Abstract
Characterization of protein crosslinking, particularly without prior knowledge of the chemical nature and site of crosslinking, poses a significant challenge due to their intrinsic structural complexity and the lack of a comprehensive analytical approach. Towards this end, we have developed a generally applicable workflow—XChem-Finder that involves four stages. (1) Detection of crosslinked peptides via 18O-labeling at C-termini. (2) Determination of the putative partial sequences of each crosslinked peptide pair using a fragment ion mass database search against known protein sequences coupled with a de novo sequence tag search. (3) Extension to full sequences based on protease specificity, the unique combination of mass, and other constraints. (4) Deduction of crosslinking chemistry and site. The mass difference between the sum of two putative full-length peptides and the crosslinked peptide provides the formulas (elemental composition analysis) for the functional groups involved in each cross- linking. Combined with sequence restraint from MS/MS data, plausible crosslinking chemistry and site were inferred, and ultimately, confirmed by matching with all data. Applying our approach to a stressed IgG2 antibody, ten cross-linked peptides were discovered and found to be connected via thioether originating from disulfides at locations that had not been previously recognized. Furthermore, once the crosslink chemistry was revealed, a targeted crosslink search yielded four additional crosslinked peptides that all contain the C-terminus of the light chain.
Protein crosslinking exists in a myriad of biological systems and protein pharmaceuticals, such as collagen, ubiquitylated proteins, and monoclonal antibodies1–6. Rich and diverse chemistry is involved as well, including disulfide1, dityrosine2, lysinoalanine7, 8, lanthionine7, 8, etc. Additionally, chemical crosslinking is widely used to probe protein structures and interactions9, 10. Due to their intrinsic structural complexity, characterization of crosslinked peptides is complex, but nonetheless tractable if the crosslink chemistry is pre-defined. For example, a database of the intact mass (precursor ion) and the tandem mass spectra (fragmentation ions) for all possible combinations of crosslinked peptides (e.g., two cysteines to form a disulfide bond) can be generated computationally, and subsequently, correlated with observed spectra to identify both the sequences and sites of modification. Such a database search strategy is the cardinal principle behind many common algorithms, including ASAP11, X!link12, BLink13, Xlink-Identifier14, 15 and MassAnalyzer16. Moreover, clever experimental tricks, such as judicious isotope labeling17, 18, can markedly simplify the process and enhance the confidence level for assignment with the assistance of software tools e.g., Pro-Cross-link19, 20, PepLynx21, xQuest22, iXLink/doXLink/XlinkViewer23. To date, the rapid advancements in mass spectrometers, data analysis algorithms, and computational capacity have made analyses of crosslinking with known chemistry much more accessible if not routine (for recent reviews, see 9, 10).
Yet the aforementioned approaches are futile if the crosslink chemistry is unknown or not pre-defined; for one thing, no theoretical mass or spectrum can be simulated. Even if crosslinked peptides have been identified, it remains a tall order to deduce the sequences and sites of crosslinking. Conceptually, de novo sequencing should provide at least partial sequences for crosslinked peptides (see review paper24). Under typical fragmentation conditions, however, a crosslinked peptide gives rise to at least five sets of b- and y-ions that are intertwined and indistinguishable (Table S1). In addition, high-charge-states (≥3+) are typically featured in the crosslinked peptides, resulting in multiple charge fragment ions (e.g. 2+ or 3+) and further complicating data interpretation12, 22, 25. High resolution mass spectrometers (e.g. Orbitrap), capable of the determination of fragment ion charge state, have become widely available only recently. As such, the drastically increased complexity in tandem spectrum renders de novo sequencing ineffective in most cases. Unknown or undefined crosslinks are typically discovered serendipitously, requiring isolation of the crosslinked peptides and “old-fashioned” protein chemistry. Even so, full characterization remains elusive for many cases. For instance, the non- reducible crosslinks between an IgG heavy chain and a light chain in a murine monoclonal antibody, OKT3, and between two heavy chains of IgG2 could not be elucidated even after intensive efforts26, 27.
To facilitate systematic and unbiased discovery of unknown crosslinks, we have developed a generally applicable workflow—XChem-Finder (Scheme 1). First, crosslinked peptides were isotopically labeled at the C-termini to facilitate their detection19–21, 28. Proteins were digested in 18O-(heavy) and 16O-(light) water, respectively, followed by LC/MS/MS analysis. At full scan, the distinct isotope pattern of the crosslinked peptides (a mass increase of 8 Da) compared to the non-crosslinked linear species (a mass increase of 4 Da) was readily detected by a spectral analysis algorithm19–21. The second and more challenging part is to determine the sequences, chemical nature and site of crosslink. The workflow breaks down the challenge into workable sub-steps. (a) The candidate ions of crosslinked peptides underwent high resolution MS/MS analysis. Based on their isotope patterns, linear and crosslinked fragment ions are divided into different groups. (b) Mass of linear fragment ions were searched against the protein sequence, yielding partial sequences (often sequence ladders) of each chain of the crosslinked peptides. In parallel, de novo sequencing of crosslinked fragment ions affords sequence tags. (c) Combining the partial sequences and sequence tags, putative full-length sequences of each chain were deduced based on protease specificity, the unique combination of mass, and other constraints. (d) The difference between the combined mass of the two putative full-length peptides and the observed mass of a crosslinked peptide provides the formula for the functional group involved in the crosslink (mass to formula). Combined with sequence restraint from MS/MS data, the crosslink chemistry and site were inferred, and ultimately, confirmed by matching with all data.
