.23 | 0.07 | 7.0E − 04 | 0.52 | 0.09 | 9.5E − 09 | 0.22 | 0.03 | 1.8E − 08 |
| 0.15 | 0.05 | 4.8E − 03 | 0.13 | 0.05 | 0.02 | −0.02 | 0.06 | 0.68 | 0.01 | 0.02 | 0.70 | 0.10 | 0.04 | 0.01 | −0.05 | 0.13 | 0.71 | 0.04 | 0.03 | 0.15 |
| – | – | – | – | – | – | −0.02 | 0.07 | 0.77 | – | – | – | – | – | – | 0.16 | 0.10 | 0.11 | 0.04 | 0.04 | 0.24 |
| 0.02 | 0.05 | 0.70 | 0.01 | 0.05 | 0.79 | 0.00 | 0.06 | 0.96 | −0.01 | 0.02 | 0.34 | 0.04 | 0.04 | 0.26 | 0.07 | 0.07 | 0.33 | 0.04 | 0.03 | 0.12 |
| – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 0.05 | 0.13 | 0.69 | 0.09 | 0.03 | 6.0E − 03 |
| 0.02 | 0.05 | 0.73 | 0.07 | 0.05 | 0.22 | −0.10 | 0.06 | 0.09 | −0.02 | 0.02 | 0.29 | 0.05 | 0.04 | 0.23 | 0.21 | 0.13 | 0.09 | – | – | – |
| 0.03 | 0.08 | 0.75 | 0.18 | 0.09 | 0.05 | 0.00 | 0.09 | 1.00 | 0.00 | 0.02 | 0.92 | −0.08 | 0.06 | 0.20 | −0.02 | 0.11 | 0.87 | −0.06 | 0.05 | 0.16 |
| −0.27 | 0.05 | 2.5E − 07 | −0.08 | 0.05 | 0.12 | 0.05 | 0.06 | 0.39 | 0.02 | 0.02 | 0.11 | −0.06 | 0.04 | 0.13 | −0.09 | 0.12 | 0.43 | −0.03 | 0.03 | 0.24 |
| −0.24 | 0.06 | 2.1E − 05 | 2.82 | 0.10 | 1.2E − 177 | −0.35 | 0.07 | 1.6E − 07 | −0.03 | 0.02 | 0.07 | −0.15 | 0.04 | 6.6E − 04 | −0.01 | 0.08 | 0.86 | −0.08 | 0.03 | 0.02 |
| 0.09 | 0.10 | 0.33 | −0.10 | 0.10 | 0.31 | 0.55 | 0.09 | 8.0E − 09 | −0.03 | 0.03 | 0.28 | 0.39 | 0.07 | 3.8E − 09 | 0.42 | 0.13 | 1.1E − 03 | 0.34 | 0.05 | 3.40E − 10 |
| – | – | – | – | – | – | 0.32 | 0.10 | 1.2E − 03 | – | – | – | – | – | – | 0.24 | 0.20 | 0.23 | 0.00 | 0.05 | 0.99 |
| −0.09 | 0.05 | 0.08 | 0.05 | 0.05 | 0.40 | 0.10 | 0.06 | 0.09 | 0.01 | 0.02 | 0.72 | 0.11 | 0.04 | 0.01 | 0.26 | 0.13 | 0.04 | 0.05 | 0.03 | 0.11 |
| −0.10 | 0.05 | 0.07 | 0.11 | 0.06 | 0.06 | −0.07 | 0.06 | 0.28 | −0.01 | 0.02 | 0.75 | 0.01 | 0.04 | 0.74 | 0.21 | 0.14 | 0.13 | −0.01 | 0.03 | 0.87 |
| −0.05 | 0.05 | 0.33 | 0.01 | 0.05 | 0.78 | 0.06 | 0.06 | 0.26 | 0.00 | 0.01 | 0.99 | −0.03 | 0.04 | 0.37 | −0.09 | 0.09 | 0.32 | −0.07 | 0.03 | 9.0E − 03 |
| 0.02 | 0.06 | 0.69 | −0.04 | 0.06 | 0.54 | 0.12 | 0.06 | 0.05 | 0.01 | 0.02 | 0.62 | 0.06 | 0.04 | 0.18 | −0.10 | 0.08 | 0.18 | 0.02 | 0.04 | 0.5 |
| 0.13 | 0.05 | 0.02 | −0.25 | 0.06 | 4.3E − 06 | 0.03 | 0.06 | 0.63 | 0.01 | 0.02 | 0.55 | −0.03 | 0.04 | 0.52 | 0.05 | 0.07 | 0.45 | 0.00 | 0.03 | 0.88 |
| −0.49 | 0.11 | 4.7E − 06 | 0.17 | 0.12 | 0.16 | 0.30 | 0.13 | 0.02 | 0.03 | 0.03 | 0.39 | −0.01 | 0.08 | 0.90 | 0.12 | 0.16 | 0.44 | −0.04 | 0.06 | 0.47 |
aThe SNP rs1805009 in the MC1R gene was not in 1000 Genome or HapMap imputed data.
bNot in 1000 Genome imputed data.
cNot in HapMap imputed data.
dNewly identified pigmentation SNPs in this present GWAS.
