Abstract
The focus of this minireview is the replication control of the 39.9-kb plasmid R6K and its derivatives. Historically, this plasmid was thought to have a narrow host range but more recent findings indicate that its derivatives can replicate in a variety of enteric and non-enteric bacterial species (Wild et al., 2004). In the four-plus decades since it was first described, R6K has proven to be an excellent model for studies of plasmid DNA replication. In part this is because of its similarities to other systems in which replication is activated and regulated by Rep protein and iteron-containing DNA. However its apparent idiosynchracies have also added to its significance (e.g., independent and co-dependent replication origins, and Rep dimers that stably bind iterons). Here, we survey the current state of knowledge regarding R6K replication and place individual regulatory elements into a proposed homeostatic model with implications for the biological significance of R6K and its multiple origins of replication.
Keywords: Plasmid R6K, DNA replication control, π monomers, π dimers, iterons, homeostasis
1. INTRODUCTION
Chromosomal and plasmid replicons have evolved strategies to assure their hereditary stability and maintenance at a specific copy number. Unraveling the regulatory mechanisms that drive these processes in plasmids is a matter of fundamental biological interest. A recurring theme in the duplication of prokaryotic replicons is the recognition of the replication origin (ori) by cis-encoded initiators that bind to repeated nucleotide sequences called iterons (reviewed in Moore et al., 1979; Chattoraj, 2000; Espinosa et al., 2000; Krüger et al., 2004b). These replication proteins (Reps) communicate amongst themselves to activate and inhibit the ori, relying on protein-protein interactions that occur in both iteron-independent and iteron-dependent fashions. Moreover, in many systems Rep can also act as an autorepressor of transcription, a function that depends on the protein binding to yet another iteron-like sequence. Defining these distinct yet related interactions has long been regarded as crucial to understanding the biology and mechanistic aspects of plasmid copy number control.
Antibiotic-resistance plasmids in general, and the Rep/iteron plasmid R6K specifically, were brought to the attention of the scientific community at a time when the connection between plasmid biology and antibiotic resistance in pathogenic bacteria was quickly emerging (reviewed in Watanabe, 1963; Kontomichalou et al., 1970). Shortly thereafter, the plasmid (and its derivatives) began taking a prominent role in studies of plasmid replication control, eventually becoming a significant model system for basic plasmid research. In the over four decades that followed R6K's introduction to the research laboratory, studies of plasmid biology have revealed many factors that allow reservoirs of resistance to emerge and rapidly spread within diverse microbial biofilms (reviewed in Madsen et al., 2012). Ecological niches that were once thought of as being distinct are increasingly recognized as being microbiologically connected. This is significant because plasmids with replication regions that are closely related to R6K have been shown to contain modules that facilitate genetic exchange in environment, and with that exchange these plasmids disseminate antibiotic resistance genes (Norman et al., 2008). The data on pOLA54 and its derivatives highlight the real-world significance of R6K-like plasmids, which can carry genes that contribute to virulence and biofilm formation (Ghigo, 2001; Burmolle et al., 2008; Madsen et al., 2012) in addition to antibiotic resistance (for review, see Mazel and Davies, 1999; de la Cruz and Davies, 2000; Heinemann et al., 2000; Giraldo and Fernandez-Tresguerres, 2004; Venkatesan and Burland, 2004; Fluit, 2005). Because of these properties, a deeper understanding of the establishment and maintenance of such plasmids is the surest route to generating solutions to a variety of growing global health crises. That goal prompted our laboratory to begin translating some of the basic knowledge we have generated about R6K replication control into practical applications, as will be described in Section 4 of this minireview.
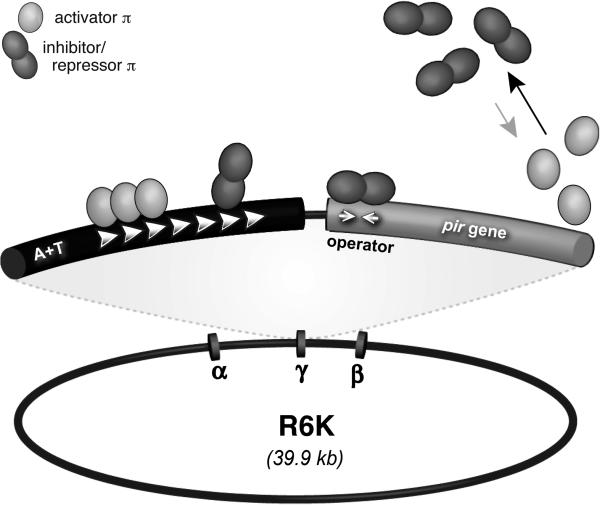
All replication in R6K relies on the two essential components of a minimal replicon, the γ ori, and its cognate Rep, π protein, encoded by the pir gene (Inuzuka and Helinski, 1978) (Fig. 1). The molecular interactions driving π/γ ori regulation have been extensively studied and reviewed (e.g., Kolter, 1981; Shafferman et al., 1981; McEachern et al., 1986; Filutowicz et al., 1994a; Filutowicz and Rakowski, 1998); and the central features (to be described in more detail, below) were found to be the different DNA sites for π binding, the disparate functions of monomers and dimers of π and the complex nucleoprotein oligomerization pathways that are driven by the concentrations of both π and iteron-containing DNA. It is of note that R6K was the first iteron-containing plasmid to provide compelling evidence that Rep-iteron interactions influence the frequency of both ori activation and inhibition (Inuzuka and Helinski, 1978; Germino and Bastia, 1983b; Germino and Bastia, 1983a; Stalker et al., 1983; Filutowicz et al., 1985b). In addition it is the first iteron-containing plasmid whose replication (γ ori) was fully reconstituted in vitro with involvement of 22 participating proteins (Abhyankar et al., 2003; Zzaman and Bastia, 2005).
Figure 1.
Salient features of plasmid R6K. Parental R6K has 3 replication origins, α, γ and β. Only γ ori and the pir gene product, π, (expanded view) are required for a minimal replicon. White arrowheads represent ‘TGAGnG-motif’ binding sites for π (in γ ori). π is multi-functional (see key.) Operator occupancy results in π autorepression while initiation is controlled, in part, by differential occupancy of the direct repeats adjacent to an A+T-rich region. Dimerization of π (black arrow) is largely irreversible (gray arrow), however π monomers have greater affinity for the 7 direct repeats than do dimers. Figure is not drawn to scale.
2. ITERONS, ITERON-LIKE SEQUENCES AND π PROTEIN
2.1 π-bound DNA sequence repeats are required for replication initiation
Iterons are the primary DNA binding sites for Rep (protein) and these sequences are typically arranged in tandem, direct repeats (DRs). Comprehensive analyses of sequence information have revealed remarkable similarities among iterons in various prokaryotic oris (Papp et al., 1993; Chattoraj and Schneider, 1997; Schneider, 2001). It is clear that of the known π binding sites in R6K, the 7 DRs of the functionally diverse γ ori core comprise the most important site of DNA binding activity for π (Fig. 1). Most nucleotides in the 22-basepair (bp) iteron sequence are conserved in each of the 7 DRs. Nonetheless, it seems possible that existing sequence differences among the (R6K) iterons could be important to the assemblies of π at γ ori and, as a result, to replication control. A comparison of the sequences of the DRs reveals that only two iterons should be considered 100% consensus, iterons number 2 and 5, whereas the least conserved iteron (number 4) has five bp changes from the consensus 22-bp sequence. These observations, when combined with π/iteron contact domain data (described below; see also Figs. 2 and 3), leave open the possibility that differences in iteron sequence may translate into differences in the DNA binding modes of π.
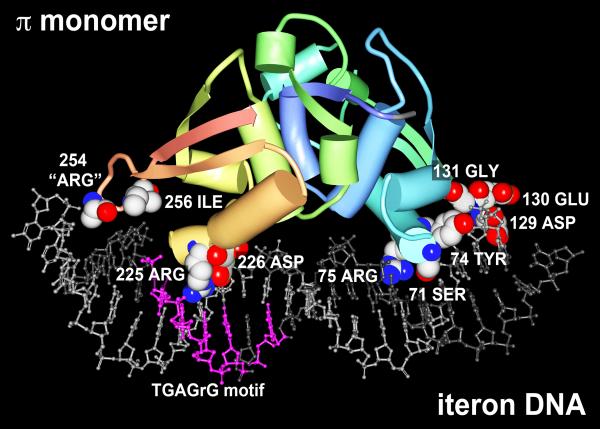
Figure 2.
A model for the interaction of a π protein monomer with R6K γ ori iteron DNA. The crystal structure for a variant π monomer bound to an iteron was solved by Swan et al. (2006) and deposited as a PDB file (PDB ID: 2NRA) to the Worldwide Protein Data Bank (www.pdb.org). The model shown was rendered using Protein Workshop 3.9 software (Moreland et al., 2005) and colors were enhanced using Adobe Photoshop 11.0.2. DNA strands (ball and stick representation) are light & dark gray with the TGAG(n)G motif in fuchsia. Amino acids predicted and/or shown to contact bases of the DNA (Swan et al., 2006; Kunnimalaiyaan et al., 2007, Saxena et al., 2010b) and shown in CPK are: Ser71, Tyr,74, Arg75, Asp129, Glu130, Gly131, Arg225, Asp226, “Arg254”, and Ile256. The PDB file does not account for Arg254 in its entirety. Atoms of CPK amino acids are colored as follows: C is white, N is blue, O is red. The remainder of the π monomer is represented as a chain color ramp from blue (start) to red (end) with helices and strands represented as cylinders and arrows, respectively.
