Abstract
Characterized by a combination of abnormalities in language, social cognition, and mental flexibility, autism is not a single disorder, but a neurodevelopmental syndrome commonly referred to as autism spectrum disorder (ASD). Several dozen ASD susceptibility genes have been identified in the past decade, collectively accounting for 10–20% of ASD cases. These findings, while demonstrating that ASD is etiologically heterogeneous, provide important clues about its pathophysiology. Diverse genetic and genomic approaches provide evidence converging on disruption of key biological pathways, many of which are also implicated in other allied neurodevelopmental disorders. Knowing the genes involved in ASD provides us with a crucial tool to probe both the specificity of ASD and the shared neurobiological and cognitive features across what are considered clinically distinct disorders, with the goal of linking gene to brain circuits to cognitive function.
Autism genetics: a decade of progress
In many ways, autism is a mysterious disorder, as it involves core abnormalities in social cognition and language, both of which are central to what makes us human. Because of this, understanding autism will have a significant impact on our basic knowledge of these fundamental cognitive processes, in addition to the obvious critical role that such mechanistic understanding has on therapeutic development. Until the last decade, virtually nothing was known about the neurobiological basis of ASD. There was no consistent neuropathology and only scant knowledge of a few causal genetic factors. The last decade has brought an explosion of genetic findings in ASD, surpassing most other common neuropsychiatric disorders, so that we now have knowledge of the etiology of ASD for between 10–20% of cases. Here, I review these exciting findings with respect to what they tell us about the genetic architecture of ASD and how this informs our mechanistic understanding of the disorder and its relation to human brain function.
Definition and evolution of ASD
Autism is a developmental neuropsychiatric syndrome with onset before the age of three. The fundamental conceptualization of the disorder is based on the initial observation of Kanner in 1943 [1], where he described 11 children with autism, mostly boys with a combination of severe social and variable language dysfunction and the presence of repetitive restrictive behaviors. Kanner made numerous interesting observations based on these case studies, including the identification of large head size in about half of the subjects and postulated a biological, genetic basis for the disorder. However, until the 1980s autism was not considered a distinct disorder in the manuals of psychiatric diagnosis, nor was it considered by most to be biologically based.
A major change in perspective came with the pioneering twin studies of Rutter and Folstein that demonstrated a genetic susceptibility to the disorder, providing incontrovertible evidence as to its biological origins [2, 3]. Over the last two decades, the concept of autism has broadened from the strict diagnosis of autistic disorder to include those with normal intelligence and language (Asperger syndrome) and those not quite meeting the strict diagnostic criteria in all three domains (PDD-NOS). These diagnoses are not based on etiology but on expert observation and assessment of behavior and cognition. So, on one hand they can be considered arbitrary –which category one fits into depends partially on subjective criteria. This is further emphasized by the current trend in the DSM-V (http://www.dsm5.org/proposedrevision/Pages/NeurodevelopmentalDisorders.aspx), in which the category of Asperger syndrome is removed and the diagnostic criteria for autism is modified under the new heading of ASD. This change in diagnostic criteria is not based on known similarities or differences in causation between these clinically defined categories, but rather on the consensus opinions of expert clinicians.
Concurrently, over the last two decades, the notion that autism represents a quantitative spectrum of impairments, rather than representing discrete disorders, has gained increasing traction for researchers [4, 5]. From this perspective, the term autism spectrum disorders (ASD) attempts to crystalize the notion that patients represent a clinically variable population that suffers from pathologic levels of quantitative variation in the major cognitive and behavioral domains that are disrupted, rather than a distinct clinical disorder. How these two varying conceptualizations, autism as a unitary disorder, versus a spectrum of dysfunction, relate to underlying etiologies is a key question facing the field. Furthermore, how the clinical domains relate to underlying dysfunction in specific cognitive domains is essentially unknown, although some clues are starting to emerge [6, 7].
Autism is a complex genetic disorder
Perhaps the biggest advance in understanding autism pathophysiology has been the appreciation of a significant genetic contribution to ASD’s etiology. Three main areas of evidence support a genetic etiology in ASD: Twin studies, comparing monozygotic twins (MZ) and dizogotic twins (DZ), family studies comparing the rate of autism in first degree relatives of affected probands versus the population, and studies of rare genetic syndromes with a comorbid autism diagnosis. Because MZ twins share 100% of their genetic material and DZ twins share 50% (similar to non-twin siblings) and both share in utero environment with their twin, higher disease co-occurrence in MZ twins than DZ twins supports a genetic etiology. This is what is observed in every twin study in ASD, overall consistent with heritability estimates of about 70–80% [8, 9]. One exception is a very recent study with a large sample of twins, which, despite showing a concordance of about 0.6 for MZ twins and 0.25 for DZ twins (consistent with a Falconer heritability of 0.7), comes to the conclusion that shared environment plays a larger role than genetic factors [10]. Given that the concordance for strict autism in DZ twins (who can be considered genetic siblings who share the same in utero environment) is not clearly different from modern infant sibling recurrence rates [11], how shared environment would have a more major role than genetics is not clear. Moreover, studies in families show that first-degree relatives of autistic probands have a markedly increased risk for autism relative to the population, consistent with a strong familial, or genetic, effect observed in twins [12]. This is not to dispute the role of the environment, but to acknowledge that genes clearly play an important role.