We did not find novel susceptibility loci for tanning ability, but replicated the previous findings, including rs12203592 in IRF4 (P = 3.3 × 10−23), rs1126809 in TYR (P = 5.0 × 10−21), rs12913832 in HERC2 (P = 1.4 × 10−22) and rs1805007 (P = 1.1 × 10−65) and rs1805008 (P = 1.3 × 10−13) in MC1R (Table 6).
Based on the knowledge that human pigmentation traits are host susceptibility factors for skin cancer (7), we further evaluated the association between the pigmentation determining loci and melanoma risk using the melanoma GWAS of the MD Anderson Cancer Center with 1804 melanoma cases and 1026 controls. As shown in Table 7, the SNP rs12520016 associated with intermediate eye color was associated with decreased risk of melanoma (G allele; OR, 0.74; 95% CI, 0.58–0.95; P = 0.02). The SNP rs12421680 associated with increased numbers of sunburns and lower expression of the gene NTM was associated with increased risk of melanoma (A allele; OR 1.12; 95% CI 1.00–1.26; P = 0.05). Consistent with the interaction between the SNP rs7173419 and the SNP rs12913832 on brown eye color, the protective effect of the SNP rs12913832 on melanoma risk was most significant among the carriers of CC genotype of rs7173419 (OR 0.64; 95% CI 0.53–0.77; P = 3.6 × 10−6), compared with the carriers of CT genotype (OR 0.76; 95% CI 0.61–0.94; P = 0.01) and TT genotype (OR 1.05; 95% CI 0.63–1.73; P = 0.86). The interaction was borderline significant (P-value for interaction = 0.07, Table 4). Besides, the SNP rs12203592 associated with increased number of NMSCs was also associated with increased risk of melanoma (T allele; OR 1.18; 95% CI 1.02–1.34; P = 0.02, Table 7).
Table 7.
Associations of the pigmentation SNPs identified in this study with melanoma riska
| SNP | Reference allele | Test allele | OR (95% CI) | P-valueb |
|---|---|---|---|---|
| rs975739 | T | G | 0.93 (0.83–1.04) | 0.25 |
| rs12421680 | G | A | 1.14 (1.01–1.27) | 0.02 |
| rs3002288 | G | A | 1.02 (0.92–1.14) | 0.63 |
| rs12520016 | T | G | 0.75 (0.59–0.96) | 0.02 |
| rs7173419 | C | T | 0.88 (0.77–1.00) | 0.06 |
| rs12203592 | C | T | 1.16 (1.01–1.34) | 0.03 |
aThe SNPs rs12202284 and rs8015138 were not available in the melanoma GWAS data.
bAdjusted for age, gender and the top two principal components.
In Table 6, we summarize the pigmentation SNPs identified in the previous GWASs (4–6) and the present study. We presented the associations of them with natural hair color, eye colors, tanning ability, the number of sunburns, mole counts, as well as the risk of melanoma, basal cell carcinoma (BCC), squamous cell carcinoma (SCC) and the number of NMSCs in our study population. In addition, we found that the associations of the SNPs newly identified in the present study remained similar after further adjusting for the pigmentation SNPs identified in the previous studies (rs16891982, rs12203592, rs1408799, rs35264875, rs3829241, rs1126809, rs1042602, rs12821256, rs12896399, rs12913832, rs1805007, rs1805008, rs4911414 and rs1015362, as listed in Table 6), suggesting they are independent from the previously known pigmentation SNPs (Supplementary Material, Table S4). We further screened the pair-wise interactions between these SNPs (including the ASIP haplotypes composed of rs4911414 and rs1015362) on pigmentation traits. None of them revealed significant interactions with each other on any pigmentation traits after the Bonferroni correction (P = 0.05/78 = 6.4 × 10−4).
DISCUSSION
Previously, we reported the GWASs of hair color and tanning response among 2287 women with 528 173 SNPs genotyped by the Illumina HumanHap550 (4,14), in which we identified novel loci associated with hair color and tanning response in the gene regions of IRF4 and SLC24A4 in addition to the known ones, MC1R, OCA2 and SLC45A2 (4). In the present study, we enlarged the sample size to 10 183 men and women and analyzed 8 221 074 autosomal SNPs imputed from 1000 Genomes Projects on hair color, tanning ability, the number of sunburns and the number of NMSCs. We additionally conducted a GWAS on eye color among 3871 males with 2 543 887 HapMap imputed autosomal SNPs. Using a multistage study design, we identified several novel genetic variants associated with pigmentary phenotypes and the risk of skin cancer. A novel region in the EDNRB gene was most significantly associated with natural hair color. The EDNRB gene encodes a G protein-coupled receptor that activates a phosphatidylinositol-calcium second messenger system. Studies suggest that mutations in EDNRB can cause Waardenburg syndrome, which is characterized by pigmentation abnormalities, including depigmented patches of the skin and hair, vivid blue eyes or heterochromia irides and sensorineural hearing loss (15). The requirement for EDNRB signaling pathway during melanocyte development has been reported (16,17). However, the previous studies on genetic variants were limited to the nonsynonymous mutations (15). In the present study, we identified a region downstream of this gene associated with natural human hair color, and the SNP rs975739 revealed the strongest association (P = 2.4 × 10−14). Our finding was consistent with a previous study presented at the 2010 meeting of the American Society of Human Genetics (18), reporting an association between another EDNRB SNP rs1668619 (in complete LD with rs975739) and hair color with a P-value of 1 × 10−8. The SNP rs1668619 reached a P-value of 6.8 × 10−9 in our hair color GWAS. We did not find significant association between variants in this region and melanoma risk, and the conclusion of the association between EDNRB and melanoma risk was still controversial (19,20). No association was identified for tanning ability or the number of sunburns either, suggesting the involvement of this gene in the basal pigmentation but not the response to sun exposure.