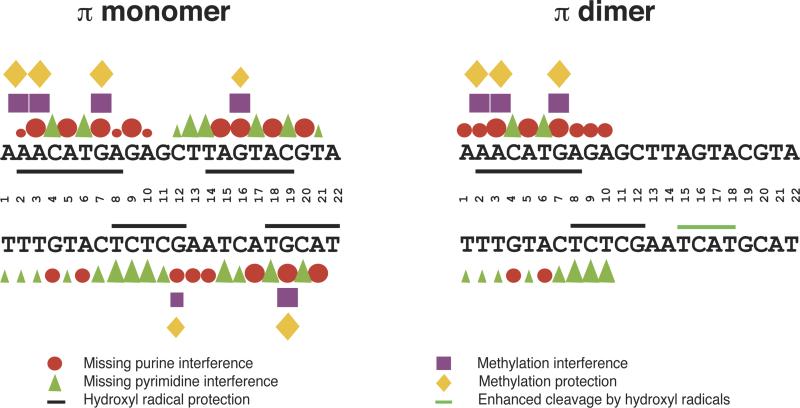
Figure 3.
Iteron DNA sequence (22-bp) and the compilation of base-specific contact probing data for π binding to the iteron. Red circles (purines) and green triangles (pyrimidines) represent bases that affect π binding when removed. Purple squares represent purines whose modification by DMS weakens π binding. Yellow diamonds represent purines that are protected, by bound π protein, from methylation by DMS. In each case, large symbols are indicative of strong effects; medium and small symbols indicate moderate and weak effects, respectively. Black lines represent protection of the DNA backbone, by π, from hydroxyl radical cleavage, and a green line represents enhanced cleavage by hydroxyl radicals. (republished from Kunnimalaiyaan et al., 2004).
π protein activates γ ori replication at low intracellular levels and inhibits replication at elevated levels (Filutowicz et al., 1985b; Filutowicz et al., 1986a; McEachern et al., 1986; McEachern et al., 1989). A model demonstrating the fundamental aspects of π/γ ori interactions (Fig. 1) draws from an abundance of data revealing that Rep monomers bound to ori DRs activate replication, yet Reps including π exist primarily as dimers in solution (e.g., Filutowicz et al., 1986b; Wickner et al., 1991; Konieczny and Helinski, 1997; Urh et al., 1998; Komori et al., 1999; Krüger et al., 2001; Giraldo et al., 2003; Gasset-Rosa et al., 2008). Dimers of π protein are remarkably stable, exhibiting little to no subunit exchange in the absence of denaturing agents (Urh et al., 1998). By mixing protein variants of different sizes (Wu et al., 1997), π was the first of three Reps shown to bind to a single DR as either a monomer or a dimer (Urh et al., 1998; Diaz-Lopez et al., 2003; Das and Chattoraj, 2004; Diaz-Lopez et al., 2006). In fact, a naturally-occurring, N-terminal truncated protein, π30.5, was observed to bind the iteron as a dimer only (not a monomer), a property which made the variant especially useful for dissecting the functional roles of π monomers and dimers (Wu et al., 1997; Urh et al., 1998; Krüger and Filutowicz, 2000). The dimer-biased π30.5 protein was shown to be inactive as a replication initiator. A full-length form of π, the double amino acid substitution variant πM36A^M38A, was similarly found to be inactive as an initiator while binding DRs exclusively as dimers (Wu et al., 1997; Krüger and Filutowicz, 2000; Krüger et al., 2001). In contrast, evocative correlations have been observed that link the destabilization of Rep dimers, with and without the assistance of chaperone proteins, and enhanced replication activity (Chen et al., 1998; Krüger et al., 2001; Zzaman et al., 2004). In the R6K system, various hyper-replicative π variants (copy-up, for short) that increase plasmid copy numbers have been found to form less stable dimers and/or display a monomer-bias when binding DRs (e.g., Urh et al., 1998; Abhyankar et al., 2004). In experiments that employed copy-up and replication-inactive mutants of π, strand opening in vitro correlated with in vivo replication activity (γ ori) and higher monomer:dimer ratios in DNA binding patterns revealed by electrophoretic mobility shift assay, i.e., EMSA (Krüger et al., 2001).
The tandem DRs are essential for replication at γ ori and certain observations suggest that cooperative DNA binding by adjacent π monomers may promote site occupancy (Filutowicz et al., 1994b; Urh et al., 1995; Krüger et al., 2004a; Bowers et al., 2007; Bowers and Filutowicz, 2008). This is not surprising in light of data suggesting that monomers of other Reps may also bind iterons cooperatively (Vocke and Bastia, 1983; Mukherjee et al., 1985; Gammie and Crosa, 1991; Perri et al., 1991; Xia et al., 1993). Conversely, the binding of π dimers is believed to inhibit replication through several likely nucleoprotein assemblies (Filutowicz et al., 1985a; Filutowicz et al., 1987; McEachern et al., 1989; Filutowicz et al., 1994a; Miron et al., 1994) and some evidence suggests that dimers may be unable to occupy consecutive iterons (e.g., Krüger et al., 2004a). This could be significant if one of the mechanisms for π function is to locally distort the DNA, thereby promoting strand opening in the near-by A+T-rich region. When the DNA bending properties of π were examined using EMSA and circular permutation assays, both monomers and dimers were found to bend a single iteron to similar degrees. Bending angles increased when two iterons were used and both sites were occupied, however two dimers were not observed to bind a two-DR probe. It seems possible then that monomers may ultimately bend the 7 DRs more than dimers do, simply by occupying more iterons. There are several possible hypotheses that can be offered to explain why π dimers failed to occupy two adjacent iterons, even when dimer-biased variants were used. The investigators proposed that dimers could cause steric interference, occluding adjacent iterons from binding π dimers (Krüger et al., 2004a). Another intriguing possibility is that perhaps not all DRs are equally proficient at binding dimers. And of course, the observations may also be evidence for the importance of the cooperative interactions of π monomers. These ideas are not mutually exclusive and remain to be explored, perhaps aided by the fortuitous discovery that electrolyte choices can mediate switches in the mode of π binding (independent vs. cooperative) (Urh et al., 1995).
2.2. Host factors that influence DNA transactions at γ ori
In addition to its various π binding sites, γ ori contains numerous host factor binding sites, at least three of which (DnaA, IHF and Fis) have been observed to affect ori activity in vivo (Filutowicz and Appelt, 1988; Kelley and Bastia, 1991; Wu et al., 1992; Dellis et al., 1996; Wu et al., 1996). Additional investigations probed the in vitro effects of two host-encoded DNA-binding proteins, DnaA and IHF, on the reactivity of γ ori DNA to KMnO4 in the presence of π protein (Krüger et al., 2001); KMnO4 reactivity is a hallmark of DNA strand opening. The data suggested that π and DnaA protein bound to iterons and DnaA box 1, respectively, might communicate with each other. Such communication could be indicative of the interactions between π and DnaA that were observed in the absence of DNA (Lu et al., 1998). Additional KMnO4 DNA probing experiments examined the effects of ATP and Mg2+ on the strand opening reaction (Krüger and Filutowicz, 2003). The results indicated that the opening of γ ori occurred in the presence of ATP as well as AMP-PCP, a non-hydrolysable ATP analog. From these observations it was concluded that ATP hydrolysis might be unnecessary for open complex formation at γ ori. In the absence of ATP or Mg2+, copy-up π yielded data suggestive of distortions in the iteron attributable to DNA bending rather than DNA melting. These results indicate that ATP and/or Mg2+ are not needed for copy-up π to bind iterons in vitro and that ATP (and perhaps Mg2+) likely effects an allosteric change in the protein bound to γ ori.
2.3. The structure of π protein
All things considered, the DNA-binding protein, π, lies at the heart of the regulation of plasmid R6K. π exhibits remarkable structural and functional plasticity in its interactions with diverse DNA target sites, and these different interactions are the basis for its different biological functions: replication initiator, replication inhibitor, transcription (auto)-repressor and origin selection (α, β or γ) factor (Germino and Bastia, 1983b; Germino and Bastia, 1983a; Urh et al., 1998; Krüger et al., 2004b; Saxena et al., 2010a; Saxena et al., 2010b). Encoded by R6K's pir gene, π is a member of the Rep family of replication initiator proteins with sequence similarities that extend even to eukaryotic proteins (Giraldo and Díaz-Orejas, 2001). As a result, data from other plasmid systems have proven useful when analyzing π protein's interactions with DNA. For example, Komori et al. (1999) determined the three-dimensional structure of monomeric RepE54, a variant of the F plasmid initiator protein, in complex with its cognate iteron DNA. It was found to be a pseudo-symmetric protein comprised of two winged-helix (WH) DNA-binding motifs. This conclusion was consistent with previous work, which had suggested that Rep proteins are composed of two domains (Chattoraj and Schneider, 1997; Giraldo et al., 1998). Early attempts to crystallize π for structural analysis, however, were thwarted by its tendency to aggregate in solution at high concentrations (Swan et al., 2006 and Filutowicz laboratory, unpublished data), a common property of Rep proteins. Thus, the information from the RepE system was used as a guide for constructing a theoretical structural model of a π monomer bound to an iteron (Kunnimalaiyaan et al., 2004; Sharma et al., 2004). In conjunction, investigations that combined genetic and biochemical approaches allowed for comparisons of nucleoprotein contact patterns of π monomers versus π dimers bound to a single iteron. The data from π monomers was then examined in the context of various solved and homology-modeled Rep/iteron structures (Giraldo et al., 2003). Roughly concurrently, researchers at the Medical University of South Carolina achieved success crystallizing a quadruple mutant of π (Fig. 2 and Swan et al., 2006).