First-degree relatives of ASD probands have an increase in behavioral or cognitive features associated with autism, such as social or language dysfunction, albeit in lesser forms, when compared with the population prevalence [13]. This has been called “the broader phenotype” and includes restrictive repetitive behaviors and sub-threshold deficits in social cognition, as well as language dysfunction [14–17]. For example, language delay is observed in a significant proportion of non-autistic siblings of autistic probands [18, 19]. Similarly, autistic-like social impairment clearly is heritable [20] and increased in unaffected parents and children of autistic probands [21]. Studies using multiple measures of sub-threshold autistic traits in population cohorts suggest that different components, separately representing language, social function and repetitive or stereotyped behaviors contribute to ASD [22–24]. On aggregate, these data suggest that different features of autism represent a quantitative continuum of function that may be inherited in distinct patterns. This is consistent with the knowledge that specific genetic factors contribute to the development and function of specific brain structures, and that distinct brain circuits may underlie different components of autism [25].
It has also been known for several decades that single rare medical or genetic conditions are associated with autism. Dozens of genetic syndromes including Joubert Syndrome, Smith Lemli Opitz, Tuberous Sclerosis, and Fragile X are known to cause autism, although many with less than 50% penetrance [26, 27]. Although this provided strong evidence in favor of a genetic cause for ASD, these syndromic forms were considered exceptional cases, and not relevant for common forms of idiopathic autism because each was rare; none account for more than 1% of ASD cases, and most are far less common [27]. Instead, similar to other common diseases with genetic contributions, autism was thought to fit a model in which multiple common variants, each with small to moderate effect sizes, interact with each other and perhaps in some cases, environmental factors, to lead to autism – a situation referred to as complex genetics [27]. In contrast to Mendelian genetics, under the complex genetic model, specific common variants in genes increase susceptibility to ASD, but each is not on its own sufficient to be causal (Figure 1). These same variants contribute to normal variation in cognition and behavior in unaffected individuals, but when mixed unfortunately as they are in the autistic proband, cause ASD. This model would explain the presence of sub-threshold traits in non-autistic first-degree relatives, such as the siblings or parents of autistic individuals, as they would be expected to harbor a subset of the genetic variants that cause autism in the affected individual.
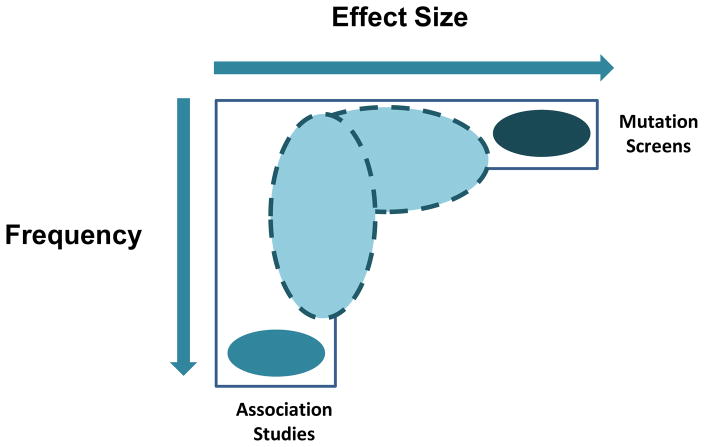
Figure 1. Common and Rare Variants.
We usually think of common and rare variants representing two extremes on the allelic spectrum. Single nucleotide polymorphisms (SNPs) with modest effects sizes (gray oval) have been identified through association studies. Rare events that segregate with disease have also been identified through re-sequencing efforts and CNV studies. Less attention has been directed at events not at either extreme, but also likely to modulate risk. Larger studies will permit consideration of lower frequency intermediate effect alleles (ovals with dotted lines). Current data do not support the presence of common alleles of large effect for affection status (bottom right corner). But, it is possible that such associations may be observed in the future when appropriate quantitative endophenotypes are used. Adapted with permission from [87].
The role of rare mutations versus common polymorphisms in ASD
A series of important findings over the last four years clearly challenges the notion that autism is mainly caused by combinations of common variants by identifying a large number of rare, recurrent, and non-recurrent mutations that lead to ASD. At the same time, whole genome association studies with common variants, while identifying a few loci with very small effect sizes, have not yielded independently replicated results [28, 29]. These rare mutations, mostly in the form of sub-microscopic chromosomal structural variation, called copy number variants (CNV), are now known to account for up to 10% of cases of idiopathic autism (those with no obvious clinical syndrome) [30–34]. Since many of these CNV have large effect sizes and thus are thought sufficient to cause ASD, they are predicted to significantly reduce reproductive fitness. Consistent with this, these causal CNV are often not transmitted from the parent, but instead occur de novo in the germline [33, 35]. However, in some cases, such as CNV at 16p11 and 15q11-13, the CNV are transmitted from an unaffected parent to cause the disorder in an offspring [33]. The genetic or epigenetic mechanism for the reduced penetrance for ASD in the mutation-carrying parent is not known. However, it is also very likely that the parent carriers of such CNV have more subtle neuropsychiatric or cognitive phenotypes that have not yet been systematically identified.