In consistent with our previous findings on the association of the IRF4 SNP rs12203592 with increased risk of all three types of skin cancer (21), we further found that this SNP was associated with an increased number of NMSCs. We also confirmed the association of this SNP with melanoma risk among an independent population from our previous report in the melanoma GWAS of the MD Anderson Cancer Center. IRF4 regulates gene expression in response to interferon and other cytokines (22) and is involved in tumor immunity (23), which is plausible to play an important role in cancer development. The other SNP rs12202284 associated with the number of NMSCs was located between IRF4 and EXOC2. We previously reported another SNP rs12210050 in this region associated with an increased risk of both BCC and SCC (24). These two SNPs (rs12202284 and rs12210050) were in modest LD (r2 = 0.77) and revealed similar effects on the number of NMSCs in the present study (rs12202284, A allele, beta = 0.15, SE = 0.03; P = 5.0 × 10−8; rs12210050, T allele, beta = 0.19, SE = 0.04, P = 8.5 × 10−8), which may represent the same causal signal. Neither of the two SNPs was in LD with the SNP rs12203592 in this study (r2 = 0.10 and 0.12, respectively).
An NTM polymorphism was associated with the number of sunburns in this study. NTM is a member of the IgLON family of immunoglobulin domain-containing glycosylphosphatidylinositol-anchored cell adhesion molecules (25), which is involved in a variety of specific cell–cell interactions in the developing nervous system (26). As for the skin, NTM was involved in the aging process as a marker of cell adhesion, and a down-regulation of NTM was detected during the replicative senescence of human dermal fibroblasts (21). Although the studies of NTM have been mostly limited to the neural system, our finding of the association between an NTM polymorphism and the number of sunburns, along with the evidence from a previous study on skin (21), suggested a promising role in cell adhesion that NTM might play in the skin besides the neural system. The lack of association between the NTM SNP with natural hair color and tanning ability further suggested a pigmentation-independent mechanism underlying sunburn reaction.
The presence of melanin pigment within the iris is responsible for the visual impression of human eye colors (12). The function of the two genes identified in eye color GWASs (VASH2 and POLS) remained unknown in terms of pigmentation. VASH2 was known to play a role in the regulation of angiogenesis (27), and POLS was involved in DNA synthesis (28). Further studies on the two genes are required to validate the involvement in pigmentation process, especially in the eyes. The association of the POLS polymorphism and melanoma risk is upon further replication, and functional studies are warranted to investigate the biological mechanisms.
The GNG2 gene was identified as a novel locus conferring susceptibility to the number of NMSCs in this study. GNG2 encodes one of the gamma subunits of a G-protein, which is involved in signaling mechanisms across membranes and has been known to play important roles in the development and progression of various cancers. The G-protein βγ subunits play an important role in melanoma cell migration by crosstalk with Epac in Ca2+ signaling (29). However, there was still lack of specific studies of GNG2 on NMSCs.
One limitation of this study is that we used self-reported pigmentation information. However, information on hair color and eye color is relatively objective, and a high reliability of self-reported tanning ability and sunburn history has been demonstrated in the previous studies (30,31). The high education level and interest in the health of cohort members further allows high quality and valid information to be collected on self-administered forms, and the self-reported pigmentation phenotypes in our cohorts significantly predicted the risk of skin cancers (including melanoma, SCC and BCC) in the previous studies (7,32,33). Another limitation on the pigmentation information was that we had categorized information only for some continuous pigmentation traits, such as hair color, which may limit our power to reveal more new pigmentation genes.
In summary, we newly identified several genes and loci associated with human pigmentation traits and skin cancer risk by conducting a series of GWASs on hair color, eye colors, tanning ability, the number of sunburns and the number of NMSCs among individuals of European ancestry. The identification of these novel genetic determinants of pigmentary phenotypes and skin cancer may help further understand the complex mechanisms of human pigmentation and the genetic basis of skin cancer. The different variants identified for different traits may explain the highly variable pigmentation characteristics in European Americans. External replication is needed to extend our finding to other populations.