As predicted by the structural models and substantiated by chemical footprinting and iteron mutagenesis, π monomers make extensive contacts with iteron DNA. Six amino acids (Tyr74, Ser71, Gly131, Gly211, Arg225 and Arg254) were deduced from the history of available biochemical data as being vital for π monomer–iteron contact (Kunnimalaiyaan et al., 2007). Four of these amino acids (Tyr74, Gly131, Gly211 and Arg254) are conserved and two are similar (Ser71 replaced by Thr68; Arg225 replaced by Lys 221) in the π protein encoded by plasmid pOLA54, mentioned earlier (Norman et al., 2008). Overall, the Rep proteins of pOLA54 and R6K display roughly 40% identity and >70% similarity. The data and conclusions presented by Kunnimalaiyaan et al. (2004; 2007) and Swan et al. (2006) are in good agreement but with some variation. For example, research by the former group did not investigate all of the amino acids implicated in DNA contact by the crystal structure, two of which (Arg75 and Asp226) received additional support in later mutation analyses (Saxena et al., 2010b). In contrast, the work of Swan et al. assigns no role in iteron binding to Gly211, which would seem to have a critical functional role based on Rep protein sequence conservation. Perhaps the amino acid only indirectly affects the binding of π monomers (as well as dimers) to iterons. Alternatively, this and other discrepancies might be attributable, at least in part, to the four mutations introduced into the pir gene to facilitate π protein crystallization.
As shown in Figure 2, several atoms of π residue Arg254 are not accounted for in the crystal structure including the 3 nitrogens of the side chain's guanidinium group. In the homology model by Kunnimalaiyaan et al., (2007), Arg254 adopts a compact form that accommodates binding with the DNA, however, its homolog in the crystallized RepE54 adopts a more extended conformation. Given the terminal location of the basic Arg254 on an unstructured loop, it is intriguing to hypothesize that the amino acid might interact with the acid-rich amino acids at the opposite end of the π monomer (e.g., Asp129 and Glu130) during cooperative binding of the protein to consecutive iterons.
Although monomers and dimers of π protein share a common iteron-contact domain, studies using DMS protection, methylation interference and EMSA, in addition to missing base interference, have indicated that the two forms of Rep differ with respect to the extent of the nucleoprotein interaction (Kunnimalaiyaan et al., 2004 and summarized in Fig. 3). Counterintuitively, a dimer of π was found to make less contact with a single DR than does a monomer, employing only one DNA binding domain (of two) from only one subunit of the dimer. Hydroxyl radical protection footprints revealed that monomers bind to one face of the DNA helix, protecting the phosphodiester backbone along the two adjacent major grooves and the central minor groove. π dimers bind to the same face of the DNA helix but only contact the backbone of the left half of the iteron. Additionally, a genetic selection scheme was devised that resulted in the isolation of iteron mutants that discriminate between the binding of π monomers and π dimers (Kunnimalaiyaan et al., 2004). In the base-substitution category, two classes of such mutants were identified; one class affects the binding of both monomers and dimers while another class affects the binding of π monomers only (not dimers). The data support a model in which π protein dimerization disturbs one of two DNA binding domains important for monomer/iteron interaction; the dimer/iteron interaction utilizes only one DNA binding domain. These results correlate with predictions based on the crystal structure of monomeric π (Swan et al., 2006) and structural analyses of other Rep dimers (Giraldo et al., 2003; Giraldo and Fernandez-Tresguerres, 2004; Nakamura et al. 2007). Importantly, however, the dimer binding motifs of Reps other than π could not be similarly mapped since dimer-bound iteron complexes are seemingly absent or unstable in vitro.
2.4. π binding sites other than DRs (in γ ori)
In addition to the 7 DRs of γ ori, two other distinct types of π binding sites have been identified in a minimal R6K replicon. One site of obvious importance occurs in the pir gene operator where repression of transcription is mediated through π binding to an inverted repeat (IR) sequence (Fig. 1 and refs. Filutowicz et al., 1985a; Kelley and Bastia, 1985; York and Filutowicz, 1993). This dimer-binding element, expected to span approximately 3 helical turns, is characterized by two TGAGnG motifs in an inverted orientation. These motifs also occur in the DRs of R6K (Fig. 2) but in a tandem arrangement. Interestingly, each 11-bp half-site of the pir operator's IR differs at one position (2 total) from the relevant portion of the consensus DR sequence (i.e., bp 1-11 of the 22-bp DR). Both of these non-consensus bp are 100% conserved amongst the 7 DRs, and changing them (in a DR probe) led to deviations in what were otherwise nearly identical contact patterns generated by π monomers and dimers. Beyond the contributions of the individual base pairs, the striking difference in the geometry of TGAGnG motifs in IRs and DRs prompted the hypothesis that isomers of π dimers may exist (Urh et al., 1998). To test this model, experiments were designed using DNA probes, each containing two TGAGnG motifs in different orientations: inverted, everted, and direct (Krüger et al., 2004a). The results showed that π subunits readily associate in head-to-head and head-to-tail fashion to form dimers/oligomers. For the π/γ ori system, a deep understanding of protein isomerization and its relevance to DNA binding and regulatory function remain goals to continue striving toward. Nonetheless, available data suggest that, like other Rep proteins (Komori et al., 1999; Giraldo et al., 2003; Nakamura et al., 2007), π can adopt more than one conformation. This characteristic likely allows the protein to: 1) assume conformations that are compatible with distinct DNA targets, 2) induce, upon binding, conformation changes in these targets (e.g., DNA bending and strand separation), and 3) promote Rep/Rep and Rep/host-factor interactions (Ratnakar et al., 1996; Lu et al., 1998).
Although typical of Rep/iteron plasmids, the ori and operator binding sites described thus far are not the only sites of Rep binding in a minimal R6K replicon. For example, an eighth DR has been identified in the operator of the pir gene, but any functional significance of the site in R6K replication or pir gene autoregulation is currently unknown. Yet another π binding site was identified in the A+T-rich region of γ ori (Filutowicz et al., 1986b; Levchenko and Filutowicz, 1996). The possible significance of this discovery was later reinforced by DNA replication studies that mapped the start sites for leading strand synthesis in a γ ori replicon to the A+T-rich segment (Chen et al., 1998). It is of note that the initiating nucleotides were shown to be the same for wt and copy-up π-dependent replication. Thus, copy-up mutants most likely utilize the same mechanism of priming, but do so more efficiently than wt π. Given what was already known regarding the multi-functionality of π, it was hypothesized that the protein might negatively modulate the priming step in replication by binding to the A+T-rich site, perhaps using a simple occupancy mechanism (Krüger and Filutowicz, 2000). One prediction of such a model would be that the over-replication of DNA initiated by copy-up mutants might be a consequence of decreased/altered binding of the variant Reps to the non-iteron site. Indeed, it was found that copy-up mutations weakened π binding to the A+T-rich site, a result that would be expected for a dimer-dependent interaction.
Data from a variant of π phenotypically distinct from the copy-up mutants also indicated that π binds to the non-iteron site as a dimer and, reminiscent of its binding to an iteron sequence (Urh et al., 1998), it appeared that a single subunit contacted the DNA. This early conclusion was drawn based on the use of a truncated, dimerization proficient protein, ΔC164π, that has been shown to lack iteron-specific DNA-binding activity. It was, therefore, surprising that this π variant could inhibit γ ori activity. Of course, the experiments using ΔC164π and the conclusions drawn from them did not have the benefit of more recent structural models of Rep, particularly the work on pPS10 which suggests dimerization need not inactivate the DNA-binding activity of WH1 (Gasset-Rosa et al., 2008). As discussed below, it is not unreasonable to expect that ΔC164π might retain some level of DNA-binding activity. The A+T-rich binding site is non-canonical since, unlike the previously identified (and discussed) DRs and IRs, it lacks the TGAGnG motif recognized by WH2 (Filutowicz et al., 1986b; Levchenko and Filutowicz, 1996). However sequence analysis does reveal some potential conservation with the half of the DR contacted by the N-terminal WH1 (Swan et al., 2006). ΔC164π retains the N-terminal amino acids of the WH1 motif. Thus, although not considered at the time, perhaps some residual DNA binding activity to the A+T-rich region could solve the puzzle of how ΔC164π was able to inhibit the activity of γ ori in vivo (Greener et al., 1990). Although this new interpretation of old data may seem rather speculative, other observations (described below) also lend their support.