Scheme 1.
Flow chart of XChem-Finder in four main stages: (1) detection of crosslink by 18O-labeling; (2) determination of partial sequences of crosslinked peptides from the mass of fragmentation ions; (3) inference of full sequence of each chain; (4) determination of crosslink site and chemistry by elemental composition analysis and chemical intuition.
Applying our XChem-Finder approach to a stressed IgG2, ten crosslinked peptides were discovered and found to be linked via thioether that originated from disulfides at locations that had not been reported. Furthermore, once the crosslinking chemistry was revealed, a targeted search yielded additional four crosslinked peptides that all contain the C-terminus of light chain.
EXPERIMENTAL SECTION
Chemicals
All chemicals were reagent grade or above. Guanidine hydrochloride (GndHCl), dithiothreitol (DTT), iodoacetic acid (IAA), trifluoroacetic acid (TFA), acetonitrile (ACN), HPLC-grade water, and bradykinin were from Sigma-Aldrich (St. Louis, MO, USA). Sequencing grade trypsin was obtained from Roche (Indianapolis, IN, USA). 18O-water (97%) was obtained from Cambridge Isotope Laboratories (Andover, MA, USA). Recombinant monoclonal antibody anti-streptavidin immunoglobulin gamma 2 (IgG2) was produced in Chinese hamster ovary (CHO) cells (Amgen, Thousand Oaks, CA, USA), purified according to standard manufacturing procedures, formulated at a concentration of 20 mg/mL in 50 mM sodium acetate pH 5.2, and stored at −70 °C.
Generation of Stressed Sample
After being buffer exchanged into 100 mM Tris at pH 8.5, the IgG2 antibody was incubated at 50 °C for 7 days in the dark.
Reduction, alkylation, tryptic digestion and 18O-labeling of the IgG2
Tryptic digestion of the stressed IgG2 was performed similarly to the procedure described by Ren et al29. Briefly, IgG2 (20 mg/mL) was diluted to 1 mg/mL in a denaturing buffer (7.5 M GndHCl, 2 mM EDTA and 0.25 M Tris-HCl, pH 7.5) to a final volume of 0.5 mL. Reduction was accomplished with the addition of 3 µL of 0.5 M DTT followed by 30 min incubation at room temperature. S-Carboxymethylation was achieved with the addition of 87 µL of 0.5 M IAA; the reaction was carried out in the dark for 15 min at room temperature. Excess IAA was quenched with the addition of 4 µL of 0.5 M DTT. The reduced and alkylated IgG2 samples were subsequently exchanged into the digestion buffer (0.1 M Tris-HCl at pH 7.5) using a NAP- 5 size-exclusion column (GE Healthcare, Piscataway, NJ, USA). After two aliquots (200 µL each) of the above buffer-exchanged antibody were completely dried via Speed Vac and reconstituted separately into the same volume of 18O-water or 16O-water, 6 µL of 1 mg/mL trypsin in 18O-water or 16O-water solution, respectively, was added to achieve a 1:25 (w/w) enzyme/substrate ratio. The reaction mixtures were incubated at 37 °C for 30 min.
HPLC
Tryptic digests of the IgG2 (25 µL) were separated on a Jupiter C5 column (250×2.0 mm, 5 µm, 300, Phenomenex, Torrance, CA, USA) at a temperature of 50 °C with a flow rate of 200 µL/min on a HPLC system (Agilent 1100, Palo Alto, CA, USA). Mobile phase A was 0.1% TFA in water (v/v) while mobile phase B contained 0.085% TFA / 90% ACN / 10%water. A gradient was applied by holding at 2% B for 2 min, increasing to 22% B in 38 min, then 42% B in 80 min, then 100% B in 25 min followed by holding at 100% B for 5 min. The column was re-equilibrated at 2% B for 30 min before next injection.