MATERIALS AND METHODS
Study population
Pigmentation GWASs
In the discovery stage of the GWASs on hair color, tanning ability, the number of sunburns and the number of NMSCs, we combined data from five case–control studies nested within the Nurses' Health Study (NHS) and the Health Professionals Follow-up Study (HPFS): a postmenopausal invasive breast cancer case–control study nested within the NHS (BC_NHS, n = 2287), a type 2 diabetes case–control study nested within the NHS (T2D_NHS, n = 3116), a type 2 diabetes case–control study nested within the HPFS (T2D_HPFS, n = 2487), a coronary heart disease case–control study nested within the NHS (CHD_NHS, n = 1146) and a coronary heart disease case–control study nested within the HPFS (CHD_HPFS, n = 1147). We excluded the breast cancer cases in BC_NHS for the GWAS on the number of NMSCs. In the replication stage, a fast-track replication was conducted among 1782 healthy controls in the skin cancer case–control study nested within the NHS and HPFS (skin cancer). Two pigmentation SNPs successfully replicated in the skin cancer set were selected for a second replication among 2722 individuals from a renal function study nested within the NHS (RF_NHS). Information on eye color was available in the HPFS only, so we combined data from three GWAS sets within the HPFS, T2D_HPFS, CHD_HPFS and a kidney stone case–control study nested within the HPFS (KS_HPFS, n = 515), with a total of 3871 men in the discovery stage of eye color GWASs. Two novel SNPs with promising association were selected for replication among 2496 participants from the prostate cancer case–control study nested within the HPFS (PrCa_HPFS). Overlapped samples among the component data sets were excluded. Detailed descriptions of the study population were published previously (34–38). The study protocol was approved by the Institutional Review Board of Brigham and Women's Hospital and the Harvard School of Public Health.
Information on pigmentation traits was collected from prospective questionnaires in both the NHS and HPFS using similar wording, except for natural eye color, which was collected in the HPFS questionnaires only. We regressed ordinal coding for natural hair color (1 = blonde, 2 = light brown, 3 = dark brown, 4 = black; excluding red hair color) and tanning ability (1 = practically none, 2 = light tan, 3 = average tan, 4 = deep tan in the NHS; 1 = pain: burn/peel, 2 = burn then tan, 3 = tan without burn in the HPFS). For the hair color phenotype, we excluded the individuals with red hair because red hair color has been known as a distinct trait from other hair colors and is predominantly determined by the MC1R gene. A total of 289 (3.9%) individuals with red hair were excluded in the hair color GWAS (77 in BC_NHS, 41 in CHD_NHS, 103 in T2D_NHS, 19 in CHD_HPFS and 49 in T2D_HPFS). The number of lifetime severe sunburns was coded as a continuous variable, summarizing those on the face, limbs and back. Different eye colors have been hypothesized to be distinct phenotypes and we treated three categories of eye colors, blue, intermediate (including green) and brown eye colors, as co-dominant outcomes, comparing each with the other two, following a previous GWAS on eye color (5).
NMSC case ascertainment
For all types of skin cancers, participants reported new cases biennially. With their permission, medical records were obtained and reviewed by physicians to confirm their self-reported diagnosis. Medical records were not obtained for self-reported cases of BCC, but the validity of BCC self-reports was >90% in validation studies in our cohorts in early years. Information on the cumulative number of NMSCs was collected through 2004 in the NHS and 2008 in the HPFS (39,40). The question asked was ‘How many squamous or basal cell carcinoma lesions have you ever had removed by surgery, cryotherapy or other means? (Include only new primary cancers. Exclude melanoma and benign lesions like moles or actinic keratoses.)’. The possible choices were never, 1, 2–4, 5–10 and 11+. To minimize the misclassification of the outcome, the eligible participants in this study were Caucasians who reported at least one pathologically confirmed diagnosis of SCC or self-reported BCC in the cohort follow-up from 1984 up to 2004. We coded the number of NMSCs as a continuous variable by using the median number in each category.
Melanoma risk association study
We evaluated the association between the pigmentary SNPs identified in this study with melanoma risk using the melanoma GWAS data from MD Anderson Cancer Center with 1804 melanoma cases and 1026 controls. Details of the study population have previously been described (41). Briefly, in this hospital-based case–control study, 1804 non-Hispanic patients with newly diagnosed, histologically confirmed, untreated melanoma were consecutively recruited at the University of Texas MD Anderson Cancer Center in Houston, TX, between April 1994 and April 2008. An additional 1026 cancer-free controls matched to the cases on age (±5 years), sex and ethnicity were recruited during the same period from among visitors to MD Anderson Cancer Center who were accompanying patients to outpatient clinics but neither seeking medical care nor biologically related to the patients included in this study. After informed consent was obtained, each participant was asked to complete a questionnaire to provide information about their demographic and known risk factors for CM. A one-time blood sample (30 ml) was drawn from each study participant. All subjects included in this analysis were US non-Hispanic European descendants. The research protocol was approved by the Institutional Review Board at MD Anderson Cancer Center.