2.5. Speculation on underappreciated non-TGAGnG π binding site motifs
Investigations of π binding sites in R6K have, to some extent, focused on the prominent TGAGnG motif, which exhibits a degree of conservation even beyond the R6K system. This 6-bp string is part of a larger but imperfect consensus that, when written as mywTGAGnG, is sufficient to encompass most but not all of the known iteron and half-iteron binding sites while only occurring once outside the replication region of R6K (Thompson and Pathogen Genomics Group, 2012). Significantly, however, only three bases in the double-strand GAG portion of the motif were identified by Swan et al. (2006) as contacting WH2 of a (mutant) π monomer. WH1 was also shown to contact three bases, but the bp involved are not consecutive as they are for the WH2 “half iteron” consensus. Relying on these potential contact nucleotides and DNA sequence patterns, a starting point for a possible consensus WH1 binding motif might be wGwnCnT; but it occurs far too frequently in R6K to seem credible as an independent binding motif. This is not surprising, however, as it has generally been thought that the TGAGnG consensus is essential for π binding. Notice that the experiments examining different orientations and spacings of “half iteron” repeats (direct, inverted and everted) only included the TGAGnG-bearing WH2 half iterons (Krüger et al., 2004a). Similar experiments have not been conducted to examine the possibility that certain configurations of π dimers (or higher order multimers) might bind DNA solely by engaging two (or more) WH1 half-iterons.
When one examines R6K for the occurrences of the wGwnCnT sequence that engages WH1, one pattern jumps out immediately. What has long been held to be a consensus DR actually contains two tandem wGwnCnT repeats, the first of which overlaps the GAG sequence that binds WH2. The duplication of the WH1 contact sequence, i.e., wGwnCnTwGwnCnT, occurs 7 times in R6K and only on one strand of the DNA. Interestingly, not all of the repeats occur in the 7DRs of γ ori. Only a singlet wGwnCnT is found in the most degenerate and centrally located DR number 4. Curiously, the seventh wGwnCnTwGwnCnT and a lone outlying WH2 motif (mywTGAGnG) occur roughly opposite to the β and α oris, respectively (Thompson and Pathogen Genomics Group, 2012). More than one hypothesis can be generated regarding the possible significance of the tandem wGwnCnT repeats. We will leave such speculation to the mind of the reader, but with the note that the apparent inability of π dimers to occupy consecutive iterons (mentioned earlier) may be informative.
The A+T-rich segment contains multiple copies of the common wGwnCnT but no tandem direct repeats of the sequence nor does it contain a WH2 binding motif. However, if the stringency of the core WH1 binding sequence is relaxed slightly, a direct repeat with a 19-bp spacer (wRwnCnT-19n-wRwnCnT) is revealed. This sequence occurs only slightly more frequently than would be expected by chance and is distributed throughout the R6K genome. But support for the significance of this putative WH1-19n-WH1 binding motif comes from it co-localizing with the mapped A+T-rich site for the binding of π dimers. In particular, π-induced enhancements of DNase I cleavage occur at positions +15 and +51 of γ ori closely flanking the wRwnCnT-19n-wRwnCnT sequence, which starts at positions +17 and ends at position +49 (Levchenko and Filutowicz, 1996; Chen et al., 1998; Thompson and Pathogen Genomics Group, 2012). Perhaps the somewhat even distribution of this sequence denotes a function in the architecture of the plasmid DNA. That said, there is at least one feature that distinguishes the wRwnCnT-19n-wRwnCnT sequence in the A+T-rich region from all the others. It is in a DNA segment in R6K with a comparatively high density of the tetranucleotide CTTA, a sequence that has also been largely conserved in the DRs of γ ori. CTTA is found in 6 of the 7 DRs, absent only from the central DR number 4 due to a C→T transition that also disrupts the tandem repeat of the WH1 motif in that iteron. Crystallography data provide no evidence of a DNA-binding role for any of the bp in the CTTA tetranucleotide. However, several strings of amino acids in π cannot be accounted for in the solved crystal structure (residues 1-8, 107-113, 268-305) or other structural models. Perhaps one of these polypeptides contacts the CTTA motif. Alternately, the conditions used for crystallization (e.g., mutant Rep) might obscure possible contacts. Most significantly, since π adopts more than one functional form, the tetranucleotide may be important for contacting a form of π not seen under the crystallization conditions used. The DNase I data from the A+T-rich binding site (Levchenko and Filutowicz, 1996) as well as hydroxyl radical probing data using a single DR (Fig. 3) both showed CTTA-adjacent (or TAAG) enhancements of DNA cleavage when dimeric π bound the DNA (Krüger and Filutowicz, 2000; Kunnimalaiyaan et al., 2004).
3. HIGHER ORDER REGULATION - INCOMPATIBILITY AND ORIGIN SELECTION
3.1. Plasmid incompatibility and titration of Rep
How were the elements that negatively influence replication initiation first identified (reviewed in Novick, 1987; Nordstrom, 1990)? In an approach referred to as incompatibility (Inc+) testing, fragments of minimal replicons were screened to identify factors/sequences that inhibit the replication of the plasmid from which they originated; the original analysis of this type was first done for mini F plasmid (Tolun and Helinski, 1981). Using similar methodology it was determined that π/γ ori interactions are the source of incompatibility in R6K (Filutowicz et al., 1985b; McEachern, 1987). In fact, across the board, iterons play a trans-acting regulatory role in Rep/iteron plasmid replication (reviewed in: McEachern et al., 1986; Helinski et al., 1996; Chattoraj, 2000; Park and Chattoraj, 2001) even though they encode no product (Papp et al., 1994; Wu et al., 1996).
Taking advantage of iteron-mediated incompatibility, Kunnimalaiyaan et al. (2004) isolated mutations in iteron DNA that adversely affected π binding. The experiment was designed to support investigations, described in Section 2.3, that characterized Rep/iteron contacts and how π monomers and dimers differ in this regard. However, the data also contribute to our understanding of the mechanisms underlying plasmid incompatibility. The success of the mutant isolation hinged on the ability of the investigators to limit the replication of a γ ori plasmid. This was accomplished by placing pir under the control of a PBAD promoter (from the araBAD operon) and keeping expression levels low (Bowers et al., 2004). When high copy vectors bearing a single wt iteron were introduced into the controlled system, the replication of a co-resident γ ori plasmid decreased or was prevented altogether. This observation was consistent with a hypothesis that the extra vector-borne iterons competed for replication-limiting π monomers. Providing additional evidence, substituting a monomer-biased copy-up π variant for wt protein restored γ ori replication. Substituting mutant iterons for the high copy wt iterons had the same effect. Thus, the inhibition of replication seemingly relied on a titration-based mechanism of incompatibility (Fig. 4, lower right).
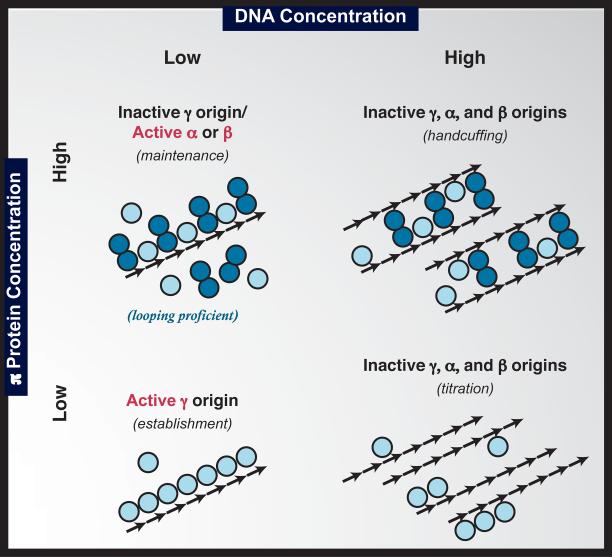
Figure 4.
Homeostasis model of R6K origin selection. Arrows represent the 7 tandem iterons (direct repeats) of γ ori. Monomers and dimers of π protein can bind iterons as indicated. Iteron concentrations regulate R6K replication in a π concentration-dependent manner. At high iteron concentrations, one of two mechanisms (minimally) would turn off R6K replication. At low iteron concentrations, replication would initiate from the establishment origin (γ ori) when low π levels promote iteron filling by monomers. At elevated π levels, γ ori is silenced by dimer binding but one of two flanking maintenance oris (α or β) can be activated by π dimer-mediated DNA looping (iteron coupling in cis rather than in trans).
3.2. Handcuffing is proposed to inhibit replication at high levels of π and iterons
Although titration of π by iterons likely accounts for plasmid incompatibility at low levels of the protein, it cannot account for observations where incompatibility persists after more π is added to the system (McEachern et al., 1989). Another proposed mechanism for iteron-mediated inhibition of replication, called handcuffing (Fig. 4), is a phenomenon in which Rep can be seen to couple iteron-bearing molecules (Mukherjee et al., 1985; McEachern, 1987; McEachern et al., 1989); see also reviews by Perri et al., (1991) and Helinski (2004). Handcuffing is not expected to be relieved by adding more π (Rep) to the system; if anything, this would likely facilitate the process. Electron microscopy (EM) has been used to visualize and characterize π-mediated handcuffing (Urh et al., 1998), however, the precise nucleoprotein composition of handcuffed structures and the mechanism(s) by which they form remain unclear. Ligation enhancement assays (see McEachern et al., 1989; Miron et al., 1992; Miron et al., 1994) were conducted to explore the role of π dimerization in the formation of handcuffed structures. The technique relies on a protein's ability to simultaneously bind two DNA fragments thereby increasing the local concentration of fragments’ ends in ligation reactions. When such assays were performed, comparing wt π with copy-up variants known to have a monomer-bias in DNA binding, the variants were less efficient in forming ligated products (Kunnimalaiyaan et al., 2005). In contrast, a dominant negative π variant, πM36A^M38A (Krüger et al., 2001) that binds iterons exclusively in dimeric form appeared to handcuff DNA more efficiently (than wt π). It was concluded that π dimers bound to iterons have a greater propensity to participate in handcuffing than do π monomers.
