Abstract
Background and Aims
Green kiwifruit (Actinidia deliciosa) retain high concentrations of chlorophyll in the fruit flesh, whereas in gold-fleshed kiwifruit (A. chinensis) chlorophyll is degraded to colourless catabolites during fruit development, leaving yellow carotenoids visible. The plant hormone group the cytokinins has been implicated in the delay of senescence, and so the aim of this work was to investigate the link between cytokinin levels in ripening fruit and chlorophyll de-greening.
Methods
The expression of genes related to cytokinin metabolism and signal transduction and the concentration of cytokinin metabolites were measured. The regulation of gene expression was assayed using transient activation of the promoter of STAY-GREEN2 (SGR2) by cytokinin response regulators.
Key Results
While the total amount of cytokinin increased in fruit of both species during maturation and ripening, a high level of expression of two cytokinin biosynthetic gene family members, adenylate isopentenyltransferases, was only detected in green kiwifruit fruit during ripening. Additionally, high levels of O-glucosylated cytokinins were detected only in green kiwifruit, as was the expression of the gene for zeatin O-glucosyltransferase, the enzyme responsible for glucosylating cytokinin into a storage form. Season to season variation in gene expression was seen, and some de-greening of the green kiwifruit fruit occurred in the second season, suggesting environmental effects on the chlorophyll degradation pathway. Two cytokinin-related response regulators, RRA17 and RRB120, showed activity against the promoter of kiwifruit SGR2.
Conclusions
The results show that in kiwifruit, levels of cytokinin increase markedly during fruit ripening, and that cytokinin metabolism is differentially regulated in the fruit of the green and gold species. However, the causal factor(s) associated with the maintenance or loss of chlorophyll in kiwifruit during ripening remains obscure.
Keywords: Actinidia deliciosa, A. chinensis, chlorophyll degradation, cytokinin, fruit ripening, kiwifruit, STAY-GREEN, transcription factor
INTRODUCTION
Kiwifruit (Actinidia deliciosa ‘Hayward’) is unusual among fleshy fruits in maintaining high chlorophyll levels throughout ripening, and appears as a bright green fruit when cut open. In contrast, chlorophyll degradation occurs in A. chinensis ‘Hort16A’, with high quality fruit having a golden yellow flesh. The immature fruit of both species are green (McGhie and Ainge, 2002). Chlorophyll is degraded in the gold fruit to colourless catabolites, leaving the yellow carotenoid pigments visible. Chlorophyll degradation occurs to a much lesser extent in green fruit. We have shown that the differential control of chlorophyll degradation in these two species is likely to be upstream of the chlorophyll degradation pathway, as the genes for chlorophyll degradation enzymes are expressed and functional in both species, but at a lower level in green kiwifruit (Pilkington et al., 2012).
While the cytokinins are now considered to be involved in diverse processes including the release of apical dominance, shoot meristem function and signalling (Jameson, 2003), they have been implicated in the delay of senescence from the time of their first discovery, where cytokinins applied to senescing leaves led to re-greening (Richmond and Lang, 1957). Subsequently, cytokinins have been shown to promote the reversal of senescence by stimulating the expression of genes for the re-differentiation of senescent plastids (gerontoplasts) into chloroplasts (Robson et al., 2004). They are known to control the activity of genes for light-harvesting chlorophyll-binding proteins (Kulaeva et al., 1996), and to stimulate chloroplast-encoded transcription in detached barley leaves (Zubo et al., 2008). However, little is known of their activity during fruit ripening. In green kiwifruit, a peak of cytokinin activity occurs during the cell division phase (Lewis et al., 1996), as also shown for other fruits (Ozga and Reinecke, 2003). In addition, a significant amount of cytokinin was detected in mature fruit (Lewis et al., 1996). We hypothesized that the differential de-greening in green and gold kiwifruit may be a consequence of differential activity associated with cytokinins.
Cytokinins are produced from isoprenoid side chains derived from the methylerythritol phosphate (MEP) pathway. The cis-forms of cytokinins associated with some tRNA moieties are derived from a second pathway, the mevalonate (MVA) pathway. Cytokinin activity in any particular organ is regulated through processes including de novo synthesis, activation, conjugation and degradation (Kudo et al., 2010), as well as the differential activity of different gene family members. The first committed step of the biosynthetic pathway is catalysed by a multigene family coding for cytokinin synthase, adenylate isopentenyltransferase (IPT). Nine IPT gene family members have been identified in Arabidopsis thaliana (Kakimoto, 2001; Takei et al., 2001). Seven of these utilize the MEP pathway, whereas AtIPT2 and AtIPT9 utilize the MVA pathway (Miyawaki et al., 2006).
Like the IPT genes, the genes coding for cytokinin oxidase/dehydrogenase (CKX) also exist as a multigene family (Schmülling et al., 2003). CKX degrades cytokinins by removal of the side chain. The preferred substrates of CKX are the free bases and corresponding ribosides, but not their dihydro derivatives (Sakakibara, 2006). However, the 9-glucosides and nucleotides have also been shown to be effectively cleaved by individual CKX isoforms. Aromatic cytokinins are also degraded, but with lower reaction rates, as reviewed in Spíchal (2012).
Conjugation can occur through O-glucosyltransferases converting, amongst others, trans-zeatin to O-glucosylzeatin (tZOG), which is resistant to degradation by CKX, and is regarded as a storage form of cytokinin able to be re-activated by β-glucosidases (Mok et al., 2000).
Cytokinin is perceived via a phosphorelay, which leads to the activation of cytokinin response regulators, the RRs. The Type-A Arabidopsis RRs (ARRs) have short C-terminal domains, are transcriptionally upregulated by cytokinin treatment and are negative regulators of cytokinin signalling. The Type-B ARRs contain DNA-binding and trans-activating domains at the C-terminus that positively regulate transcription of cytokinin-activated targets, including the Type-A ARRs (To and Kieber, 2008). Emerging evidence indicates that complex transcriptional cascades play an important role in the response to cytokinin, including changes in the expression of the components of the cytokinin signalling, biosynthetic and metabolic pathways, as well as the induction of various transcription factors (Argueso et al., 2010). It has recently been proposed that in species other than Arabidopsis, the Type-A response regulators should be referred to as RRAs and the Type-B response regulators as RRBs (Heyl et al., 2013), where the kiwifruit genes are named Ax for Actinidia species where appropriate.
In this study, we examined not only the endogenous cytokinin content in green and gold kiwifruit but also the expression of key cytokinin biosynthesis and metabolism genes during fruit development. Additionally, expression of genes encoding RRs was measured throughout fruit development, and the interaction of selected RR genes with the promoter of STAY-GREEN2 (SGR2) was assayed to determine whether the cytokinin signalling pathway interacted with a key step in the chlorophyll degradation pathway within the fruit. SGR2 was selected as it had been shown to be the same in sequence but somewhat less expressed in ripening green fruit compared with ripening yellow fruit (Pilkington et al., 2012), and was thus a potential candidate for differential regulation by RR genes.