SNP selection for replication
Four novel regions revealed the strongest associations with pigmentation traits in the discovery stage of this study besides the known pigmentation loci and were selected for replication: one region on chromosome 13 with hair color (smallest P = 5.4 × 10−9), one region on chromosome 11 with the number of sunburns (smallest P = 5.3 × 10−5), one region on chromosome 1 with brown eye color (smallest P = 4.2 × 10−6) and one region on chromosome 5 with intermediate (including green) eye color (smallest P = 3.1 × 10−6). For the region on chromosome 13 associated with natural hair color GWAS, we found 117 SNPs with P < 1 × 10−7 in two LD blocks in this region (chr13:77 259 946–77 370 845). We forced in the top SNP in the regional Manhattan plot (rs975739) and selected other two tag-SNPs (rs1146927 and rs58188699) for replication using the Tagger program in Haploview 4.2 (r2 > 0.8) (42,43). A proxy SNP was selected for each of them to ensure the validity of genotyping. For each region associated with the number of sunburns, brown eye color and intermediate eye color, we selected the SNP with the strongest association as the representative. Besides, two novel regions revealed promising associations with the number of NMSCs in the discovery stage: one on chromosome 6 and the other on chromosome 14. We selected the five SNPs with the strongest associations as the representatives (two on chromosome 6 and three on chromosome 14). Additionally, the well-known pigmentation SNP rs12913832 in HECT and RLD domain containing E3 ubiquitin protein ligase 2 (HERC2) revealed the strongest association with brown eye color in our study and also a significant interaction with the SNP rs7173419 in OCA2. To validate this interaction, we further selected rs12913832 and rs7173419 for replication.
Genotyping, imputation and quality control
We previously used either the Illumina/Affymetrix SNP chip for the genotyping of the component GWAS sets (34–36). We used Illumina HumanHap550 array to perform genotyping in the BC_NHS (34), used Illumina 610Q array to perform genotyping in the KS_HPFS and used Affymetrix 6.0 array in the other four GWAS sets, including T2D_NHS, T2D_HPFS (35), as well as CHD_NHS, CHD_HPFS (36). For five of the GWAS sets (BC_NHS, T2D_NHS, T2D_HPFS, CHD_NHS and CHD_HPFS), we used the MACH program (44) to impute 8 221 074 autosomal SNPs based on haplotypes from the 1000 Genomes Project (using the August 2009 release of the Sanger Institute SNP calls based on the pilot 1 project). For the KS_HPFS set, we used the same program to impute 2 543 887 autosomal SNPs based on haplotypes from the HapMap database phase II data build 35 (CEU) (45). The HapMap imputed data of T2D_HPFS and CHD_HPFS were used for eye color GWASs to be combined with KS_HPFS. Samples from the six GWASs were imputed separately. We observed high imputation quality in each cohort's imputation. In particular, the numbers of SNPs with MACH r2 > 0.4 were 7 438 552 (90.5%) for NHS_BC, 7 399 703 (90.0%) for NHS_T2D, 7 437 076 (90.5%) for NHS_CHD, 7 370 611 (89.7%) for HPFS_T2D and 7 417 449 (90.2%) for HPFS_CHD. Most of the imputed SNPs were common [for all five cohorts, >98% SNPs have a minor allele frequency (MAF) > 0.01 and >79% SNPs have an MAF > 0.05]. SNPs with imputation r2 > 0.4 and an MAF > 0.01 in each study were included in meta-analysis. Finally, 7 588 169 SNPs were analyzed for natural hair color, tanning ability, the number of sunburns and the number of NMSCs, and 2 469 762 SNPs were analyzed for eye colors.
The promising SNPs selected from the discovery stage were genotyped in the replication sets using TaqMan/BioTrove assays at the Dana Farber/Harvard Cancer Center Polymorphism Detection Core. Laboratory personnel were blinded to the case–control status, and blinded quality control samples were inserted to validate genotyping procedures; concordance for the blinded samples was 100%. Primers, probes and conditions for genotyping assays are available upon request. All these SNPs were successfully genotyped with call rate >95%, except for the SNP rs1146926 (a proxy SNP of rs1146927), which failed in genotyping.
As for the melanoma risk association study, the samples were previously genotyped by the Illumina Omni 1M Quad V1-0_B SNP chip (41). An imputation was conducted based on the genotyped SNPs and haplotype information in the HapMap phase II data build 35 (CEU) (45). For the SNPs used in this study, the SNPs rs12913832, rs12421680, rs12520016 and rs12203592 were directly genotyped, and the SNPs rs975739, rs3002288 and rs7173419 were imputed with imputation r2 of 1.00, 0.92 and 0.98, respectively.
Statistical analysis
We used linear regression to test the association between minor allele counts and phenotypes of hair color, tanning ability, the number of sunburns and the number of NMSCs, and used logistic regression to test association separately for brown, intermediate and blue eye colors as co-dominant outcomes (comparing one with the other two). The imputed dosage data were used for the analysis. The model was adjusted for the first four principal components. Age was also adjusted for the GWASs of the number of sunburns and the number of NMSCs, but not for the other pigmentation traits, which were not likely to be confounded by age. We used ProbABEL for the GWAS analysis in this study. Associations in each component GWAS set were combined in an inverse-variance-weighted meta-analysis using the METAL software. We additionally adjusted for gender in the skin cancer replication set, which was mixed of males and females. The same meta-analysis was conducted to combine the component replication sets and to combine the discovery set and the replication set. We used logistic regression for the melanoma risk association study and adjusted for age, gender and the first two principal components. These principal components were calculated for all individuals on the basis of approximately 10 000 unlinked markers using the EIGENSTRAT software (46). We used the algorithm developed by Kraft et al. (47) to infer the haplotypes from the unphased genotypic data. The description of this algorithm was previously published, and this approach has been proved by simulation studies as well as subsequent applications in several other studies (47–49).