In each subunit of a Rep dimer, the C-terminal DNA-binding motif (or WH2) is thought to be available for DNA binding. However, for a period of several years, π appeared to be unique among Reps in its ability to bind iterons as a dimer (Urh et al., 1998; Diaz-Lopez et al., 2003; Das and Chattoraj, 2004; Diaz-Lopez et al., 2006). The unusually high stability of nucleoprotein complexes involving dimeric π and iteron-bearing DNA probes led to the hypothesis that Rep dimers might be both necessary and sufficient to couple DNA segments into handcuffing structures (Urh et al., 1998). This proposed mechanism for handcuffing involved simpler nucleoprotein complexes than those invoked in several other systems (Toukdarian and Helinski, 1998; Das and Chattoraj, 2004; Zzaman and Bastia, 2005). Although π may employ a mechanism of handcuffing distinct from these other Reps, that seems unlikely. What seems more likely is that most Rep dimers bind iterons more weakly than π (and RepA of pPS10, discussed below) and the formation of a handcuffed structure helps stabilize the dimer/iteron association by coupling binding sites and bridging them with multiple dimers. As we have discussed, π exhibits both similarities to, and differences from, other Reps. It was π's unusual ability to stably bind an iteron as a dimer that allowed investigators to generate the contact probing data, described earlier (Kunnimalaiyaan et al., 2004), which so strikingly support crystallography predictions from other Rep molecules (Rep dimers can only use one of two DNA binding domains). Thus, the facility with which R6K researchers can manipulate the forms of Rep (monomer and/or dimer) that bind to iterons appears to offer a unique tool for gaining insight into the composition of Rep/iteron handcuffing complexes.
Another handcuffing model that invoked simple nucleoprotein structures was proposed for the F plasmid system (Uga et al., 1999). The main difference between the models of F and R6K centers on the question of whether preformed dimers mediate handcuffing or, alternatively, a dimeric association between iteron-bound monomers. Recent work on the pPS10 model system supports the involvement of a “simple” nucleoprotein complex in handcuffing while adding the twist that the dimerization interface used by a handcuffing Rep dimer may be related to but not identical with the interface employed by other Rep dimers (Chattoraj, 2000; Gasset-Rosa et al., 2008). As noted above, the ability of ΔC164π to bind the A+T-rich region as a dimer despite lacking a WH2 domain would be consistent with a dimerization interface that allows WH1 interaction with the DNA. Of course unlike most other systems, R6K also presents the possibility that competition by dimers for iteron binding could supersede handcuffing as the primary mode of shutting down the replication origin. Rather than discounting one or more competing models, we would propose that dimers of π employ multiple mechanisms for shutting down replication at γ ori or the entire R6K plasmid (Filutowicz et al., 1986a), and that the concentration of iterons and π protein dictate which mechanism(s) will be employed (Fig. 4). Adding another layer to the regulation of R6K, at the same time that dimers are inhibiting γ ori, they may be activating one of its flanking replication origins as will be described, below.
3.3. Replication origins α and β, and their relationships to γ ori
How does the minimal π/γ ori replicon fit into the life cycle of the parental R6K plasmid? Unlike most Rep/iteron plasmids, an important characteristic of R6K (a.k.a., RSF1040) is that it has, not 1, but 3 functional origins of replication and, typically, the oris do not fire concurrently (Crosa, 1980). Research designed to elucidate the mechanisms that regulate ori selection has recently been undertaken (Saxena et al., 2010a; Saxena et al., 2010b) and more work in this area will be necessary for a full appreciation of the biology of this antibiotic resistance plasmid. It will also contribute to our understanding of replication in a broader context, as multiple replication origins are typical of eukaryotic replicons and are also seen outside of Eukarya (e.g., Huberman and Riggs, 1968; Blumenthal et al., 1974; Newlon et al., 1974; Bourguignon et al., 1976; Robinson et al., 2004). Although the γ ori of R6K has been the most studied, its other origins, α and β are preferentially utilized in vivo (Crosa et al., 1976; Crosa, 1980). Curiously, however, the γ ori core is necessary for the activation of the other two oris. π protein facilitates interaction between the iterons in γ ori and iteron-like sequences in oris α and β (Mukherjee et al., 1988b; Miron et al., 1992; Saxena et al., 2010a; Saxena et al., 2010b). This results in a looping of the plasmid DNA, which is believed to transmit the replication signal from the internal iteron cluster (7 DRs) to the outlying R6K oris (Mukherjee et al., 1988a; Mukherjee et al., 1988b). Studies of the interaction between γ and α oris suggest that both monomers and dimers of π protein participate in the looping process (Saxena et al., 2010a; Saxena et al., 2010b). Thus, related yet distinct protein-protein and protein-DNA interactions appear to be crucial to the function of each replication origin. The looping itself, however, does not adequately explain the purpose of γ ori if it is nearly silent under typical laboratory conditions.
R6K is a self-transmissible plasmid (Kontomichalou et al., 1970). Its conjugative lifestyle means that a newly transferred replicon will find itself in a very different intracellular environment than an established replicon. A conjugation-proficient donor cell would be expected to have roughly 13-38 copies of the R6K and 4,000 molecules of π (Filutowicz et al., 1986a). In contrast, a brand new recipient cell will have acquired a single copy of the plasmid upon conjugational transfer into an initially π-free environment. As we have noted, the α and β oris appear to be more active than γ ori in vivo under presumably homeostatic conditions. Thus, it has been proposed that the γ ori might be an establishment origin, preferentially firing in recipients immediately after plasmid transfer when levels of available π protein are low (Filutowicz et al., 1994a). As π accumulates in the cell, its auto-regulatory circuitry becomes active (Filutowicz et al., 1985a). Concomitantly, a shift in ori usage is predicted with γ ori becoming relatively less active while more activity would occur at the maintenance origins - α and β. Activation of γ ori is known to be dependent on π monomers while evidence suggests that dimers play a role in activating the flanking oris (Saxena et al., 2010a; Saxena et al., 2010b), which would fit a ‘maintenance vs. establishment’ hypothesis. If the hypothesis is correct, it could explain why a two orders-of-magnitude decrease in π protein levels has only a modest effect on plasmid copy number while a two-fold increase inhibits replication of γ ori (Filutowicz et al., 1986a; Dellis et al., 1996). Additional support for the idea comes from investigations in which the intracellular π levels that activate α and β oris were found to be considerably higher than the levels activating γ ori (Filutowicz et al., 1994a).
Plasmid establishment and maintenance are two aspects of the model of R6K copy number control presented in Figure 4. This model combines several identified regulatory mechanisms into a homeostatic system arising from what are predicted to be distinct yet inter-related roles for π and iteron concentrations in R6K regulation. Supporting evidence is strongly suggestive but not yet conclusive. Nonetheless, the interplay of the multiple negative regulatory mechanisms proposed is in many ways similar to the homeostasis model described for another Rep/iteron plasmid, P1 (Das et al., 2005).
The concentration-dependent effects of π and iterons on the replication of R6K derivatives have been well documented (Stalker et al., 1981; Filutowicz et al., 1986a; McEachern et al., 1986). What has not been systematically investigated are the possible ori-specific effects these replication regulators may have, how these variables might influence ori selection, and how the differential ori activities are integrated into the overall replication phenomena observed. Pertinent to resolving the roles of π concentration on global R6K regulation, M. McEachern was able to generate a plasmid collection with deletions in the 5' non-coding region of pir (McEachern et al., 1985; McEachern, 1987) that alter the intracellular levels of π (Filutowicz et al., 1986a). Together, these mutant pir genes cover a range of π expression from <1% of wt levels to an 8-fold increase in the protein level. Variants producing the smallest amounts of protein resulted in reduced γ ori plasmid copy number whereas the 8-fold over-expression variant did not allow establishment of either a γ ori plasmid or an α–γ–β ori plasmid. Moderate reduction of π levels led to elevated γ ori copy number (Filutowicz et al., 1986a). Thus, these π expression variants appear to cover the Rep concentration range illustrated in Fig 4, from π levels low enough for titration to be observed (lower right) to levels high enough to shut down all 3 oris (upper right). This collection of pir expression plasmids (wt and copy up variants) should be a useful tool for further examinations of how the intracellular levels of π, and the ratios of its monomers vs. dimers, influence the process of origin switching.
4. NEW FRONTIERS IN PLASMID R6K RESEARCH
4.1. Applying basic research to develop new classes of antibiotics
Research in many laboratories has demonstrated that the fundamental principles of vegetative and conjugative DNA replication apply to all kinds of plasmids, whether they are benign “models” used for basic research or virulence and antibiotic resistance plasmids found in bacterial pathogens. This realization inspired a group of plasmid biologists to propose that plasmids can be modified to become new classes of antibiotics and that plasmid replication can be utilized as a new antibacterial target (Filutowicz, 2004; Peng et al., 2006; Filutowicz et al., 2008). The latter idea, known as “plasmid curing”, dates back to pioneering work by M. Yoshikawa (1974), and a true renaissance of this idea was recently reviewed by J. Williams and P. Hergenrother (Williams and Hergenrother, 2008). The homeostasis model of Figure 4 illustrates how R6K might be maintained at a fixed copy number inside the bacterial cell. Well-planned interference with regulatory elements might cause R6K to reproduce too fast, intoxicating the host bacteria with copious amounts of plasmid DNA, a phenomenon first described for plasmid R1 (Uhlin et al., 1979; Molin et al., 1980). This was the goal of a set of experiments in which site-directed mutagenesis was used to ‘delete codons’ of the pir gene that corresponded to known copy-up amino acid substitutions (Peng et al., 2006). The approach was guided by the knowledge that the phenotypes of copy-up pir mutants can be enhanced by combining them (Filutowicz et al., 1986b; Krüger et al., 2001) and one, lethal, over-replication form of π protein had been previously described (Stalker et al., 1983). Moreover, it was anticipated that deleting amino acid residues in the known ‘copy control domain’ might have a more severe phenotype than the corresponding, known, copy-up substitutions. Indeed, the small pir deletions generated by Peng et al., (2006) displayed copy-up phenotypes, making them, to our knowledge, the first recorded instances of amino acid deletions in any Rep protein to confer a hyper-replication phenotype.