MATERIALS AND METHODS
Measurement of outer pericarp flesh colour
In 2009 and 2010, ten fruit were collected at random from one vine from each species [green Actinidia deliciosa (A.Chev.) C.F.Liang & A.R.Ferguson ‘Hayward’ and gold A. chinensis Planch. ‘Hort16A’] grown under normal commercial management at the Plant & Food Research orchard, Te Puke, New Zealand. Sampling dates for each cultivar are given as days after full bloom (90 % flowers open; DAFB). Kiwifruit are considered to be maturing before the harvest date as determined by the industry standard for colour in gold kiwifruit and for soluble sugar content in green kiwifruit, and considered to be ripening after the harvest date as marked by arrows in Supplementary Data Fig. S1. Each fruit was measured for the fruit characteristics of colour, firmness, soluble sugars, dry matter and weight, as previously described in Pilkington et al. (2012). Fruit data from the 2010 series have previously been published (Pilkington et al., 2012), and are presented with the unpublished fruit data from 2009 in Supplementary Data Fig. S1. The outer pericarp tissues of the same fruit were cut from the skin and locules, to ensure that only the outer pericarp tissue was harvested, snap-frozen in liquid nitrogen and stored at –80 °C. The outer pericarp tissue of ten fruit was pooled for each sampling point, and a second year of fruit was collected as a biological replicate.
Sample extraction for high-performance liquid chromatography–tandem mass spectometry (HPLC-MS/MS)
The extraction method was employed as described previously in Quesnelle and Emery (2007) with minor modifications. The biological samples were homogenized to a powder with liquid nitrogen using a mortar and pestle and freeze-dried. The samples were weighed (approx 100 g d. wt) and 1 mL of cold modified Bieleski's solvent (methanol/water/formic acid: 75/20/5, v/v/v) was added. The tissue samples were then spiked with 10 ng of labelled internal cytokinin standard mix (2H7BA, 2H7[9R]cBA, 2H5ZOG, 2H3DHZOG, 2H5[9R]ZOG, 2H3[9R]DHZOG, 2H6iP7G, 2H5Z9G, 2H5MeSZ, 2H6MeSiP, 2H5[9R]MeZ, 2H5[9R]MeSiP, 2H6[9R]iP, 2H5[9R]Z, 2H3[9R]DHZ, 2H6iP, 2H3DHZ, 2H5Z, 2H6iPMP, 2H5ZRMP and 2H3DHZRMP) (OlchemIm Ltd, Olomouc, Czech Republic). Samples were vortexed, sonicated for 1 min and allowed to extract further passively overnight (approx. 12 h) at –20 °C. After extraction, samples were centrifuged at 8400 g for 10 min and the supernatants were transferred to 1·5 mL microcentrifuge tubes. Supernatants were dried using a speed vacuum concentrator at ambient temperature (UVS400, Thermo Fisher Scientific, FL, USA).
Column purification and HPLC-MS/MS conditions
Extraction residues were reconstituted in 1 mL of 1 m formic acid to ensure complete protonation of all cytokinins. Each extract was purified on a mixed mode, reverse-phase, cation-exchange cartridge (Oasis MCX 6cc; Waters, Ontario, Canada). Cartridges were activated using 5 mL of methanol and equilibrated using 5 mL of 1 m formic acid. After equilibration, the sample was loaded and washed with 5 mL of 1 m formic acid. First, the samples were washed using 5 mL of methanol. Nucleotide forms of cytokinin were eluted next using 5 mL of 0·35 m ammonium hydroxide. Finallly, cytokinin free base, ribosides and glucosides were eluted using 5 mL of 0·35 m ammonium hydroxide in 60 % (v/v) methanol. All samples were evaporated to dryness in a speed vacuum concentrator at ambient temperature and subsequently stored at –20 °C.
Because it was not possible to measure cytokinin nucleotides directly, they were dephosphorylated to form ribosides using 3·4 U of bacterial alkaline phosphatase (Sigma, Ontario, Canada) in 1 mL of 0·1 m ethanolamine-HCl (pH 10·4) for 12 h at 37 °C. The resulting cytokinin ribosides were brought to dryness in a speed vacuum concentrator at ambient temperature. The dephosphorylated nucleotides were reconstituted in 1·5 mL of water for further purification on a reversed-phase C18 column (Oasis C18 3 cc; Waters). Columns were activated using 3 mL of methanol and equilibrated with 6 mL of water. The samples were loaded onto the C18 cartridge and passed through the column under gravity. The sorbent was washed with 3 mL of water and analytes were eluted using 1·5 mL of methanol:water (80:20, v/v). All sample eluents were dried in a speed vacuum concentrator at ambient temperature and stored at –20 °C until analysis. Prior to mass spectrometric analysis, cytokinin nucleotide, cytokinin riboside and free base fractions were reconstituted with 1 mL of HPLC mobile phase starting conditions [95:5 water:methanol with 0·08 % (v/v) acetic acid] and transferred to glass autosampler vials. Samples were stored at 4 °C until analysis. The nucleotide analysis did not differentiate between -triphosphates, -diphosphates and/or -monophosphates.
HPLC-MS/MS
All metabolites were separated and analysed using a Dionex Ultimate 3000 HPLC system coupled to a Qtrap 5500 triple quadrupole hybrid ion-trap mass spectrometer (MDS Sciex, Ontario, Canada) equipped with a turbo V-spray source in positive ion mode. A 20 µL aliquot of each sample was injected on a Genesis C18 reversed-phase column (4 µm, 150 × 2·1 mm; Jones Chromatography, Foster City, CA, USA) and the cytokinins were eluted with an increasing gradient of acetonitrile (A) mixed with 0·1 % formic acid in 20 mm ammonium acetate (v/v) at a pH adjusted to 4·0 (B) at a flow rate of 0·2 mL min−1. The initial conditions were 8 % A and 92 % B, changing linearly after 5 min to 15 % A and 85 % B for 2 min, followed by 100 % A for 2 min, then linearly returning back to initial conditions for 2 min.
The HPLC effluents were introduced into the turbo V-spray source using conditions specific for each analyte, where quantification was obtained by multiple reaction monitoring of the protonated intact precursor molecule [M + H]+ and a specific product ion (Schoor et al., 2011). All data were analysed and processed using Analyst version 1·5 software. Concentrations were calculated on the basis of the peak areas for the endogenous compounds relative to those determined for the internal standards.
Reverse transcription–quantitative PCR (RT–qPCR)
Kiwifruit flesh was sampled as described above where, for each cultivar, the tissue from the ten fruit from each sampling point was pooled and RNA was isolated from kiwifruit samples using the pinetree method (Chang et al., 1993). The RNA was DNase treated (Ambion, Applied Biosciences, Auckland, New Zealand) and first-strand cDNA synthesis was carried out using oligo(dT) according to the manufacturer's instructions (Transcriptor; Roche Diagnostics, Auckland, New Zealand).