WEB RESOURCES
The URLs for data presented herein are as follows:
1000 Genomes Project, http://www.1000genomes.org
International HapMap Project, http://www.hapmap.org
METAL software, http://www.sph.umich.edu/csg/abecasis/Metal/index.html
SUPPLEMENTARY MATERIAL
FUNDING
This work is supported by National Institutes of Health grants CA122838, CA87969, CA055075, CA49449, CA100264 and CA093459.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr David Hunter, Dr Frank B. Hu, Dr Eric B. Rimm and Dr Gary Curhan for their work on the component GWAS sets. We thank Dr Mousheng Xu and Constance Chen for programming support, and thank Pati Soule and Dr Hardeep Ranu of the Dana Farber/Harvard Cancer Center High-Throughput Polymorphism Detection Core for sample handling and genotyping of the NHS and HPFS samples. We are grateful to Merck Research Laboratories for the funding of the GWAS of coronary heart disease. We thank Dr Jinyan Huang for improving the figure quality. We are also indebted to the participants in all of these studies. We thank the cancer registries in the following states for their help: Alabama, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Idaho, Illinois, Indiana, Iowa, Kentucky, Louisiana, Maine, Maryland, Massachusetts, Michigan, Nebraska, New Hampshire, New Jersey, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Rhode Island, South Carolina, Tennessee, Texas, Virginia, Washington and Wyoming.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Clark P., Stark A.E., Walsh R.J., Jardine R., Martin N.G. A twin study of skin reflectance. Ann. Hum. Biol. 1981;8:529–541. doi: 10.1080/03014468100005371. doi:10.1080/03014468100005371. [DOI] [PubMed] [Google Scholar]
- 2.Frisancho A.R., Wainwright R., Way A. Heritability and components of phenotypic expression in skin reflectance of Mestizos from the Peruvian lowlands. Am. J. Phys. Anthropol. 1981;55:203–208. doi: 10.1002/ajpa.1330550207. doi:10.1002/ajpa.1330550207. [DOI] [PubMed] [Google Scholar]
- 3.Gerstenblith M.R., Shi J., Landi M.T. Genome-wide association studies of pigmentation and skin cancer: a review and meta-analysis. Pigment Cell Melanoma Res. 2010;23:587–606. doi: 10.1111/j.1755-148X.2010.00730.x. doi:10.1111/j.1755-148X.2010.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han J., Kraft P., Nan H., Guo Q., Chen C., Qureshi A., Hankinson S.E., Hu F.B., Duffy D.L., Zhao Z.Z., et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. doi:10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Magnusson K.P., Manolescu A., Karason A., Palsson A., Thorleifsson G., et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. doi:10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 6.Sulem P., Gudbjartsson D.F., Stacey S.N., Helgason A., Rafnar T., Jakobsdottir M., Steinberg S., Gudjonsson S.A., Palsson A., Thorleifsson G., et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 2008;40:835–837. doi: 10.1038/ng.160. doi:10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 7.Han J., Colditz G.A., Hunter D.J. Risk factors for skin cancers: a nested case-control study within the Nurses' Health Study. Int. J. Epidemiol. 2006;35:1514–1521. doi: 10.1093/ije/dyl197. doi:10.1093/ije/dyl197. [DOI] [PubMed] [Google Scholar]
- 8.Stacey S.N., Gudbjartsson D.F., Sulem P., Bergthorsson J.T., Kumar R., Thorleifsson G., Sigurdsson A., Jakobsdottir M., Sigurgeirsson B., Benediktsdottir K.R., et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat. Genet. 2008;40:1313–1318. doi: 10.1038/ng.234. doi:10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- 9.Stacey S.N., Sulem P., Masson G., Gudjonsson S.A., Thorleifsson G., Jakobsdottir M., Sigurdsson A., Gudbjartsson D.F., Sigurgeirsson B., Benediktsdottir K.R., et al. New common variants affecting susceptibility to basal cell carcinoma. Nat. Genet. 2009;41:909–914. doi: 10.1038/ng.412. doi:10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nan H., Xu M., Kraft P., Qureshi A.A., Chen C., Guo Q., Hu F.B., Curhan G., Amos C.I., Wang L.E., et al. Genome-wide association study identifies novel alleles associated with risk of cutaneous basal cell carcinoma and squamous cell carcinoma. Hum. Mol. Genet. 2011;20:3718–3724. doi: 10.1093/hmg/ddr287. doi:10.1093/hmg/ddr287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang R.S., Duan S., Bleibel W.K., Kistner E.O., Zhang W., Clark T.A., Chen T.X., Schweitzer A.C., Blume J.E., Cox N.J., et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. doi:10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturm R.A., Larsson M. Genetics of human iris colour and patterns. Pigment Cell Melanoma Res. 2009;22:544–562. doi: 10.1111/j.1755-148X.2009.00606.x. doi:10.1111/j.1755-148X.2009.00606.x. [DOI] [PubMed] [Google Scholar]