One key issue in genotype-phenotype mapping in ASD is reconciling the role of causal rare de novo variants that occur in probands and are not inherited by the unaffected siblings, with the observations of broader sub-threshold traits in siblings. The latter suggests the role of common variation, but as mentioned above, the genetic evidence for specific common variants in ASD so far has been sparse. The former are major-effect genetic mutations that are mainly observed in probands, and are therefore not expected to contribute to the broader phenotype. In this regard, two observations may be informative. First, de novo CNV are observed in 5–10% of ASD probands, but they are also observed in 1–2% of unaffected controls. So all such CNV are not necessarily causal or fully penetrant [36]. From this perspective, some CNV may be acting as complex genetic risk factors, with intermediate effect sizes, and variable penetrance and expressivity. Such is certainly the case with 16p11 duplications, which are observed in ASD, schizophrenia, and various forms of developmental delay [37–39] as well as Neurexin 1 CNV, which are observed in about 50% of unaffected carriers [36], and observed across several neurodevelopmental disorders (Table 1). Such pleiotropy and variable penetrance and expressivity is the rule, rather than the exception (Table 1). Second, the frequency of de novo mutations is significantly lower in multiplex (familial) cases of autism versus those with only one affected proband, so called simplex families [30, 31, 34]. This difference in the frequency of rare de novo CNV in families with different structures (simplex vs. multiplex) suggests that the contribution from different types of genetic variation between simplex and multiplex autism may differ. Recent family data based on the social responsiveness scale (SRS), which is a quantitative measure of autism-related features heavily weighted on social factors, suggests that this is indeed the case [40].
Table 1. Pleiotropic effects of major genes/mutations associated with ASD and allied neurodevelopmental disorders.
Here we list genes, form of genetic risk variant, clinical disorders where the mutation has been observed, and some representative references. This table is not meant to be exhaustive, but illustrative of the pleiotropic effects of known ASD genes or loci with relatively large effect sizes (OR > 5–10 for ASD).
| Gene/region | Mechanism | Disorders | References |
|---|---|---|---|
| NRXN1 | CNV, PM | ASD, SZ | [36], [50, 72, 73] |
| CNTNAP2 | CNV, PM, CP | ASD, ID, epilepsy, LD/SLI, TS | [59, 61, 74, 75] |
| 16pdel | CNV | ASD, SZ, DD, LD, normal carrier | [33, 37, 42, 76–78] |
| 16pdup | CNV | SZ, ID, DD, LD, ADHD, normal carrier | [33, 39, 42, 78] |
| 15q13.3del | CNV | SZ, epilepsy, ASD, normal carrier | [42, 73, 79] |
| 17q12del | CNV | SZ, ASD, ID | [80] |
| 15q11-13dup | CNV | ASD, SZ/psychosis | [33, 81] |
| 22q11 | CNV | ASD, ADHD, SZ, ID, epilepsy | [73, 82–84] |
| 1q21 | CNV | ASD, SZ, ID, epilepsy | [42, 85, 86] |
Abbreviation are as follows: SZ, schizophrenia; ID, intellectual disability, DD, developmental delay, TS, Tourette syndrome, LD, language delay, SLI, specific language impairment, PM, point mutation, CP, common polymorphism.
This discussion of rare and common genetic variants highlights an area of major tension in autism genetic research. On one hand, the causal nature of Mendelian (recessive or dominantly acting) mutations is fairly easy to establish, as there is essentially a strong correspondence between the presence of a mutation and a disease phenotype. Certainly, dominant or recessive mutations are easier to model in vitro and in vivo. At the same time, few such variants cause autism alone, but instead may cause a wide variety of phenotypes (variable expressivity) with or without autism, ranging from intellectual disability (ID), epilepsy, psychosis, and global developmental delay to neurotypical [27, 41]. Thus, even for the simpler Mendelian mutations, understanding which aspects of the phenotype observed in a model system or a human patient relate to autism, and which may relate to more global aspects of cognitive dysfunction, needs to be carefully established. This provides a great opportunity for detailed phenotypic study of patients with and without autism who harbor such rare causal mutations, so as to connect specific aspects of brain structure and function with cognitive and behavioral phenotypes that underlie the clinical features of the disease.
Common variants, while not causal for the disease, seem to have more subtle effects, making them harder to model in vitro or in animal models. However, perhaps because they are common in human populations and may have more subtle or specific effects on particular aspects of cognition and behavior, common risk variants may be easier to relate to particular brain phenotypes or circuits in humans. This issue will be discussed below in more detail with respect to imaging-genetic findings.