Reverse transcription–qPCR and primer design was carried out as previously described in Pilkington et al. (2012). Oligonucleotide primers were designed and tested by end-point PCR to ensure identity with kiwifruit source sequence and identity between species. Actinidia expressed sequence tags (ESTs) with homology to Arabidopsis (or other species) cytokinin enzymes and ARRs were mined by BLAST match from the Plant & Food Research EST database (Table 1) (Crowhurst et al., 2008). The nine members of the Arabidopsis IPT family are shown in the phylogenetic tree along with kiwifruit IPT genes (Supplementary Data Fig. S2). AtIPT1, AtIPT4 and AtIPT8 have the same kiwifruit EST as their best BLAST match. This EST was, therefore, subsequently referred to as IPT. AtIPT2 had a different kiwifruit EST as its best BLAST match. However, this gene was not included in the RT–qPCR work as the cytokinins detected in the present study were predominantly trans-isomers and AtIPT2 and AtIPT9 are known to be involved with the production of the cis-isomers (Miyawaki et al., 2006). The expression of IPT3 was also measured (Supplementary Data Fig. S3). Error bars shown in RT–qPCR data are for technical replicates, representing the means ± s.e. of four replicate RT–qPCRs.
Table 1.
Kiwifruit gene candidates for the control of chlorophyll levels
| Gene | Gene name | Kiwifruit source | Reference BLAST match | GenBank accession no. |
|---|---|---|---|---|
| Cytokinin genes | ||||
| IPT | Isopentenyl transferase/cytokinin synthase | A. deliciosa ripe fruit | At3g23630 | FG525359 |
| CKX | Cytokinin oxidase/dehydrogenase | A. eriantha young fruit | At1g75450 | FG422203 |
| ZOG | Zeatin O-glucosidase | A. deliciosa ripe fruit | At1g22380 | Plant & Food Research EST 99598 |
| BGLU | β-Glucosidase | A. deliciosa ripe fruit | At5g36890 | FG441373 |
| RRs | ||||
| ARR2 | Arabidopsis response regulator | A. chinensis young fruit | At4g16110 | FG519734 |
| ARR4 | Arabidopsis response regulator | A. chinensis flower | At1g10470 | Plant & Food Research EST 1833785 |
| ARR6 | Arabidopsis response regulator | A. chinensis young fruit | At5g62920 | FG518227 |
| ARR9 | Arabidopsis response regulator | A. deliciosa flower | At3g57040 | FG468906 |
| ARR11 | Arabidopsis response regulator | A. chinensis ripe fruit | At1g67710 | FG455801 |
| ARR12 | Arabidopsis response regulator | A. chinensis meristems | At2g25180 | FG497476 |
| ARR120 | Arabidopsis response regulator | A. chinensis meristems | At2g25180 | FG489257 |
| ARR13 | Arabidopsis response regulator | A. chinensis young fruit | At2g27070 | Plant & Food Research EST 2232475 |
| ARR17 | Arabidopsis response regulator | A. polygama petal | At3g56380 | FG484527 |
Genes, gene names, tissue source from which the primary expressed gene was isolated, best Arabidopsis BLAST match and GenBank accession numbers are shown.
Construction of sequence alignments and phylogenetic trees
Phylogenetic and molecular evolutionary studies were carried out using MEGA 4 (Tamura et al., 2007). Bootstrap values from 100 bootstraps were included when >50 %.
Transformation of Agrobacterium
Agrobacterium tumefaciens GV3101 (MP90) was transformed by electroporation with pHEX2 containing ESTs of interest (Hellens et al., 2005). Competent cells (50 µL aliquots) were thawed on ice and plasmid DNA (50–200 ng in 1 µL water) was added, mixed gently, and pipetted into a pre-chilled electroporation cuvette (0·2 cm gap, Bio-Rad Laboratories, Philadelphia, PA, USA). Electroporation was carried out in a GenePulser (Bio-Rad Laboratories) at a voltage of 2·5 kV (capacitance, 25 µFd; resistance, 400 Ω) with a typical pulse time of 7–9 ms. The cells were recovered by addition of 1 mL of LB, transferred to 1·5 mL tubes and incubated at room temperature, with shaking (60 rpm), for 2 h. Aliquots of 10 µL were spread onto separate LB plates containing the appropriate antibiotics. Plates were grown at 30 °C for 48 h.
SGR promoter isolation
Upstream DNA, immediately adjacent to the start codon (ATG) of AcSGR, was isolated from kiwifruit genomic DNA by PCR genome walking based on the GenomeWalker™ kit (BD Biosciences Clontech, CA, USA) protocol, with the following method. Genomic DNA was isolated as described above. Seven libraries were constructed from high quality genomic DNA preparations by digestion with seven restriction enzymes (DraI, Ecl13611, EcoRV, ApaI, ScaI, SspI and StuI), leaving blunt ends, to which the GenomeWalker™ adaptor was ligated. Nested primers to the coding region of AcSGR were designed and used in conjunction with adaptor primers for primary and secondary PCR, following the manufacturer's method for GenomeWalker™. A second genome walk was performed, with nested primers based on the sequence from the first walk. The PCR products were cloned using the pGEM-T easy cloning vector as described above, and the sequences were aligned using Vector NTI version 9·0 (Invitrogen, Auckland, New Zealand). This resulted in approx. 1 kb of confirmed upstream sequence from the transcription start site. The plasmid DNA was then digested, and the gene of interest ligated into pGREEN 0800-LUC (Hellens et al., 2005) using a rapid ligation kit (Roche Diagnostics, Auckland, New Zealand).
Dual luciferase assay of transiently transformed N. benthamiana leaves
Transient assays were carried out using the methods described previously in Hellens et al. (2005). Nicotiana benthamiana plants were grown under glasshouse conditions at 22 °C using natural light with daylight extension to 16 h, until at least four leaves were available for infiltration with Agrobacterium. Plants were maintained in the glasshouse for the duration of the experiment. Transformed Agrobacterium cultures containing pGREENII 0800-LUC reporter cassettes with a candidate promoter insert, as described in Hellens et al. (2005), or pHEX2S with a candidate transcription factor insert were cultured on LB plates containing the appropriate antibiotics and incubated at 28 °C. A 10 µL loop of bacteria was re-suspended in 10 mL of infiltration buffer (10 mm MgCl2, 0·5 µm acetosyringone) to an OD600 of 0·6–0·8, and incubated at room temperature without shaking for 2 h before infiltration. Infiltrations were performed according to the methods described by Voinnet et al. (2003). Approximately 150 µL of this Agrobacterium mixture was infiltrated at two points per young, fully expanded leaf of N. benthamiana. Two leaves were infiltrated per gene, and randomized to different plants.