In a series of experiments, three of the four mutant pir-containing constructs could not be established in E. coli unless a form of π retaining inhibitor function was also expressed. This result, which required the mutant pir genes to be placed in cis to the ori, was consistent with the idea that plasmid over-replication can be lethal (or bacteriostatic). Since the synthesis of plasmids and chromosomes share components of DNA replication machinery, perhaps runaway plasmid replication siphons off rate-limiting replication components (Peng et al., 2006; Filutowicz et al., 2008). Relevant to the potential antibacterial effects, in the RK2/RP4 plasmid system, some copy up mutants have been found to be bacteriocidal in a species dependent fashion (Haugan et al., 1995). This observation led the authors to propose that the maximal tolerable plasmid content could vary from one bacterial species to another. But regardless of the mechanism that causes cell death as a function of plasmid over-replication, the phenomenon deserves to be examined rigorously as it may reveal novel target(s) for antimicrobial control of plasmid-harboring pathogens.
5. CONCLUDING REMARKS
The contributions of plasmids to the development of modern molecular biology are unparalleled and have been summarized elsewhere (Cohen, 1993). But the need for ongoing and vigorous research into plasmid biology remains as strong as ever. Detailed analyses of vegetative and conjugative plasmid replication are expected to continue providing valuable information about the mechanisms controlling the vertical and horizontal inheritance of extra-chromosomal DNA. Only when armed with multiple tools can scientists hope to probe deeply into the mechanisms that regulate these processes, and plasmid R6K is particularly bountiful in this regard. Already available to researchers are large collections of pir mutants, assortments of informative iteron mutants, useful ori constructs and a promising model for the 3 dimensional structure of π bound to an iteron. Moreover, R6K both contributes to and benefits from a vast and ever-expanding knowledge-base of Rep/iteron plasmid replication. Thoughtful interventions that disrupt the regulation of plasmid copy number could, conceivably, be employed to kill bacteria or displace the determinants of antibiotic resistance, virulence, and biofilm formation in the innumerable cases where bacterial plasmids contribute to these phenomena. As evidenced by the body of published work described here, plasmid R6K is superbly positioned to test creative approaches for managing bacteria and their less desirable traits through manipulating their plasmid content.
HIGHLIGHTS.
* A survey of investigations into the replication control of plasmid R6K is presented.
* Special emphasis is placed on the functional diversity of π / π and π /DNA interactions.
* A simple homeostatic model is offered to account for the regulated usage of R6K's three replication origins.
* Prospects for the use of the plasmid in antibiotic discovery research are described.
ACKNOWLEDGEMENTS
This work was supported by the NIAID at the National Institutes of Health (Grant AI081087), the Alfred P. Sloan Foundation (Grant B2007-10), and the Cooperative State Research Service, U.S. Department of Agriculture (Project No. WIS01412). Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the funding organizations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT
Marcin Filutowicz is required by the U.W Conflict of Interest Committee to disclose a financial interest in ConjuGon, Inc. and PlasmiGon, LLC, Madison-based companies that he founded.
REFERENCES
- Abhyankar MM, et al. Biochemical investigations of control of replication initiation of plasmid R6K. J Biol Chem. 2004;279:6711–6719. doi: 10.1074/jbc.M312052200. [DOI] [PubMed] [Google Scholar]
- Abhyankar MM, et al. Reconstitution of R6K DNA replication in vitro using 22 purified proteins. J Biol Chem. 2003;278:45476–45484. doi: 10.1074/jbc.M308516200. [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, et al. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Bourguignon GJ, et al. Multiple origins and circular structures in replicating T5 bacteriophage DNA. J Virol. 1976;18:245–259. doi: 10.1128/jvi.18.1.245-259.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers LM, Filutowicz M. Cooperative binding mode of the inhibitors of R6K replication, π dimers. J Mol Biol. 2008;377:609–615. doi: 10.1016/j.jmb.2008.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers LM, et al. Mechanism of origin activation by monomers of R6K-encoded π protein. J Mol Biol. 2007;368:928–938. doi: 10.1016/j.jmb.2007.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers LM, et al. Bacterial expression system with tightly regulated gene expression and plasmid copy number. Gene. 2004;340:11–18. doi: 10.1016/j.gene.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Burmolle M, et al. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology. 2008;154:187–195. doi: 10.1099/mic.0.2007/010454-0. [DOI] [PubMed] [Google Scholar]
- Chattoraj DK. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- Chattoraj DK, Schneider TD. Replication control of plasmid P1 and its host chromosome: the common ground. Prog Nucleic Acid Res Mol Biol. 1997;57:145–186. doi: 10.1016/s0079-6603(08)60280-9. [DOI] [PubMed] [Google Scholar]
- Chen D, et al. Replication of R6K γ origin in vitro: discrete start sites for DNA synthesis dependent on π and its copy-up variants. J Mol Biol. 1998;282:775–787. doi: 10.1006/jmbi.1998.2055. [DOI] [PubMed] [Google Scholar]
- Cohen SN. Bacterial plasmids: their extraordinary contribution to molecular genetics. Gene. 1993;135:67–76. doi: 10.1016/0378-1119(93)90050-d. [DOI] [PubMed] [Google Scholar]
- Crosa JH. Three origins of replication are active in vivo in the R plasmid RSF1040. J Biol Chem. 1980;255:11075–11077. [PubMed] [Google Scholar]
- Crosa JH, et al. Mode of replication of the conjugative R-plasmid RSF1040 in Escherichia coli. J Bacteriol. 1976;126:454–466. doi: 10.1128/jb.126.1.454-466.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N, Chattoraj DK. Origin pairing (‘handcuffing’) and unpairing in the control of P1 plasmid replication. Mol Microbiol. 2004;54:836–849. doi: 10.1111/j.1365-2958.2004.04322.x. [DOI] [PubMed] [Google Scholar]
- Das N, et al. Multiple homeostatic mechanisms in the control of P1 plasmid replication. Proc Natl Acad Sci U S A. 2005;102:2856–2861. doi: 10.1073/pnas.0409790102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- Dellis S, et al. Replication of plasmid R6K γ origin in vivo and in vitro: dependence on IHF binding to the ihf1 site. J Mol Biol. 1996;257:550–560. doi: 10.1006/jmbi.1996.0184. [DOI] [PubMed] [Google Scholar]
- Diaz-Lopez T, et al. Early events in the binding of the pPS10 replication protein RepA to single iteron and operator DNA sequences. J Mol Biol. 2006;364:909–920. doi: 10.1016/j.jmb.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Diaz-Lopez T, et al. Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J Biol Chem. 2003;278:18606–18616. doi: 10.1074/jbc.M212024200. [DOI] [PubMed] [Google Scholar]
- Espinosa M, et al. Plasmid replication and copy number control. In: Thomas CM, editor. The Horizontal Gene Pool : Bacterial Plasmids and Gene Spread. Harwood Academic; Amsterdam: 2000. [Google Scholar]
- Filutowicz M. United States Patent and Trademark Office, editor. Displacing a plasmid in a bacterial population. 2004 http://www.google.com/patents/US20040224340 US 2004/0224340 A1, United States.