Last, it should also be emphasized that although they are often presented as dichotomous models, the contributions of rare and common variants to ASD in an individual are not mutually exclusive [25]. For example, although rare de novo CNV are considered causal mutations, it is clear that mutations may have intermediate effect sizes and variable penetrance, as mentioned above [36, 42] so such mutations must be interacting with other factors. Penetrance less than 50% for ASD is also observed in many Mendelian forms of ASD, such as Fragile X, Jouberts Syndrome, and Tuberous Sclerosis, so other genetic, epigenetic, or environmental factors must be contributing to autism causation in such cases as well [27]. Thus, different rare variants may act as either susceptibility or strongly causal alleles, and, in conjunction with common variation, may contribute in differing proportions to ASD risk in individual patients [25]. Fortunately, over the next several years, whole genome or exome sequencing if performed in large enough populations, will settle this issue by providing an empiric assessment of the relative contributions of alleles of different effect sizes, rare or common [43, 44].
The molecular diversity of ASD
Several recent reviews summarize the growing list of dozens of common and rare genetic variants that have been associated with ASD at varying levels of statistical evidence [27, 41, 45–47], so I refer the reader to these reviews for comprehensive gene lists. Here, instead, I will try to synthesize what these findings may be telling us about ASD pathophysiology at what is still an early juncture in the search for ASD susceptibility genes.
The most obvious general conclusion from all of the published genetic studies is the extraordinary etiological heterogeneity of ASD. No specific gene accounts for the majority of ASD, rather, even the most common genetic forms account for not more than 1–2% of cases [27]. Further, these genes, including those mentioned earlier, represent a diversity of specific molecular mechanisms, ranging from cell adhesion, synaptic vesicle release and neurotransmission, synaptic structure, RNA processing/splicing, and activity-dependent protein translation. On one hand, this should not be surprising, as autism is defined based on observation of cognition and behavior, not etiology. On the other hand, the diversity of potential mechanisms and the apparent lack of specificity of mutations for ASD begs the question as to whether ASD should even be viewed as a unitary disorder. Therefore, asking whether the diverse genes implicated in autism might converge on common pathways becomes an important question for understanding autism and developing new therapeutics.
Converging circuits and common pathways
Convergence of pathways may be assessed at many levels, ranging from molecular mechanisms to brain circuitry. The earliest synthetic molecular model was based on the notion that the primary area of convergence in ASD was the postnatal, experience-dependent development of the synapse [48]. This was a highly productive model based on several forms of Mendelian mutations in ASD, which has led to successful exploration of synaptic scaffolding molecules and other synaptic genes as ASD susceptibility genes [27, 46]. Interestingly, because many mRNA for synaptic genes are alternatively spliced, and the proteins regulated by ubiquitination, this paper was prescient in predicting alterations in mRNA splicing and ubiquitination pathways that have since been identified [49, 50]. Nevertheless, since the synapse is integral to the function of all neurons, how the cognitive and behavioral specificity of ASD emerges, distinct from ID or other synaptic disorders, is not yet explained by this molecularly focused model.
Belmonte and colleagues have articulated a cogent unifying model based on imaging, electrophysiological and anatomical data, which implicates abnormal neural connectivity disrupting timing and information processing [51, 52]. This general model was subsequently anatomically refined and modified to focus primarily on reduced long-range, and increased short range frontal lobe connectivity [53]. Since the general connectivity model was proposed, many subsequent studies provide strong supporting data, including a recent study showing weak interhemispheric functional connectivity in young toddlers with ASD [54]. This study is particularly important, as it shows altered connectivity in developing language circuits to be an early (and thus potentially causal) feature of the disorder, which could not be inferred from studies of older children or adults.
A more recent model, developed by Geschwind and Levitt [55], synthesizes several key features of the synaptic and connectivity models above, and is based on developmental disconnection of frontal and temporal cortical regions. This model strives to explain the diversity of genetic findings without necessitating a particular molecular mechanism. Instead, it relies on a behavioral neurologic framework to integrate many potential levels of molecular dysfunction, from synapse formation and maturation, vesicle trafficking and signaling, to defective neuronal migration, interneuron development, dendritic maturation, or axon path finding, all of which could contribute to such a functional disconnection [55] (Figure 2). Indeed several of the monogenic causes of ASD, including TSC [56] and Joubert Syndrome [57, 58] show manifestations of abnormal axon path finding or migration abnormalities that are known to lead to alterations in brain structural connectivity. The study of common variants, for example in the gene CNTNAP2, also supports disconnection as a causal phenomenon based on Initial imaging-genetic evidence. Two common polymorphisms in a relatively circumscribed area of CNTNAP2 have been shown to modulate language function in ASD and other conditions [59–61]. Van Zeeland and colleagues [62] showed long-range disconnection of medial pre-frontal cortex and the precuneus, and short-range over-connectivity of the prefrontal region in carriers of the CNTNAP2 risk allele, regardless of whether they were autistic or neurotypical. This finding is particularly important, as the genetic mediation of this disconnection supports its causal nature.