Transient expression was assayed 3 d after inoculation. Two replicates were taken per leaf of 2 mm diameter leaf discs and crushed in 50 µL of phosphate-buffered saline (PBS) (6·3 mm Na2PO4, 0·2 mm Na2HPO4, 0·2 mm KH2PO4, 2·6 mm KCl, 138 mm NaCl, pH 7·4). Plate-based assays were conducted using a Victor Luminometer, according to the manufacturer's instructions for the dual luciferase assay, using the Dual Glow assay reagents for firefly luciferase (LUC) and Renilla luciferase (REN) (Targeting Systems, San Diego, CA, USA). The fluorescence of LUC was measured relative to REN, where an increase in the LUC:REN ratio indicates an activation of the candidate promoter by candidate transcription factors. A one-way analysis of variance (ANOVA) was performed, followed by a Tukey post-hoc test, to determine if there were significant differences between the means.
RESULTS
Endogenous cytokinins in green and gold fruit
The endogenous cytokinins were measured in the outer pericarp tissue of green and gold fruit in both 2009 and 2010 (Fig. 1). A wide range of different cytokinin forms was detected including the nucleotides of zeatin (tZ), dihydrozeatin (DZ) and isopentenyladenine (iP) (Fig. 2; Supplementary Data Table S1). The free base and ribosides of tZ, DZ and iP were also detected (Fig. 2). The O-glucosides of tZR and DZR (Fig. 2) were detected, as were tZ, DZ and tZ9G (Supplementary Data Table S1). Very low levels of cisZR were detected, the cis-isomer of tZ was not detected, and only low levels of cisZNT were detected (Supplementary Data Table S1).
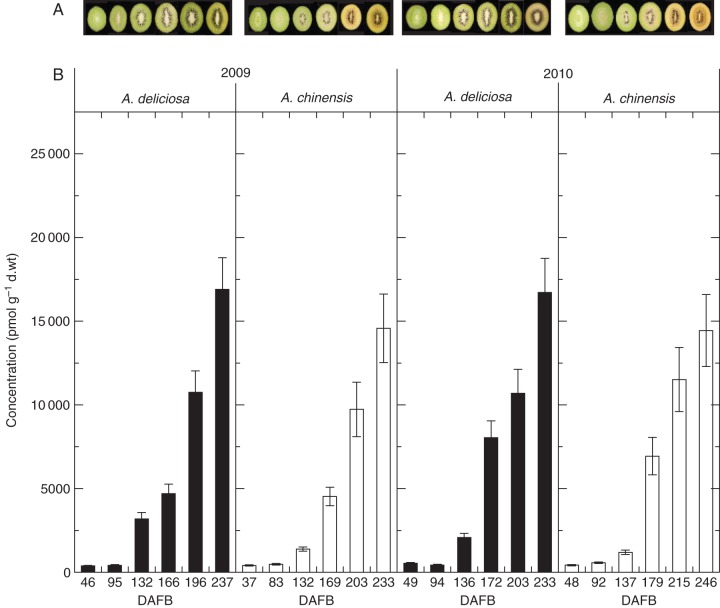
Fig. 1.
Colour change and total cytokinin concentration in green and gold kwifruit during fruit development in 2009 and 2010. (A) In 2009, the photographs of green fruit were taken at 46, 95, 132, 166, 196 and 237 days after full bloom (DAFB) (left to right) and the photographs of gold fruit were taken at 37, 83, 132, 169, 203 and 233 DAFB (left to right). In 2010, the photographs of green fruit were taken at 49, 94, 136, 172, 203 and 238 DAFB (left to right) and the photographs of gold fruit were taken at 48, 92, 137, 179, 215 and 246 DAFB (left to right) in 2010. (B) Total cytokinin complement in green and gold fruit in 2009 (left) and 2010 (right). Green fruit samples are shown as black columns and gold fruit samples are shown as white columns. Bars are one standard error above and below the mean of technical replicates (n = 3).
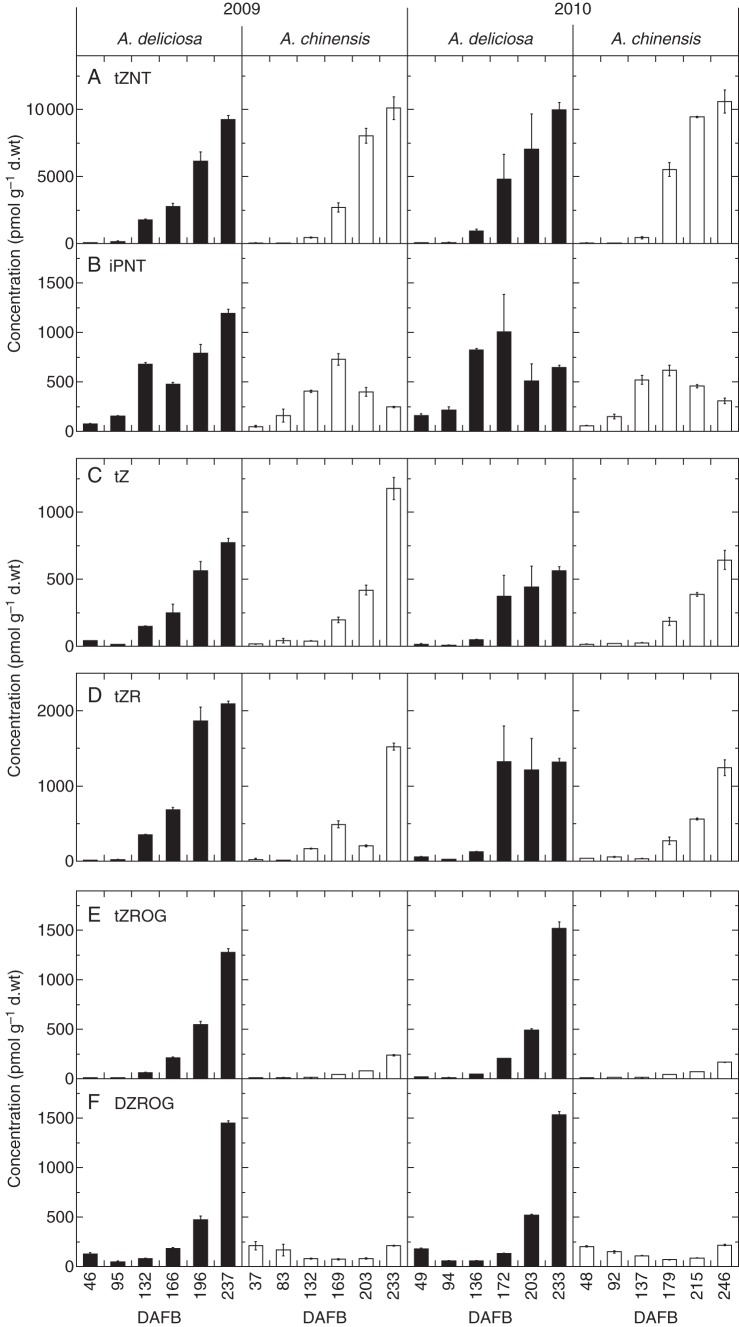
Fig. 2.