- 13.Sturm R.A., Duffy D.L., Zhao Z.Z., Leite F.P., Stark M.S., Hayward N.K., Martin N.G., Montgomery G.W. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 2008;82:424–431. doi: 10.1016/j.ajhg.2007.11.005. doi:10.1016/j.ajhg.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nan H., Kraft P., Qureshi A.A., Guo Q., Chen C., Hankinson S.E., Hu F.B., Thomas G., Hoover R.N., Chanock S., et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. doi:10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pingault V., Ente D., Dastot-Le Moal F., Goossens M., Marlin S., Bondurand N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 31:391–406. doi: 10.1002/humu.21211. doi:10.1002/humu.21211. [DOI] [PubMed] [Google Scholar]
- 16.Hou L., Pavan W.J., Shin M.K., Arnheiter H. Cell-autonomous and cell non-autonomous signaling through endothelin receptor B during melanocyte development. Development. 2004;131:3239–3247. doi: 10.1242/dev.01193. doi:10.1242/dev.01193. [DOI] [PubMed] [Google Scholar]
- 17.Stanchina L., Baral V., Robert F., Pingault V., Lemort N., Pachnis V., Goossens M., Bondurand N. Interactions between Sox10, Edn3 and Ednrb during enteric nervous system and melanocyte development. Dev. Biol. 2006;295:232–249. doi: 10.1016/j.ydbio.2006.03.031. doi:10.1016/j.ydbio.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 18.Hinds D.A., Macpherson J.M., Tung J., Naughton B., Kiefer A., Do C., Wojcicki A., Mountain J., Eriksson N. Genetics of infectious disease susceptibility and morphological traits in a participant driven cohort. Abstract presented at the 60th Annual Meeting of The American Society of Human Genetics; 2–6 November 2010; Washington, DC: 2010. http://www.ashg.org/2010meeting/abstracts/fulltext/f22581.htm . [Google Scholar]
- 19.Fernandez L.P., Milne R.L., Pita G., Floristan U., Sendagorta E., Feito M., Aviles J.A., Martin-Gonzalez M., Lazaro P., Benitez J., et al. Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp. Dermatol. 2009;18:634–642. doi: 10.1111/j.1600-0625.2009.00846.x. doi:10.1111/j.1600-0625.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- 20.Soufir N., Meziani R., Lacapere J.J., Bertrand G., Fumeron F., Bourillon A., Gerard B., Descamps V., Crickx B., Ollivaud L., et al. Association between endothelin receptor B nonsynonymous variants and melanoma risk. J. Natl Cancer. Inst. 2005;97:1297–1301. doi: 10.1093/jnci/dji253. doi:10.1093/jnci/dji253. [DOI] [PubMed] [Google Scholar]
- 21.Yoon I.K., Kim H.K., Kim Y.K., Song I.H., Kim W., Kim S., Baek S.H., Kim J.H., Kim J.R. Exploration of replicative senescence-associated genes in human dermal fibroblasts by cDNA microarray technology. Exp. Gerontol. 2004;39:1369–1378. doi: 10.1016/j.exger.2004.07.002. doi:10.1016/j.exger.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Lu R. Interferon regulatory factor 4 and 8 in B-cell development. Trends Immunol. 2008;29:487–492. doi: 10.1016/j.it.2008.07.006. doi:10.1016/j.it.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaffer A.L., Emre N.C., Romesser P.B., Staudt L.M. IRF4: Immunity. Malignancy! Therapy? Clin. Cancer Res. 2009;15:2954–2961. doi: 10.1158/1078-0432.CCR-08-1845. doi:10.1158/1078-0432.CCR-08-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Struyk A.F., Canoll P.D., Wolfgang M.J., Rosen C.L., D'Eustachio P., Salzer J.L. Cloning of neurotrimin defines a new subfamily of differentially expressed neural cell adhesion molecules. J. Neurosci. 1995;15:2141–2156. doi: 10.1523/JNEUROSCI.15-03-02141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellar G.C., Watt K.P., Rabiasz G.J., Stronach E.A., Li L., Miller E.P., Massie C.E., Miller J., Contreras-Moreira B., Scott D., et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat. Genet. 2003;34:337–343. doi: 10.1038/ng1183. doi:10.1038/ng1183. [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Li G., Peng X., Liu B., Yin B., Tan X., Fan M., Fan W., Qiang B., Yuan J. The cloning and preliminarily functional analysis of the human neurotrimin gene. Sci. China C Life Sci. 2004;47:158–164. doi: 10.1360/03yc0072. doi:10.1360/03yc0072. [DOI] [PubMed] [Google Scholar]
- 27.Kimura H., Miyashita H., Suzuki Y., Kobayashi M., Watanabe K., Sonoda H., Ohta H., Fujiwara T., Shimosegawa T., Sato Y. Distinctive localization and opposed roles of vasohibin-1 and vasohibin-2 in the regulation of angiogenesis. Blood. 2009;113:4810–4818. doi: 10.1182/blood-2008-07-170316. doi:10.1182/blood-2008-07-170316. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z., Castano I.B., De Las Penas A., Adams C., Christman M.F. Pol kappa: a DNA polymerase required for sister chromatid cohesion. Science. 2000;289:774–779. doi: 10.1126/science.289.5480.774. doi:10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- 29.Baljinnyam E., Umemura M., De Lorenzo M.S., Xie L.H., Nowycky M., Iwatsubo M., Chen S., Goydos J.S., Iwatsubo K. Gβγ subunits inhibit Epac-induced melanoma cell migration. BMC Cancer. 2011;11:256. doi: 10.1186/1471-2407-11-256. doi:10.1186/1471-2407-11-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westerdahl J., Anderson H., Olsson H., Ingvar C. Reproducibility of a self-administered questionnaire for assessment of melanoma risk. Int. J. Epidemiol. 1996;25:245–251. doi: 10.1093/ije/25.2.245. doi:10.1093/ije/25.2.245. [DOI] [PubMed] [Google Scholar]