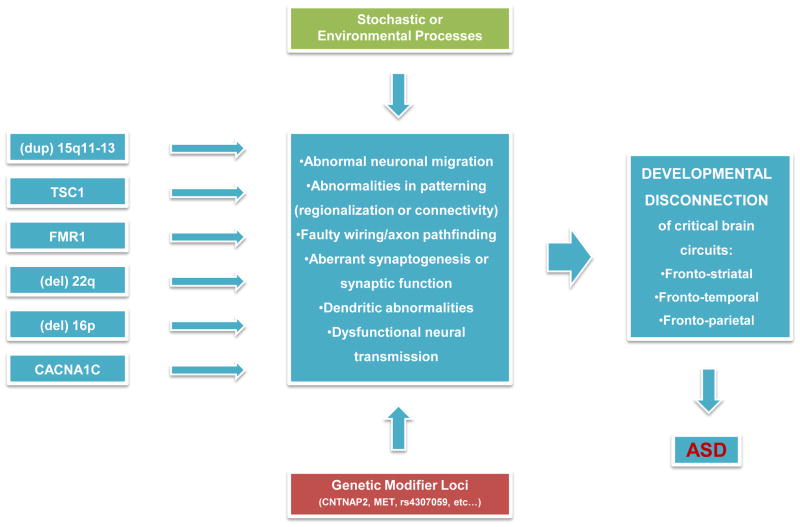
Figure 2. Convergence of genes on neural systems.
Here we illustrate a working model for how major affect autism risk alleles, such as those shown at the left, might act to lead to autism. Because these Mendelian conditions are not specific for ASD and typically lead to ID, either environmental or genetic factors must modify their affects on brain development. The biological pathways through which such genes are known to act (shown in the middle box) are myriad. Although the gene expression and proteomic studies identify molecular and biological pathways that provide a source of convergence, the ultimate convergence must lie in neural systems. At a neural systems level, the convergent process will likely be disconnection of the circuits outlined in the far right box, since these systems are thought to underlie the core deficits of ASD. Adapted with permission from [88].
Endophenotypes, common variants and domain specificity in ASD
The relationship of specific genetic variants with specific cognitive processes, such as language, highlight the notion that the broad syndrome of ASD can be broken down into many component, or intermediate phenotypes, referred to as endophenotypes. The familial segregation of endophenotypes provides a genetic basis for the broader phenotype described earlier. A logical extension of this concept is that these endophenotypes represent one end of the continuum of the normal spectrum of behavior and cognition [25]. Several groups have demonstrated that this is indeed the case with respect to CNTNAP2 [60, 61, 63], as the same allele that increases risk for language delay in ASD, increases risk for specific language impairment (SLI) and modifies language ability in the general population. Similarly, common variation in different ASD-associated CNTNAP2 single nucleotide polymorphism (SNP) [64], has been shown to modulate brain morphology in several ASD related cortical regions in normal controls [65].
However, just as specificity for ASD is an issue for rare, Mendelian variants, it is also an issue for common variation [66]. Since genes’ effects on behavior are mediated through neural circuits that mediate a multiplicity of functions (e.g. frontal-striatal circuits), their pleiotropic effects on behavior and cognition should not be surprising. In fact, pleiotropy should be the rule, rather than the exception. Recently, CNTNAP2 variants have been associated with mutism and social anxiety [67]. This complex relationship between genetic variation and phenotype highlights the importance of in depth exploration of cognitive and neurobehavioral phenotypes in ASD patients, their family members and in the general population. Phenotype discovery and the relationship of core ASD-related endophentoypes to each other and specific brain circuits in patients with ASD and related neurodevelopmental disorders remains a key, but under-explored area.
Evidence for converging molecular pathways
Several recent studies have suggested that in addition to convergent brain pathways, that there may as well be convergence at the level of molecular mechanisms in ASD. One class of such studies has asked whether putative ASD susceptibility genes are enriched in members for specific molecular or biological processes more than expected by chance. The value of this approach depends on the level of experimental support for the specific genes tested and the degree to which current pathway annotations represent reality [25, 68]. For genes identified within CNV this can be particularly problematic, as most known pathological CNV contain more than one gene and it is not expected that all genes within the CNV contribute to ASD, potentially increasing noise in this analysis. One recent study [69] reduced such background by using a new phenotype-driven method to group genes within high confidence de novo CNV [34, 69], identifying significant enrichment for several categories of genes, including axon outgrowth, synaptogenesis, cell-cell adhesion, GTPase signaling, and the actin cytoskeleton. These results replicate and extend earlier composite pathway analysis of putative ASD susceptibility genes compiled from the literature [68], and CNV pathway analysis in the Autism Genetic Resource Exchange (AGRE) and other cohorts [32, 36]. Still, these studies place ASD genes within a multiplicity of pathways, several of which are broad and do not necessarily demonstrate convergence on final common molecular processes in individuals.
In this regard, two recent studies use quite different systems biology approaches to provide a new perspective on the concept of molecular convergence. The first, an analysis of gene expression in post-mortem autism brain, provides the strong evidence for a shared set of molecular alterations in a majority of cases of ASD. This included disruption of the normal gene expression pattern that differentiates frontal and temporal lobes (consistent with an early developmental patterning defect), and two groups of genes dysregulated in ASD brains: one related to neuronal function, and the other to immune/inflammatory responses [49]. The neuronal function genes were enriched in genetic association signals, providing evidence that these changes were causal, rather than the consequence of the disease [49], while the immune/inflammatory changes did not show a strong genetic signal, implicating environmental or epigenetic factors instead. It is also notable that the several of the same biological pathways identified in this gene expression study overlapped with the pathway analysis of CNV described above. This analysis of post-mortem autism brain also showed down-regulation of several markers of GABAergic interneurons, suggesting potential inhibitory interneuron dysfunction. These results provide the first strong evidence for both a shared genetic and an environmental/epigenetic basis for ASD and the presence of an early developmental patterning defect. It is tempting to speculate that the abnormalities in cortical patterning and interneuron function provide a molecular basis for cortical-cortical and cortical-fugal disconnection, linking molecular abnormalities with anatomical, physiological and imaging findings.