Concentration of major cytokinin metabolites in green and gold kiwifruit during fruit development in 2009 (left) and 2010 (right). Green fruit samples are shown as black columns and gold fruit samples as white columns. (A) Cytokinin nucleotides, (B) free bases, (C) ribosides and (D) O-glucosides. Bars are one standard error above and below the mean of technical replicates (n = 3). DAFB, days after full bloom.
While the total concentration of cytokinin in the fruit was similar between species (Fig. 1), the nature of some of the constituent cytokinins differed distinctly between species. The nucleotide tZNT reached the highest concentration of any of the cytokinins detected; its levels were similar between species and increased during ripening (Fig. 2). The DZNT and iPNT concentrations were also high, but at a concentration >10-fold lower (Supplementary Data Table S1). The DZNT concentration also increased during ripening, whereas the amount of iPNT decreased in gold fruit, while the levels of iPNT either continued to increase in green fruit as they ripened (2009) or were decreasing (2010). At this late stage of ripening, gold A. chinensis fruit had de-greened and green A. deliciosa fruit were beginning to de-green in 2010, as shown by the decreasing hue angle (Supplementary Data Fig. S1).
The pattern of concentration of the free bases is mirrored in the concentrations of the cytokinin ribosides (Fig. 2; Supplementary Data Table S1). Both tZ and tZR occurred at greater concentrations compared with iPR > DZR ≫ cisZR > DZ > iP, with tZR accumulating at an earlier stage of ripening in green compared with gold fruit. This pattern is mirrored by DZR, albeit at a lower level (Supplementary Data Table S1). The concentration of tZ was high and increased during ripening in both green and gold kiwifruit.
The most notable difference is in the conjugation of cytokinins, with both tZROG and DZROG concentrations increasing in green fruit to very high levels during ripening (Fig. 2). In contrast, gold fruit contained very little tZROG or DZROG. tZOG also followed this trend, but at concentrations 10-fold lower than DZROG and tZROG (Supplementary Data Table S1). Small amounts of DZOG accumulated in both species and, interestingly, tZ9G increased late in ripening but in gold fruit only (Supplementary Data Table S1).
Expression of cytokinin metabolic pathway genes in green and gold kiwifruit
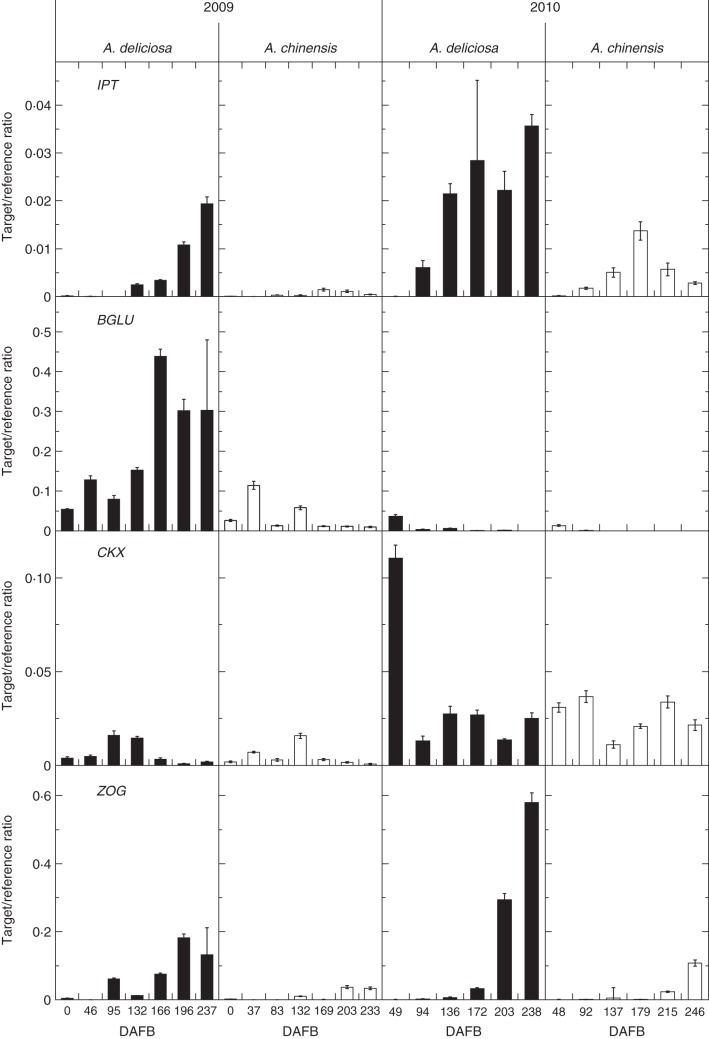
An IPT phylogenetic tree, constructed to select an IPT gene candidate for the RT–qPCR analysis, showed that published kiwifruit ESTs (Crowhurst et al., 2008) contained five IPT genes, clustering into three clades (Supplementary Data Fig. S2). One IPT family member was selected from each of Clades I and II, as Clade III, containing AtIPT2 and AtIPT9, is likely to be involved in cisZ biosynthesis (Miyawaki et al., 2006). The expression profile for IPT from Clade I showed an increase towards ripening in green fruit, but this was not seen in gold fruit, where expression remained low (Fig. 3). In 2010, the expression of IPT had a higher target/reference ratio when compared with the 2009 data but had a similar trend, where the expression increased during ripening in green fruit (Fig. 3). The second IPT family member was selected from Clade II, and showed a similar expression pattern in fruit harvested in 2009 (Supplementary Data Fig. S3).
Fig. 3.
Expression of cytokinin biosynthetic and metabolism genes in green and gold fruit during development in 2009 (left) and 2010 (right) measured as a target/reference ratio. Green fruit samples are shown as black columns and gold fruit samples as white columns. Bars are one standard error above and below the mean (n = 4). DAFB, days after full bloom.
Like IPT expression, ZOG showed higher expression in green fruit, increasing during ripening, with the strongest expression in 2010 (Fig. 3). In contrast, BGLU expression was detected in fruit of both species at the beginning of development in 2009, and showed increased expression during ripening in green fruit but only in 2009 (Fig. 3). Apart from one strong peak early in development in green fruit, CKX had a relatively similar expression profile in fruit of both species, with expression at similar levels throughout ripening, but elevated during the 2010 season (Fig. 3).