- 31.Berwick M., Chen Y.T. Reliability of reported sunburn history in a case-control study of cutaneous malignant melanoma. Am. J. Epidemiol. 1995;141:1033–1037. doi: 10.1093/oxfordjournals.aje.a117367. [DOI] [PubMed] [Google Scholar]
- 32.Qureshi A.A., Zhang M., Han J. Heterogeneity in host risk factors for incident melanoma and non-melanoma skin cancer in a cohort of US women. J. Epidemiol. 2011;21:197–203. doi: 10.2188/jea.JE20100145. doi:10.2188/jea.JE20100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho E., Rosner B.A., Feskanich D., Colditz G.A. Risk factors and individual probabilities of melanoma for whites. J. Clin. Oncol. 2005;23:2669–2675. doi: 10.1200/JCO.2005.11.108. doi:10.1200/JCO.2005.11.108. [DOI] [PubMed] [Google Scholar]
- 34.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. doi:10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi L., Cornelis M.C., Kraft P., Stanya K.J., Linda Kao W.H., Pankow J.S., Dupuis J., Florez J.C., Fox C.S., Pare G., et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum. Mol. Genet. 2010;19:2706–2715. doi: 10.1093/hmg/ddq156. doi:10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rimm E.B., Giovannucci E.L., Willett W.C., Colditz G.A., Ascherio A., Rosner B., Stampfer M.J. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–468. doi: 10.1016/0140-6736(91)90542-w. doi:10.1016/0140-6736(91)90542-W. [DOI] [PubMed] [Google Scholar]
- 37.Taylor E.N., Stampfer M.J., Curhan G.C. Obesity, weight gain, and the risk of kidney stones. JAMA. 2005;293:455–462. doi: 10.1001/jama.293.4.455. doi:10.1001/jama.293.4.455. [DOI] [PubMed] [Google Scholar]
- 38.Forman J.P., Fisher N.D., Schopick E.L., Curhan G.C. Higher levels of albuminuria within the normal range predict incident hypertension. J. Am. Soc. Nephrol. 2008;19:1983–1988. doi: 10.1681/ASN.2008010038. doi:10.1681/ASN.2008010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin W., Qureshi A.A., Kraft P., Nan H., Guo Q., Hu F.B., Jensen M.K., Han J. ASIP genetic variants and the number of non-melanoma skin cancers. Cancer Causes Control. 2011;22:495–501. doi: 10.1007/s10552-010-9724-1. doi:10.1007/s10552-010-9724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei-Passanese E.X., Han J., Lin W., Li T., Laden F., Qureshi A.A. Geographical variation in residence and risk of multiple nonmelanoma skin cancers in US women and men. Photochem. Photobiol. 2012;88:483–489. doi: 10.1111/j.1751-1097.2012.01077.x. doi:10.1111/j.1751-1097.2012.01077.x. [DOI] [PubMed] [Google Scholar]
- 41.Amos C.I., Wang L.E., Lee J.E., Gershenwald J.E., Chen W.V., Fang S., Kosoy R., Zhang M., Qureshi A.A., Vattathil S., et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet. 2011;20:5012–5023. doi: 10.1093/hmg/ddr415. doi:10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlson C.S., Eberle M.A., Rieder M.J., Yi Q., Kruglyak L., Nickerson D.A. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 2004;74:106–120. doi: 10.1086/381000. doi:10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stram D.O., Leigh Pearce C., Bretsky P., Freedman M., Hirschhorn J.N., Altshuler D., Kolonel L.N., Henderson B.E., Thomas D.C. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum. Hered. 2003;55:179–190. doi: 10.1159/000073202. doi:10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Ann. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. doi:10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchini J., Donnelly P., Cardon L.R. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat. Genet. 2005;37:413–417. doi: 10.1038/ng1537. doi:10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 46.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 47.Kraft P., Cox D.G., Paynter R.A., Hunter D., De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet. Epidemiol. 2005;28:261–272. doi: 10.1002/gepi.20061. doi:10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 48.Zhai R., Gong M.N., Zhou W., Thompson T.B., Kraft P., Su L., Christiani D.C. Genotypes and haplotypes of the VEGF gene are associated with higher mortality and lower VEGF plasma levels in patients with ARDS. Thorax. 2007;62:718–722. doi: 10.1136/thx.2006.069393. doi:10.1136/thx.2006.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feigelson H.S., Cox D.G., Cann H.M., Wacholder S., Kaaks R., Henderson B.E., Albanes D., Altshuler D., Berglund G., Berrino F., et al. Haplotype analysis of the HSD17B1 gene and risk of breast cancer: a comprehensive approach to multicenter analyses of prospective cohort studies. Cancer Res. 2006;66:2468–2475. doi: 10.1158/0008-5472.CAN-05-3574. doi:10.1158/0008-5472.CAN-05-3574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