The second study on molecular convergence in ASD identified protein interactors of known ASD or ASD-associated genes [70]. This interactome revealed several novel interactions, including between two ASD candidate genes, Shank3 and TSC1. The biological pathways identified in this study include synapse, cytoskeleton, and GTPase signaling, demonstrating a remarkable overlap with those identified by the gene expression and CNV pathway studies discussed above. This study differs from other genome-wide studies (such as mRNA expression) as it begins with known ASD genes and asks about their relationships. So although it is “ –-omic” in nature [71], it is not quite as unbiased as methods that survey the genome more agnostically. Despite the significant heterogeneity in ASD, these diverse studies identify several common areas of molecular convergence in ASD. Understanding how these pathways relate to individual differences now becomes an important research priority.
Concluding remarks
Many genes have been identified for ASD and some biologically coherent functional pathways that link these genes are emerging. Few of the genes, whether contributing by common or rare variation, are specific to ASD, but instead variably contribute to genetic risk for ASD, ID, schizophrenia, SLI, epilepsy, and even attention deficit hyperactivity disorder (ADHD). Therefore, understanding the specificity of specific variants to ASD is a crucial challenge that requires several advances to be made (see also Box 1). Diseases of cognition and behavior have their basis in brain circuit dysfunction. Therefore, we need to understand how specific genetic risk leads to changes in neural circuitry and function in those with and without a specific diagnosis. This will likely involve understanding gene – gene and gene- environment interactions, as well as epigenetics, areas that have not yet been explored in ASD. We need to understand how the implicated neural circuitry relates to specific cognitive and behavioral elements, and we need to link the neural circuitry and behaviors to disease. Additionally, such circuits are related to specific aspects of human higher cognition and a more detailed notion of the autism cognitive phenotype is required. Components of this have been achieved in a preliminary manner for only a few elements, but to understand ASD from a systems level mechanistic perspective, we will need to integrate all of these levels of understanding [71]. This will no doubt involve the use of model systems, including mouse and invertebrates, so it will be important to be cognizant of evolutionary similarity and divergence in the interpretation of such data with respect to a disorder of human higher cognition. Regardless of the challenges, the identification of causal genetic variants provides a starting point for a new mechanistic understanding of ASD, enabling connection of genes to brain to cognition and behavior.
Box 1. Priorities for future genetic research in ASD.
Delineate areas of molecular convergence between diverse mutational mechanisms underlying ASD susceptibility using unbiased, systems biology methods.
Perform whole genome sequencing in large numbers of patients with a variety of neurodevelopmental and psychiatric disorders to clearly define genetic overlap and differences in susceptibility.
Define appropriate endophenotypes through family case-control designs, so as to have a sound basis for understanding cross disorder genotype-phenotype relationships.
Translate the knowledge of mutational basis of ASD into mechanistic understanding at the cellular and circuit levels.
Characterize subtypes of ASD based on molecular or genetic signatures and relate them to trajectory and treatment response.
Identify environmental modifiers and gene by environment interactions.
Define potential epigenetic contributions to ASD.
Acknowledgments
I thank Lauren Kawaguchi for her editorial assistance. Our laboratory’s work in autism and the molecular basis of human higher cognition is supported by two Autism Center of Excellence grants from NIMH: ACE Network 5R01MH081754-03 to D.H.G and ACE Center 5P50HD055784-03 (S. Bookheimer PI, D.H.G. co-PI), a Merit award from NIMH to D.H.G., 4R37MH060233-10, as well as funding from Autism Speaks and The Simons Foundation.