Selection of candidate kiwifruit RR genes
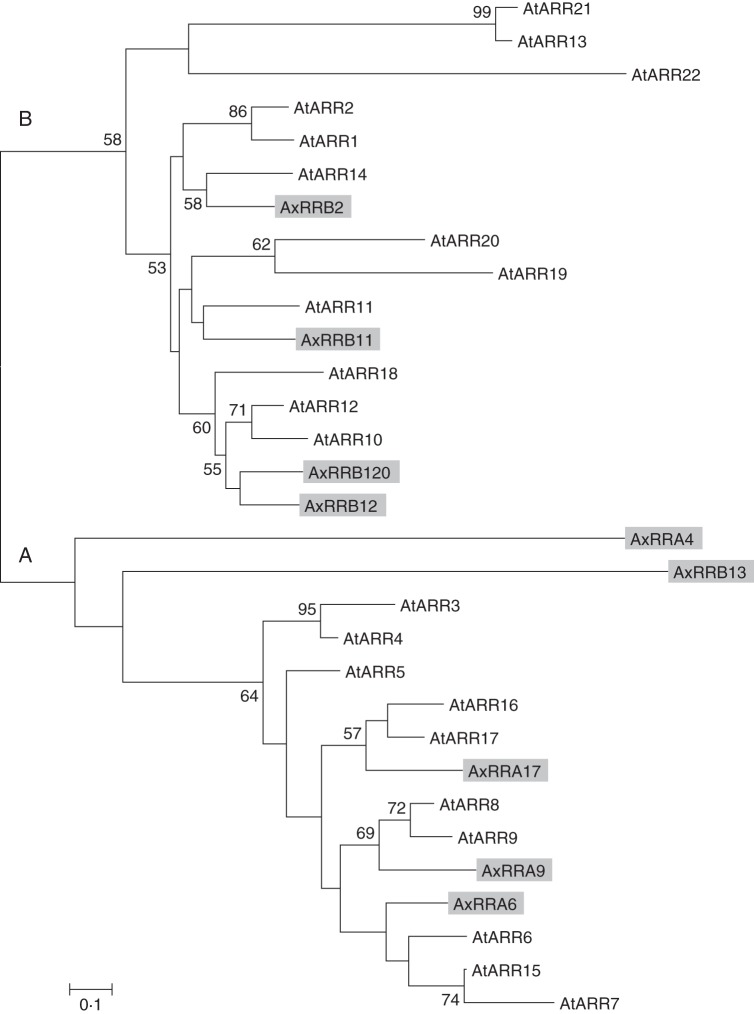
A phylogenetic tree of kiwifruit and Arabidopsis RRs was generated (Fig. 4). Nine putative RR proteins, translated from the Plant & Food Research EST database, showed high sequence homology to the 22 Arabidopsis RRs. Kiwifruit RRB2, 11, 12 and 120 clustered in the Type-A Arabidopsis clade, and kiwifruit RRA6, 9 and 17 clustered in the Type-B Arabidopsis clade. Kiwifruit RRA4 and RRB13 clustered in the Type-A clade, but are not closely related.
Fig. 4.
Phylogenetic tree of Actinidia and Arabidopsis RRs. The unrooted phylogenetic tree of full-length protein sequences aligned using Clustal W. Branch support values from 100 bootstraps are indicated when higher than 50 %. Two clades, Type-A and Type-B RRs (A and B, respectively), are distinguished. Arabidopsis gene names are as in Heyl and Schmülling (2003).
Expression of RR transcription factor genes in kiwifruit
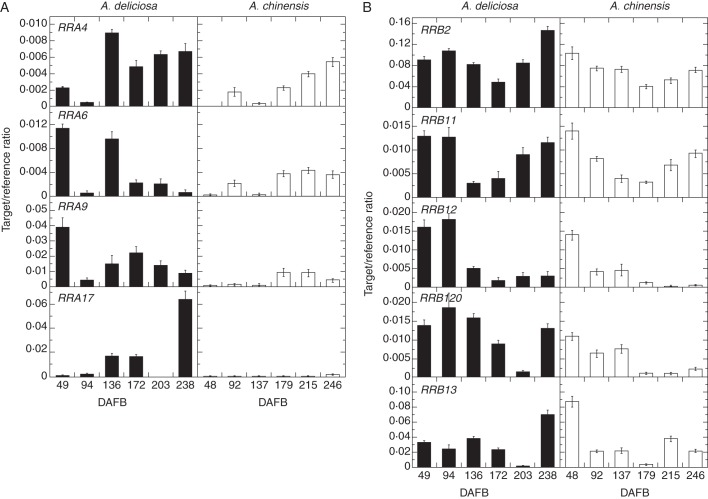
The expression of RR genes was measured by RT–qPCR in green and gold fruit in 2010 (Fig. 5). The majority of the RR genes had similar expression in green and gold fruit in 2010. RRA6 and RRA9 showed greater expression in green fruit at the beginning of development, but the expression was similar in green and gold fruit during ripening. RRA17 expression was noticeably greater in green fruit, especially in those fruit that were de-greening late in ripening (Fig. 5; Supplementary Data Fig. S1). The expression of RRB2, RRB11 and RRB120 was very similar in both species of kiwifruit. RRB120 and RRB13 showed similar patterns during development, but both had increased expression in de-greening green fruit.
Fig. 5.
Expression of RR genes in green and gold fruit during development in 2010. Expression of kiwifruit Type-A (left) and Type-B (right) RRs in green fruit (left panels, black columns) and gold fruit (right panels, white columns) during fruit development (days after full bloom; DAFB), measured as a target/reference ratio. RRA4 and RRB13 are ESTs from deep sequencing. Bars are one standard error above and below the mean (n = 4).
Dual luciferase transient assay screen for interacting transcription factors
Analysis of the SGR2 promoter revealed several motifs that predicted interaction with a number of MYB-related elements and a number of transcription factors involved with light regulation (data not shown). The interaction of the SGR2 promoter with kiwifruit RR genes was investigated to elucidate possible relationships between the promoters and transcription factors (Fig. 6). The one-way ANOVA showed that there were significant differences between the group means. The Tukey post-hoc test showed that there was a significant difference between the means of pHEX2 and RRB120 (P < 0·05) and RRA17 (P < 0·01), indicating an activation of the SGR2 promoter by RRB120 and particularly by RRA17 compared with the pHEX control.
Fig. 6.