- Filutowicz M, Appelt K. The integration host factor of Escherichia coli binds to multiple sites at plasmid R6K γ origin and is essential for replication. Nucleic Acids Res. 1988;16:3829–3843. doi: 10.1093/nar/16.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M, et al. Bacterial conjugation-based antimicrobial agents. Plasmid. 2008;60:38–44. doi: 10.1016/j.plasmid.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Filutowicz M, et al. Autorepressor properties of the π-initiation protein encoded by plasmid R6K. Nucleic Acids Res. 1985a;13:103–114. doi: 10.1093/nar/13.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M, et al. Regulation of replication of an iteron-containing DNA molecule. In: Cohn W, Moldave K, editors. Progress in Nucleic Acid Research and Molecular Biology. Vol. 48. Academic Press; San Diego: 1994a. [DOI] [PubMed] [Google Scholar]
- Filutowicz M, et al. Role of the π initiation protein and direct nucleotide sequence repeats in the regulation of plasmid R6K replication. In: Helinski DR, et al., editors. Plasmids in Bacteria. Vol. 30. Plenum Press; New York: 1985b. [DOI] [PubMed] [Google Scholar]
- Filutowicz M, et al. Positive and negative roles of an initiator protein at an origin of replication. Proc Natl Acad Sci U S A. 1986a;83:9645–9649. doi: 10.1073/pnas.83.24.9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M, et al. DNA and protein interactions in the regulation of plasmid replication. J Cell Sci Suppl. 1987;7:15–31. doi: 10.1242/jcs.1987.supplement_7.2. [DOI] [PubMed] [Google Scholar]
- Filutowicz M, Rakowski SA. Regulatory implications of protein assemblies at the γ origin of plasmid R6K - a review. Gene. 1998;223:195–204. doi: 10.1016/s0378-1119(98)00367-9. [DOI] [PubMed] [Google Scholar]
- Filutowicz M, et al. Binding of purified wild-type and mutant π initiation proteins to a replication origin region of plasmid R6K. J Mol Biol. 1986b;187:225–239. doi: 10.1016/0022-2836(86)90230-5. [DOI] [PubMed] [Google Scholar]
- Filutowicz M, et al. Cooperative binding of initiator protein to replication origin conferred by single amino acid substitution. Nucleic Acids Res. 1994b;22:4211–4215. doi: 10.1093/nar/22.20.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluit AC. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol Med Microbiol. 2005;43:1–11. doi: 10.1016/j.femsim.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Gammie AE, Crosa JH. Roles of DNA adenine methylation in controlling replication of the REPI replicon of plasmid pColV-K30. Mol Microbiol. 1991;5:495–503. doi: 10.1111/j.1365-2958.1991.tb02133.x. [DOI] [PubMed] [Google Scholar]
- Gasset-Rosa F, et al. Negative regulation of pPS10 plasmid replication: origin pairing by zipping-up DNA-bound RepA monomers. Mol Microbiol. 2008;68:560–572. doi: 10.1111/j.1365-2958.2008.06166.x. [DOI] [PubMed] [Google Scholar]
- Germino J, Bastia D. Interaction of the plasmid R6K-encoded replication initiator protein with its binding sites on DNA. Cell. 1983a;34:125–134. doi: 10.1016/0092-8674(83)90142-3. [DOI] [PubMed] [Google Scholar]
- Germino J, Bastia D. The replication initiator protein of plasmid R6K tagged with beta- galactosidase shows sequence-specific DNA-binding. Cell. 1983b;32:131–140. doi: 10.1016/0092-8674(83)90503-2. [DOI] [PubMed] [Google Scholar]
- Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- Giraldo R, et al. Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. EMBO J. 1998;17:4511–4526. doi: 10.1093/emboj/17.15.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo R, Díaz-Orejas R. Similarities between the DNA replication initiators of Gram-negative bacteria plasmids (RepA) and eukaryotes (Orc4p)/archaea (Cdc6p). Proc Natl Acad Sci U S A. 2001;98:4938–4943. doi: 10.1073/pnas.081079298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo R, et al. A conformational switch between transcriptional repression and replication initiation in the RepA dimerization domain. Nat Struct Biol. 2003;10:565–571. doi: 10.1038/nsb937. [DOI] [PubMed] [Google Scholar]
- Giraldo R, Fernandez-Tresguerres ME. Twenty years of the pPS10 replicon: insights on the molecular mechanism for the activation of DNA replication in iteron-containing bacterial plasmids. Plasmid. 2004;52:69–83. doi: 10.1016/j.plasmid.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Greener A, et al. N-terminal truncated forms of the bifunctional π initiation protein express negative activity on plasmid R6K replication. Mol Gen Genet. 1990;224:24–32. doi: 10.1007/BF00259447. [DOI] [PubMed] [Google Scholar]
- Haugan K, et al. The host range of RK2 minimal replicon copy-up mutants is limited by species-specific differences in the maximum tolerable copy number. Plasmid. 1995;33:27–39. doi: 10.1006/plas.1995.1004. [DOI] [PubMed] [Google Scholar]
- Heinemann JA, et al. Do antibiotics maintain antibiotic resistance? Drug Discov Today. 2000;5:195–204. doi: 10.1016/s1359-6446(00)01483-5. [DOI] [PubMed] [Google Scholar]
- Helinski D. Introduction to plasmids: A selective review of their history. In: Funnell BE, Phillips GJ, editors. Plasmid Biology. ASM Press; Washington, D.C.: 2004. [Google Scholar]
- Helinski DR, et al. Replication control and other stable maintenance mechanisms of plasmids. In: Neidhardt RCIF, Ingraham JL, Lin ECC, Brooks Low K, Magasanik B, Reznikoff W, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella Cellular and Molecular Biology. Vol. 2. ASM Press; Washington D.C.: 1996. [Google Scholar]
- Huberman JA, Riggs AD. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968;32:327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Inuzuka M, Helinski DR. Replication of antibiotic resistance plasmid R6K DNA in vitro. Biochemistry. 1978;17:2567–2573. doi: 10.1021/bi00606a017. [DOI] [PubMed] [Google Scholar]
- Kelley W, Bastia D. Replication initiator protein of plasmid R6K autoregulates its own synthesis at the transcriptional step. Proc Natl Acad Sci U S A. 1985;82:2574–2578. doi: 10.1073/pnas.82.9.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley WL, Bastia D. Conformational changes induced by integration host factor at origin γ of R6K and copy number control. J Biol Chem. 1991;266:15924–15937. [PubMed] [Google Scholar]
- Kolter R. Replication properties of plasmid R6K. Plasmid. 1981;5:2–9. doi: 10.1016/0147-619x(81)90073-1. [DOI] [PubMed] [Google Scholar]
- Komori H, et al. Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 Å resolution. Embo J. 1999;18:4597–4607. doi: 10.1093/emboj/18.17.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny I, Helinski DR. The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc Natl Acad Sci U S A. 1997;94:14378–14382. doi: 10.1073/pnas.94.26.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontomichalou P, et al. Circular R-factor molecules controlling penicillinase synthesis, replicating in Escherichia coli under either relaxed or stringent control. J Bacteriol. 1970;104:34–44. doi: 10.1128/jb.104.1.34-44.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger R, Filutowicz M. Dimers of π protein bind the A+T-rich region of the R6K γ origin near the leading-strand synthesis start sites: regulatory implications. J Bacteriol. 2000;182:2461–2467. doi: 10.1128/jb.182.9.2461-2467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger R, Filutowicz M. π protein- and ATP-dependent transitions from ‘closed’ to ‘open’ complexes at the γ ori of plasmid R6K. Nucleic Acids Res. 2003;31:5993–6003. doi: 10.1093/nar/gkg809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger R, et al. Monomer/dimer ratios of replication protein modulate the DNA strand- opening in a replication origin. J Mol Biol. 2001;306:945–955. doi: 10.1006/jmbi.2000.4426. [DOI] [PubMed] [Google Scholar]
- Krüger R, et al. Isomerization and apparent DNA bending by π, the replication protein of plasmid R6K. Biochem Biophys Res Commun. 2004a;313:834–840. doi: 10.1016/j.bbrc.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Krüger R, et al. Participating elements in the replication of iteron-containing plasmids. In: Funnell BE, Phillips GJ, editors. Plasmid Biology. ASM Press; Washington, D.C.: 2004b. [Google Scholar]
- Kunnimalaiyaan S, et al. Role of π dimers in coupling (“handcuffing”) of plasmid R6K's γ ori iterons. J Bacteriol. 2005;187:3779–3785. doi: 10.1128/JB.187.11.3779-3785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnimalaiyaan S, et al. Binding modes of the initiator and inhibitor forms of the replication protein π to the γ ori iteron of plasmid R6K. J Biol Chem. 2004;279:41058–41066. doi: 10.1074/jbc.M403151200. [DOI] [PubMed] [Google Scholar]
- Kunnimalaiyaan S, et al. Structure-based functional analysis of the replication protein of plasmid R6K: Key amino acids at the π/DNA Interface. J Bacteriol. 2007;189:4953–4956. doi: 10.1128/JB.00109-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Filutowicz M. Initiator protein π can bind independently to two domains of the γ origin core of plasmid R6K: the direct repeats and the A+T-rich segment. Nucleic Acids Res. 1996;24:1936–1942. doi: 10.1093/nar/24.10.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YB, et al. Mechanistic studies of initiator-initiator interaction and replication initiation. Embo J. 1998;17:5192–5200. doi: 10.1093/emboj/17.17.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JS, et al. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012;65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- Mazel D, Davies J. Antibiotic resistance in microbes. Cell Mol Life Sci. 1999;56:742–754. doi: 10.1007/s000180050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ. Regulation of plasmid replication by the interaction of an initiator protein with multiple DNA binding sites. University of California-San Diego; LaJolla, Ca: 1987. [Google Scholar]
- McEachern MJ, et al. Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc Natl Acad Sci U S A. 1989;86:7942–7946. doi: 10.1073/pnas.86.20.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, et al. Mutations in direct repeat sequences and in a conserved sequence adjacent to the repeats result in a defective replication origin in plasmid R6K. Proc Natl Acad Sci U S A. 1985;82:1480–1484. doi: 10.1073/pnas.82.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, et al. Elements involved in the copy control regulation of the antibiotic resistance plasmid R6K. In: Levy SB, Novick RP, editors. Banbury Report 24: Antibiotic Resistance Genes: Ecology, Transfer, and Expression. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1986. [Google Scholar]