Glossary
- Complex genetics
A term used in contrast to Mendelian patterns of inheritance, whereby instead of being dominantly or recessively inherited a phenotype is caused by multiple genes that may interact with each other and the environment
- Concordance
The rate at which a second individual has the same phenotype as the first, for example in the second sibling of a pair, or the second twin
- Copy Number Variation
A greater than one Kb change in chromosomal complement so that a particular region or chromosome is diverges from the normal diploid human copy number (aneuploidy) over that region. A deletion would yield one copy, and a duplication of the region would yield 3 copies. Copy number changes can be inherited or de novo
- De novo
A genetic alteration that arises either in the gamete or in the fertilized egg during its early development, but is not observed in the somatic cells of the parent
- Endophenotype
A heritable phenotypic feature that is closely related to a disorder, but represents a more narrow or simple component than the disorder itself. An endophenotype should be observed in first degree relatives more frequently than in the general population and may be quantitative rather than binary
- Epigenetic
Non-sequence based changes to DNA that affect transcription, leading to changes in the expression of the gene product. Known major epigenetic modifications include DNA methylation and histone modifications, such as acetylation or methylation
- Heritability
In general this refers to the extent to which a particular trait is due to inherited DNA sequence variation. A heritability of 1.0 indicates that a trait is 100% heritable and no environmental component contributes. However, heritability is an estimate based on the specific measurements taken in a particular population, and therefore is only a rough guide
- Mendelian inheritance
A dominant or recessive pattern of inheritance first defined by Gregor Mendel
- Penetrance
The extent to which a particular phenotype is observed, given a specific genotype. Penetrance is always relative to the phenotype in question
- Pleiotropy
When a gene influences more than one phenotype
- Polymorphism
A particular variant that is observed in more than 1% (common) in the population. A single nucleotide polymorphism (SNP) is a single DNA base pair change
- Proband
The index case being studied, usually based on their having a particular phenotype or disorder
- Whole genome or whole- exome sequencing
The use of new high-throughput Next Generation Sequencing to sequence whole genomes or the protein coding portion of the genome (exome) in patients and controls
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kanner L. Autistic disturbances of affective contact. Nerv Child. 1943;2:217–250. [PubMed] [Google Scholar]
- 2.Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 3.Folstein S, Rutter M. Genetic influences and infantile autism. Nature. 1977;265:726–728. doi: 10.1038/265726a0. [DOI] [PubMed] [Google Scholar]
- 4.Wing L. The continuum of autistic characteristics. In: Schopler EMG, editor. Diagnosis and Assessment in Autism. 1988. pp. 91–110. Plenum. [Google Scholar]
- 5.Constantino JN, Todd RD. Autistic traits in the general population: a twin study. Arch Gen Psychiatry. 2003;60:524–530. doi: 10.1001/archpsyc.60.5.524. [DOI] [PubMed] [Google Scholar]
- 6.Mundy P, et al. Self-referenced processing, neurodevelopment and joint attention in autism. Autism. 2010;14:408–429. doi: 10.1177/1362361310366315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charman T, et al. Defining the cognitive phenotype of autism. Brain Res. 2011;1380:10–21. doi: 10.1016/j.brainres.2010.10.075. [DOI] [PubMed] [Google Scholar]
- 8.Bailey A, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg RE, et al. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med. 2009;163:907–914. doi: 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- 10.Hallmayer J, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011 doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brian J, et al. Clinical assessment of autism in high-risk 18-month-olds. Autism. 2008;12:433–456. doi: 10.1177/1362361308094500. [DOI] [PubMed] [Google Scholar]
- 12.Bolton P, et al. A case-control family history study of autism. J Child Psychol Psychiatry. 1994;35:877–900. doi: 10.1111/j.1469-7610.1994.tb02300.x. [DOI] [PubMed] [Google Scholar]
- 13.Losh M, et al. Neuropsychological profile of autism and the broad autism phenotype. Arch Gen Psychiatry. 2009;66:518–526. doi: 10.1001/archgenpsychiatry.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren ZE, et al. Neurocognitive and behavioral outcomes of younger siblings of children with Autism Spectrum Disorder at age five. J Autism Dev Disord. 2011 doi: 10.1007/s10803-011-1263-4. [DOI] [PubMed] [Google Scholar]
- 15.Constantino JN. The quantitative nature of autistic social impairment. Pediatr Res. 2011;69:55R–62R. doi: 10.1203/PDR.0b013e318212ec6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gamliel I, et al. Developmental trajectories in siblings of children with autism: cognition and language from 4 months to 7 years. J Autism Dev Disord. 2009;39:1131–1144. doi: 10.1007/s10803-009-0727-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickles A, et al. Variable expression of the autism broader phenotype: findings from extended pedigrees. J Child Psychol Psychiatry. 2000;41:491–502. [PubMed] [Google Scholar]
- 18.Gamliel I, et al. The development of young siblings of children with autism from 4 to 54 months. J Autism Dev Disord. 2007;37:171–183. doi: 10.1007/s10803-006-0341-5. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Yizhak N, et al. Pragmatic language and school related linguistic abilities in siblings of children with autism. J Autism Dev Disord. 2011;41:750–760. doi: 10.1007/s10803-010-1096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Constantino JN, Todd RD. Intergenerational transmission of subthreshold autistic traits in the general population. Biol Psychiatry. 2005;57:655–660. doi: 10.1016/j.biopsych.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Constantino JN, et al. Autistic social impairment in the siblings of children with pervasive developmental disorders. Am J Psychiatry. 2006;163:294–296. doi: 10.1176/appi.ajp.163.2.294. [DOI] [PubMed] [Google Scholar]
- 22.Steer CD, et al. Traits contributing to the autistic spectrum. PLoS One. 2010;5:e12633. doi: 10.1371/journal.pone.0012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronald A, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 24.Ronald A, et al. A twin study of autism symptoms in Sweden. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.82. [DOI] [PubMed] [Google Scholar]
- 25.Geschwind DH. Autism: many genes, common pathways? Cell. 2008;135:391–395. doi: 10.1016/j.cell.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillberg C, Coleman M. The Biology of the Autistic Syndromes. Greenwood Publishing Group; 1985. Incorporated. [Google Scholar]