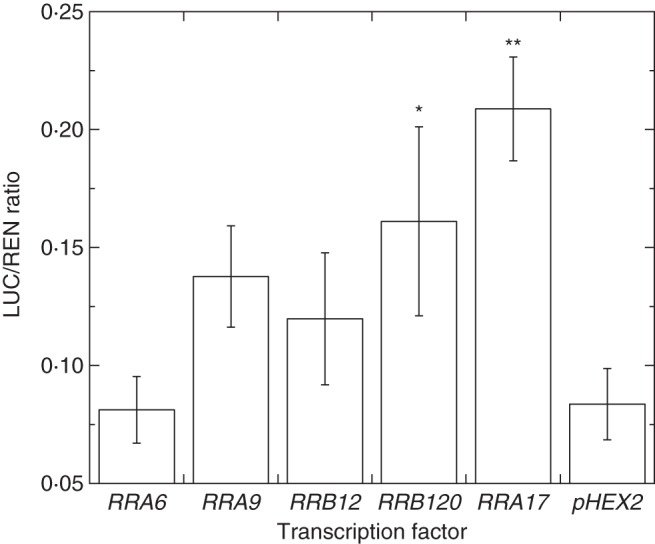
Interaction of the SGR promoter and selected RR transcription factors. Luminescence measurements are expressed as a ratio of luciferase to renilla (LUC/REN ratio) for the SGR promoter and selected transcription factors. Bars are one standard error above and below the mean (n = 4); *P <0·05; **P < 0·01.
DISCUSSION
Measuring cytokinin in plant tissues is challenging due to their relatively low levels and multiple forms among the bulk of other substances in plant extracts (Dobrev and Kamínek, 2002; Novák et al., 2008). Kiwifruit cytokinins were measured using triple quadrupole LC-MS/MS analysis. This revealed surprising trends in green and gold fruit in terms of both the differential nature and the quantity of cytokinins during development. Confirming and extending observations of high cytokinin content in green fruit at harvest ripeness by Lewis et al. (1996), the cytokinin concentrations were shown to increase throughout ripening in both species and to be high in fully ripe fruit, rather than limited to peak activity during the phase of cell division. Interestingly, while the expression of IPT (which codes for the rate-limiting enzyme in the cytokinin biosynthetic pathway) showed an increase during ripening in green fruit, which was not seen in gold fruit (Fig. 3; Supplementary Data Fig. S3), both species had high levels of nucleotides, free bases and ribosides (Fig. 2).
The most striking difference between species occurred with the O-glucosides – putative storage forms of cytokinins: both tZROG and DZROG concentrations increased during development in green fruit to very high levels, whereas the gold fruit contained very little tZROG or DZROG (Fig. 2).
Cytokinin activity is regulated by de novo synthesis, activation, conjugation and degradation (Kudo et al., 2010). A key enzyme in maintaining cytokinin homeostasis is CKX. While IPT expression is mirrored by CKX expression in a number of species during development, such as N. tabacum, Brassica rapa and wheat (Motyka et al., 2003; O'Keefe et al., 2011; Song et al., 2012), this did not occur in green fruit (Fig. 3), suggesting that there could be a breakdown in homeostatic regulation, although the higher CKX expression in 2010 in green fruit may have been reflected in the level of de-greening that did occur. Conversely, CKX may not be a key factor in moderating the cytokinin levels in ripening fruit.
Another key mechanism involved with cytokinin homeostasis is O-glucosylation. O-Glucosylation converts active cytokinin to inactive storage forms, resistant to CKX, but able to be reactivated by β-glucosidases (Jameson, 1994). ZOG showed substantially higher expression in green fruit compared with gold and, moreover, this difference in expression increased during ripening. This indicates that the homeostatic mechanism of O-glucosylation is operational in green kiwifruit but considerably less so in gold, as seen in the endogenous profile. This suggests that there is a greater perceived amount of cytokinin in green kiwifruit that has activated the conjugation mechanism. It is possible that the greater quantity of tZR in green fruit as they ripen could have activated this mechanism.
Seasonal differences were apparent both in the late de-greening of green fruit in 2010 and with a lesser accumulation of iPNT, tZ and tZR in 2010 compared with 2009. Moreover, expression of ZOG was higher in 2010 (potentially deactivating active forms via conjugation), while in 2009 the continued expression of a gene coding for β-glucosidase in green fruit could be counteracting that of ZOG, thereby maintaining a higher level of active cytokinin throughout ripening.
The kiwifruit SGR2 was shown to be more highly expressed in gold fruit, had an identical protein sequence in both green and gold fruit, and was able to induce chlorophyll degradation upon transient infiltration in N. benthamiana, suggesting that SGR2 is differentially regulated in ripening kiwifruit (Pilkington et al., 2012). Cytokinins have been shown to stimulate chloroplast transcription in detached barley leaves (Zubo et al., 2008). However, the effect of cytokinin levels on the transcription of chlorophyll degradation genes, which are nuclear encoded, has not been elucidated. The transcriptional control of chlorophyll de-greening was investigated using transient assays of selected RRs and the kiwifruit SGR2 promoter.
The dual luciferase system has been shown to provide a rapid method of transient gene expression analysis (Hellens et al., 2005). Trans-activation of the luciferase gene was measured as the ratio of LUC to 35S:Renilla after transient transformation in N. benthamiana. A transcription factor introduced into the plant cell by one Agrobacterium population can bind recognition sites in a target promoter introduced into the same plant cell from another Agrobacterium population, which promotes the expression of LUC, relative to 35S:Renilla.
RRA17 showed the strongest activation of the SGR2 promoter, suggesting a possible role in regulation of chlorophyll levels. Both RRA17 (Fig. 5) and SGR2 (Pilkington et al., 2012) were expressed in the fully ripe green fruit that were de-greening but, as expression of RRA17 was barely detectable in gold fruit as they yellowed, it is unlikely that RRA17 plays a role in the regulation of de-greening in gold kiwifruit.
RRB120 was also able to activate the SGR2 promoter. This gene showed a similar expression pattern in both fruit but, as with RRA17, showed the highest expression in green fruit that were de-greening. As RRB120 was expressed similarly in green and gold fruit throughout development, it is again unlikely that RRB120 regulates de-greening in gold kiwifruit.
The elevated cytokinin levels in the fruit of both species would be expected to delay yellowing, as cytokinin has been postulated to protect cell membranes and the photosynthetic machinery from oxidative damage during senescence (Zavaleta-Mancera et al., 2007). It is possible, despite its high cytokinin levels, if indeed the gold fruit do perceive the cytokinin, that other ripening signals (e.g. sugar, ethylene or removal of auxin) over-ride the effect of the cytokinin in gold but not green fruit. In this case, if gold fruit no longer have functional chloroplasts, cytokinin would be unable to stimulate chloroplast gene expression.