- Miron A, et al. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance [published erratum appears in EMBO J 1992 May;11(5):2002]. Embo J. 1992;11:1205–1216. doi: 10.1002/j.1460-2075.1992.tb05161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A, et al. Multiple pathways of copy control of γ replicon of R6K: mechanisms both dependent on and independent of cooperativity of interaction of π protein with DNA affect the copy number. Proc Natl Acad Sci U S A. 1994;91:6438–6442. doi: 10.1073/pnas.91.14.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S, et al. Runaway replication of plasmid R1 is not caused by loss of replication inhibitor activity of gene cop. J Bacteriol. 1980;143:1046–1048. doi: 10.1128/jb.143.2.1046-1048.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DD, et al. Dissection and comparative anatomy of the origins of replication of lambdoid phages. Cold Spring Harb Symp Quant Biol. 1979;43:155–163. doi: 10.1101/sqb.1979.043.01.022. [DOI] [PubMed] [Google Scholar]
- Moreland JL, et al. The Molecular Biology Toolkit (MBT): a modular platform for developing molecular visualization applications. BMC Bioinformatics. 2005;6:21. doi: 10.1186/1471-2105-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, et al. Detection of DNA looping due to simultaneous interaction of a DNA- binding protein with two spatially separated binding sites on DNA. Proc Natl Acad Sci U S A. 1988a;85:6287–6291. doi: 10.1073/pnas.85.17.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, et al. Enhancer-origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell. 1988b;52:375–383. doi: 10.1016/s0092-8674(88)80030-8. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, et al. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985;43:189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Nakamura A, et al. Structural basis for regulation of bifunctional roles in replication initiator protein. Proc Natl Acad Sci U S A. 2007;104:18484–18489. doi: 10.1073/pnas.0705623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon CS, et al. Replication of yeast chromosomal DNA. Nature. 1974;247:32–35. doi: 10.1038/247032a0. [DOI] [PubMed] [Google Scholar]
- Nordstrom K. Control of plasmid replication--how do DNA iterons set the replication frequency? Cell. 1990;63:1121–1124. doi: 10.1016/0092-8674(90)90405-4. [DOI] [PubMed] [Google Scholar]
- Norman A, et al. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid. 2008;60:59–74. doi: 10.1016/j.plasmid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp PP, et al. Information analysis of sequences that bind the replication initiator RepA. J Mol Biol. 1993;233:219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- Papp PP, et al. Negative control of plasmid DNA replication by iterons. Correlation with initiator binding affinity. J Biol Chem. 1994;269:23563–23568. [PubMed] [Google Scholar]
- Park K, Chattoraj DK. DnaA boxes in the P1 plasmid origin: the effect of their position on the directionality of replication and plasmid copy number. J Mol Biol. 2001;310:69–81. doi: 10.1006/jmbi.2001.4741. [DOI] [PubMed] [Google Scholar]
- Peng Y, et al. Small deletion variants of the replication protein, π, and their potential for over-replication-based antimicrobial activity. FEMS Microbiol Lett. 2006;261:245–252. doi: 10.1111/j.1574-6968.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- Perri S, et al. Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J Biol Chem. 1991;266:12536–12543. [PubMed] [Google Scholar]
- Ratnakar PV, et al. The replication initiator protein π of the plasmid R6K specifically interacts with the host-encoded helicase DnaB. Proc Natl Acad Sci U S A. 1996;93:5522–5526. doi: 10.1073/pnas.93.11.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, et al. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- Saxena M, et al. Replication initiation at a distance: determination of the cis- and trans-acting elements of replication origin α of plasmid R6K. J Biol Chem. 2010a;285:5705–5712. doi: 10.1074/jbc.M109.067348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M, et al. Investigations of π initiator protein-mediated interaction between replication origins α and γ of the plasmid R6K. J Biol Chem. 2010b;285:5695–5704. doi: 10.1074/jbc.M109.067439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD. Strong minor groove base conservation in sequence logos implies DNA distortion or base flipping during replication and transcription initiation. Nucleic Acids Res. 2001;29:4881–4891. doi: 10.1093/nar/29.23.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafferman A, et al. Structure-function relationships in essential regions for plasmid replication. In: Levy SB, et al., editors. Molecular Biology, Pathogenicity, and Ecology of Bacterial Plasmids. Plenum Publishing Corporation; 1981. [Google Scholar]
- Sharma S, et al. Plasmid P1 RepA is homologous to the F plasmid RepE class of initiators. J Biol Chem. 2004;279:6027–6034. doi: 10.1074/jbc.M310917200. [DOI] [PubMed] [Google Scholar]
- Stalker DM, et al. Release of initiation control by a mutational alteration in the R6K π protein required for plasmid DNA replication. Proc Natl Acad Sci U S A. 1983;80:5500–5504. doi: 10.1073/pnas.80.18.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalker DM, et al. Direct repeats of nucleotide sequences are involved in plasmid replication and incompability. In: Ray DS, editor. The Initiation of DNA Replication. XXII. Academic Press; New York: 1981. [Google Scholar]
- Swan MK, et al. Crystal structure of π initiator protein-iteron complex of plasmid R6K: implications for initiation of plasmid DNA replication. Proc Natl Acad Sci U S A. 2006;103:18481–18486. doi: 10.1073/pnas.0609046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N, Pathogen Genomics Group . Data: E. coli reference plasmids. Plasmid:R6K. Sequence. Wellcome Trust Sanger Institute; 2012. http://www.sanger.ac.uk/resources/downloads/plasmids/ [Google Scholar]
- Tolun A, Helinski DR. Direct repeats of the F plasmid incC region express F incompatibility. Cell. 1981;24:687–694. doi: 10.1016/0092-8674(81)90095-7. [DOI] [PubMed] [Google Scholar]
- Toukdarian AE, Helinski DR. TrfA dimers play a role in copy-number control of RK2 replication. Gene. 1998;223:205–211. doi: 10.1016/s0378-1119(98)00370-9. [DOI] [PubMed] [Google Scholar]
- Uga H, et al. Regulation of DNA replication by iterons: an interaction between the ori2 and incC regions mediated by RepE-bound iterons inhibits DNA replication of mini-F plasmid in Escherichia coli. EMBO J. 1999;18:3856–3867. doi: 10.1093/emboj/18.13.3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin BE, et al. Plasmids with temperature-dependent copy number for amplification of cloned genes and their products. Gene. 1979;6:91–106. doi: 10.1016/0378-1119(79)90065-9. [DOI] [PubMed] [Google Scholar]
- Urh M, et al. Assemblies of replication initiator protein on symmetric and asymmetric DNA sequences depend on multiple protein oligomerization surfaces. J Mol Biol. 1998;283:619–631. doi: 10.1006/jmbi.1998.2120. [DOI] [PubMed] [Google Scholar]
- Urh M, et al. Buffer composition mediates a switch between cooperative and independent binding of an initiator protein to DNA. Gene. 1995;164:1–7. doi: 10.1016/0378-1119(95)00493-p. [DOI] [PubMed] [Google Scholar]
- Venkatesan MM, Burland V. Genome-scale analysis of virulence plasmids: the contribution of plasmid-borne virulence genes to enterobacterial pathogenesis. In: Funnell BE, Phillips GJ, editors. Plasmid Biology. ASM Press; Washington, D.C.: 2004. [Google Scholar]
- Vocke C, Bastia D. DNA-protein interaction at the origin of DNA replication of the plasmid pSC101. Cell. 1983;35:495–502. doi: 10.1016/0092-8674(83)90183-6. [DOI] [PubMed] [Google Scholar]
- Watanabe T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, et al. Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc Natl Acad Sci U S A. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J, et al. γ origin plasmids of R6K lineage replicate in diverse genera of Gram-negative bacteria. Annals Microbiol. 2004;54:471–480. [Google Scholar]
- Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles’ heel of drug-resistant bacteria. Curr Opin Chem Biol. 2008;12:389–399. doi: 10.1016/j.cbpa.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, et al. Roles of a 106-bp origin enhancer and Escherichia coli DnaA protein in replication of plasmid R6K. Nucleic Acids Res. 1992;20:811–817. doi: 10.1093/nar/20.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, et al. Preponderance of Fis-binding sites in the R6K γ origin and the curious effect of the penicillin resistance marker on replication of this origin in the absence of Fis [published erratum appears in J Bacteriol 1997 Apr;179(7):2464]. J Bacteriol. 1996;178:4965–4974. doi: 10.1128/jb.178.16.4965-4974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, et al. Two forms of replication initiator protein: positive and negative controls. Proc Natl Acad Sci U S A. 1997;94:13967–13972. doi: 10.1073/pnas.94.25.13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, et al. In vivo and in vitro studies of a copy number mutation of the RepA replication protein of plasmid pSC101. J Bacteriol. 1993;175:4165–4175. doi: 10.1128/jb.175.13.4165-4175.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York D, Filutowicz M. Autoregulation-deficient mutant of the plasmid R6K-encoded π protein distinguishes between palindromic and nonpalindromic binding sites. J Biol Chem. 1993;268:21854–21861. [PubMed] [Google Scholar]
- Yoshikawa M. Screening method of agents against the R factor by the use of an Hfr made by integrative suppression with an R factor. Antimicrob Agents Chemother. 1974;5:362–365. doi: 10.1128/aac.5.4.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zzaman S, Bastia D. Oligomeric initiator protein-mediated DNA looping negatively regulates plasmid replication in vitro by preventing origin melting. Mol Cell. 2005;20:833–843. doi: 10.1016/j.molcel.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Zzaman S, et al. The DnaK-DnaJ-GrpE chaperone system activates inert wild type π initiator protein of R6K into a form active in replication initiation. J Biol Chem. 2004;279:50886–50894. doi: 10.1074/jbc.M407531200. [DOI] [PubMed] [Google Scholar]