- 27.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anney R, et al. A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet. 2010;19:4072–4082. doi: 10.1093/hmg/ddq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams Syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Beaudet AL. Autism: highly heritable but not inherited. Nat Med. 2007;13:534–536. doi: 10.1038/nm0507-534. [DOI] [PubMed] [Google Scholar]
- 36.Bucan M, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez BA, et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2010;47:195–203. doi: 10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- 38.Bijlsma EK, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy SE, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virkud YV, et al. Familial aggregation of quantitative autistic traits in multiplex versus simplex autism. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:328–334. doi: 10.1002/ajmg.b.30810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 42.Vassos E, et al. Penetrance for copy number variants associated with schizophrenia. Hum Mol Genet. 2010;19:3477–3481. doi: 10.1093/hmg/ddq259. [DOI] [PubMed] [Google Scholar]
- 43.Gibb BC. Teetering towards chaos and complexity. Nat Chem. 2009;1:17–18. doi: 10.1038/nchem.148. [DOI] [PubMed] [Google Scholar]
- 44.Zeggini E. Next-generation association studies for complex traits. Nat Genet. 2011;43:287–288. doi: 10.1038/ng0411-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speicher MR, Antonarakis Stylianos E, Motulsky Arno G, editors. Vogel and Motulsky’s Human Genetics: Problems and Approaches. Springer; 2009. [Google Scholar]
- 46.State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 48.Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- 49.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belmonte MK, et al. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belmonte MK, et al. Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol Psychiatry. 2004;9:646–663. doi: 10.1038/sj.mp.4001499. [DOI] [PubMed] [Google Scholar]
- 53.Courchesne E, Pierce K. Why the frontal cortex in autism might be talking only to itself: local over-connectivity but long-distance disconnection. Curr Opin Neurobiol. 2005;15:225–230. doi: 10.1016/j.conb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Dinstein I, et al. Disrupted neural synchronization in toddlers with autism. Neuron. 2011;70:1218–1225. doi: 10.1016/j.neuron.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 56.Tsai P, Sahin M. Mechanisms of neurocognitive dysfunction and therapeutic considerations in tuberous sclerosis complex. Curr Opin Neurol. 2011;24:106–113. doi: 10.1097/WCO.0b013e32834451c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poretti A, et al. Joubert Syndrome and related disorders: Spectrum of neuroimaging findings in 75 patients. AJNR Am J Neuroradiol. 2011 doi: 10.3174/ajnr.A2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JE, Gleeson JG. Cilia in the nervous system: linking cilia function and neurodevelopmental disorders. Curr Opin Neurol. 24:98–105. doi: 10.1097/WCO.0b013e3283444d05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alarcon M, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitehouse AJ, et al. CNTNAP2 variants affect early language development in the general population. Genes Brain Behav. 2011;10:451–456. doi: 10.1111/j.1601-183X.2011.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vernes SC, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scott-Van Zeeland AA, et al. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2:56ra80. doi: 10.1126/scitranslmed.3001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peter B, et al. Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J Neurodev Disord. 2011;3:39–49. doi: 10.1007/s11689-010-9065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arking DE, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan GC, et al. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. Neuroimage. 2010;53:1030–1042. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.State MW. The erosion of phenotypic specificity in psychiatric genetics: emerging lessons from CNTNAP2. Biol Psychiatry. 2011;69:816–817. doi: 10.1016/j.biopsych.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 67.Stein MB, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 is associated with increased risk for selective mutism and social anxiety-related traits. Biol Psychiatry. 2011;69:825–831. doi: 10.1016/j.biopsych.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bill BR, Geschwind DH. Genetic advances in autism: heterogeneity and convergence on shared pathways. Curr Opin Genet Dev. 2009;19:271–278. doi: 10.1016/j.gde.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakai Y, et al. Protein interactome reveals converging molecular pathways among autism disorders. Sci Transl Med. 2011;3:86ra49. doi: 10.1126/scitranslmed.3002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gauthier J, et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011 doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levinson DF, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strauss KA, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 75.Verkerk AJ, et al. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 76.Kumar RA, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 77.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 78.Shinawi M, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Bon BWM, et al. In: 15q13.3 Microdeletion. Pagon RABT, Dolan CR, Stephens K, editors. University of Washington; Seattle: 1993. [Google Scholar]
- 80.Moreno-De-Luca D, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mefford HC, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fine SE, et al. Autism spectrum disorders and symptoms in children with molecularly confirmed 22q11.2 deletion syndrome. J Autism Dev Disord. 2005;35:461–470. doi: 10.1007/s10803-005-5036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niklasson L, et al. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil. 2009;30:763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 84.Vorstman JA, et al. The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child Adolesc Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 85.Mefford HC, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abrahams BS, Geschwind DHG. Genetics of Autism. In: Speicher MR, et al., editors. Vogel and Motulsky’s Human Genetics. Springer; Heidelberg: 2010. pp. 699–714. [Google Scholar]
- 88.Geschwind DHG. Autism Genetics and Genomics: S Brief Overview and Synthesis. In: Amaral DG, et al., editors. Autism Spectrum Disorders. Oxford University Press; Oxford: 2011. pp. 812–824. [Google Scholar]