CONCLUSIONS
The cytokinin biosynthetic and metabolic enzymes, receptors and RRs are all encoded by multigene families. Primers used in the expression studies were designed for the single best kiwifruit gene family member candidate based on high transcript levels, tissue source and putative function based on Arabidopsis BLAST matches. Further analysis, now more possible with high-throughput genome sequencing, might reveal other cytokinin gene family members with differential expression during ripening of these kiwifruit species. Both green and gold kiwifruit have increasing levels of cytokinin during ripening, although where the cytokinin derives from in the gold fruit remains a mystery. Whatever the case, there were obvious differences in the metabolism of the cytokinins in ripening fruit from the two species, and differences between the two seasons. These differences are consistent with expression of cytokinin metabolic genes. The tight link between ZOG expression and O-glucosylation in green fruit strongly suggests that these fruit are perceiving the cytokinin whereas the gold fruit are not, and that this in some way leads to the maintenance of chlorophyll in green fruit and its loss in gold. Future work will include the identification of gene variants in kiwifruit populations segregating for gold and green flesh colour.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was supported by the New Zealand Foundation for Research Science and Technology [grant no. C06X0812]. We would like to thank the Tertiary Education Commission for an Enterprise Scholarship with Plant & Food Research to S.M.P.
LITERATURE CITED
- Argueso CT, Raines T, Kieber JJ. Cytokinin signaling and transcriptional networks. Current Opinion in Plant Biology. 2010;13:533–539. doi: 10.1016/j.pbi.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Crowhurst R, Gleave A, MacRae E, et al. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics. 2008;9:351. doi: 10.1186/1471-2164-9-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev PI, Kamínek M. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. Journal of Chromatography A. 2002;950:21–29. doi: 10.1016/s0021-9673(02)00024-9. [DOI] [PubMed] [Google Scholar]
- Hellens R, Allan A, Friel E, et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Schmülling T. Cytokinin signal perception and transduction. Current Opinion in Plant Biology. 2003;6:480–488. doi: 10.1016/s1369-5266(03)00087-6. [DOI] [PubMed] [Google Scholar]
- Heyl A, Brault M, Frugier F, et al. Nomenclature for members of the two-component signaling pathway of plants. Plant Physiology. 2013;161:1063–1065. doi: 10.1104/pp.112.213207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson PE. Cytokinin metabolism and compartmentation. In: Mok DWS, Mok MC, editors. Cytokinins: chemistry, activity, and function. Boca Raton, FL: CRC Press Inc; 1994. pp. 113–128. [Google Scholar]
- Jameson PE. Regulators of growth: cytokinins. In: Thomas B, Murphy D, Murray B, editors. Encyclopedia of applied plant sciences. Oxford: Academic Press; 2003. pp. 1000–1011. [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant and Cell Physiology. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Kudo T, Kiba T, Sakakibara H. Metabolism and long-distance translocation of cytokinins. Journal of Integrative Plant Biology. 2010;52:53–60. doi: 10.1111/j.1744-7909.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- Kulaeva ON, Karavaiko NN, Selivankina SY, et al. Cytokinin signalling systems. Plant Growth Regulation. 1996;18:29–37. [Google Scholar]
- Lewis DH, Burge GK, Schmierer DM, Jameson PE. Cytokinins and fruit development in the kiwifruit (Actinidia deliciosa). I. Changes during fruit development. Physiologia Plantarum. 1996;98:179–186. [Google Scholar]
- McGhie TK, Ainge GD. Color in fruit of the genus Actinidia: carotenoid and chlorophyll compositions. Journal of Agricultural and Food Chemistry. 2002;50:117–121. doi: 10.1021/jf010677l. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, et al. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proceedings of the National Academy of Sciences, USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DWS, Martin RC, Shan X, Mok MC. Genes encoding zeatin O-glycosyltransferases. Plant Growth Regulation. 2000;32:285–287. [Google Scholar]
- Motyka V, Vaňková R, Čapková V, Petrášek J, Kamínek M, Schmülling T. Cytokinin-induced upregulation of cytokinin oxidase activity in tobacco includes changes in enzyme glycosylation and secretion. Physiologia Plantarum. 2003;117:11–21. [Google Scholar]
- Novák O, Hauserová E, Amakorová P, Dolezal K, Strnad M. Cytokinin profiling in plant tissues using ultra-performance liquid chromatography–electrospray tandem mass spectrometry. Phytochemistry. 2008;69:2214–2224. doi: 10.1016/j.phytochem.2008.04.022. [DOI] [PubMed] [Google Scholar]
- O'Keefe D, Song J, Jameson P. Isopentenyl transferase and cytokinin oxidase/dehydrogenase gene family members are differentially expressed during pod and seed development in rapid-cycling Brassica. Journal of Plant Growth Regulation. 2011;30:92–99. [Google Scholar]
- Ozga J, Reinecke D. Hormonal interactions in fruit development. Journal of Plant Growth Regulation. 2003;22:73–81. [Google Scholar]
- Pilkington SM, Montefiori M, Jameson PE, Allan AC. The control of chlorophyll levels in maturing kiwifruit. Planta. 2012;236:1615–1628. doi: 10.1007/s00425-012-1723-x. [DOI] [PubMed] [Google Scholar]
- Quesnelle PE, Emery RJN. cis-Cytokinins that predominate in Pisum sativum during early embryogenesis will accelerate embryo growth in vitro. Canadian Journal of Botany. 2007;85:91–103. [Google Scholar]
- Richmond AE, Lang A. Effect of kinetin on protein content and survival of detached Xanthium leaves. Science. 1957;125:650–651. [Google Scholar]
- Robson PRH, Donnison IS, Wang K, et al. Leaf senescence is delayed in maize expressing the Agrobacterium IPT gene under the control of a novel maize senescence-enhanced promoter. Plant Biotechnology Journal. 2004;2:101–112. doi: 10.1046/j.1467-7652.2004.00054.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annual Review of Plant Biology. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- Schmülling T, Werner T, Riefler M, Krupková E, Bartrina y Manns I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. Journal of Plant Research. 2003;116:241–252. doi: 10.1007/s10265-003-0096-4. [DOI] [PubMed] [Google Scholar]
- Schoor S, Farrow S, Blaschke H, et al. Adenosine kinase contributes to cytokinin interconversion in Arabidopsis. Plant Physiology. 2011;157:659–672. doi: 10.1104/pp.111.181560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Jiang L, Jameson P. Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biology. 2012;12:78. doi: 10.1186/1471-2229-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spíchal L. Cytokinins – recent news and views of evolutionally old molecules. Functional Plant Biology. 2012;39:267–284. doi: 10.1071/FP11276. [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. Journal of Biological Chemistry. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software Version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- To JPC, Kieber JJ. Cytokinin signaling: two-components and more. Trends in Plant Science. 2008;13:85–92. doi: 10.1016/j.tplants.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the pp 19 protein of tomato bushy stunt virus. The Plant Journal. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Zavaleta-Mancera HA, López-Delgado H, Loza-Tavera H, et al. Cytokinin promotes catalase and ascorbate peroxidase activities and preserves the chloroplast integrity during dark-senescence. Journal of Plant Physiology. 2007;164:1572–1582. doi: 10.1016/j.jplph.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, et al. Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiology. 2008;148:1082–1093. doi: 10.1104/pp.108.122275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.