Mass Spectrometry
An LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) was used in-line with the HPLC system for the analyses of the IgG2 tryptic digests. A full MS scan (with 60,000 resolution at m/z 400 and an automatic gain control (AGC) target value of 2x105) followed by data-dependent MS/MS scans of the three most abundant precursor ions was set up to acquire both the peptide mass and sequence information. The spray voltage was 5.5 kV, and the capillary temperature was 250 °C. The instrument was tuned using the doubly-charged ion of a synthetic peptide, bradykinin. The MS/MS spectra were obtained using collision-induced dissociation (CID) with normalized collision energy of 35%. For MS/MS with ion detection in the Orbitrap, the AGC target was set to 3x106, resolution to 7,500, and the precursor isolation width to 4 m/z unit. Peptides were identified by MassAnalyzer by comparing experimental MS/MS to theoretically predicted MS/MS16, 30–32. Peak alignment between 16O- and 18O-digest runs was automatically performed by MassAnalyser33. A new function was implemented in MassAnalyzer to calculate the level of 18O-labeling in each peptide. The number of incorporated 18O in a peptide is calculated from the following equation:
| Eq. 1 |
where Mlabeled and Munlabeled are the average masses of the 18O-labeled and unlabeled peptides, respectively, as calculated by the centroids of their respective isotope envelopes. The value of 2.004 Da is the mass difference between an 18O atom and an 16O atom.
RESULTS AND DISCUSSION
Stage 1: Identification of Crosslinked Peptides
18O-labeling combined with mass spectrometry is commonly used to identify crosslinked peptides 19–21, 28. As shown in Scheme 1, newly created C-termini of tryptic peptides from digestion in 18O-water were completely labeled by 18O. The distinct isotope pattern for the labeled crosslinked peptides (a mass increase of 8 Da) compared to linear (non-crosslinked) species (a mass increase of 4 Da) can be automatically detected by common spectral analysis algorithms, such as an in-house isotopic screening algorithm (MassAnalyzer16, 30–32).
18O-Labeling
A general strategy to label the C-termini of peptides in 18O-water catalyzed by proteases for peptide identification and quantification is well documented34–40. Under our experimental conditions, near complete (four) 18O-incorporation for crosslinked peptides was evident from the isotopic distributions (see Figure S2-1). The small amount of 16O- water (3%) in 18O-water had no significant impact in their isotopic patterns and the subsequent data analysis. 18O-labeling during tryptic digestion is only applied for newly created C-termini, not the C-termini of proteins. Hence, a crosslinked tryptic peptide that contains the C-terminus of the protein only has a mass shift of 4 Da, therefore cannot be differentiated from the linear peptides. This limitation can be overcome by using proteases with different substrate specificity or labeling N-termini (e.g., fromaldehyde-d2 and sodium cyanoborohydride or succinic anhydride-d4)41–43. In this paper, this was satisfactorily addressed via a targeted mass search after the crosslink chemistry was elucidated. In addition, the deamidation of asparagine and isomerization of aspartic acid could potentially introduce 18O into peptides44–46; under our conditions, no isoaspartic acid was detected in the candidate peptides.
Screening of Crosslinked Peptides in Full Scan
An 18O incorporation value (Eq. 1) of 4.0 ± 0.3 was set as cut-off in our screening. The initial screening results for the stressed IgG2 are shown in Table S2. Each peak was evaluated for false positive. For instance, gas phase dimerization, commonly observed in mass spectrometry47, 48, was readily determined based on retention time (same as the monomers) and mass (exactly double that of monomers). In addition, weak precursor ions (typically with peak intensity of 50,000 count or lower) with poor or no MS/MS data were excluded. Based on these criteria, ten candidates shown in red in Table S2 were selected for subsequent high resolution MS/MS analysis.
Stage 2: Deduce partial sequence for each chain
As illustrated in Scheme 1, this stage involves (a) grouping fragment ions based on their isotope patterns imparted by their corresponding structural features (e.g., linear or cross-linked), (b) deducing partial peptide sequences via a database search (match mass with partial peptide sequences using FindPept) and de novo sequencing, and (c) determining most likely candidate peptides.
Deconvolution of Fragment Ions
Most precursor ions for cross-linked peptides were highly charged (e.g., 3+ or 4+), thus doubly and triply charged fragment ions abound, e.g. ion m/z 839.49 (2+) and 1300.47 (3+) in Figure S2-2. The high resolution of the tandem mass spectrum allowed us to measure the isotope envelope and hence determine the charge state. Also considering fragment ion type (b- vs y-ion), monoisotopic neutral mass of each fragment ion from a crosslinked peptide was calculated manually. For example, +17.0033 Da (the mass of OH−) and -1.0073 Da (the mass of H+) were added to a singly charged b-and y-ion, respectively, to obtain their neutral peptide mass. The high-resolution for the tandem spectra was crucial in determining the correct charge state and hence neutral mass; otherwise, incorrect monoisotopic mass would lead to false hits and even erroneous assignment.
Grouping Fragment Ions by 18O Incorporation
The fragment ions containing zero, one, and two C-termini displayed a mass shift of 0, 4, and 8 Da, respectively, in the corresponding MS/MS spectra obtained from 18O-water vs 16O-water (referred as 18O/16O rule in this paper) and accordingly, are divided into different groups (Table S1). For each crosslinked peptide, two sets of linear fragment ions that contain no crosslink site do exist for each chain. One set is the b-ions prior to the crosslink site, which show no mass shifts with 18O-labeling and thus are separated from other fragment ions (group 1 in Table S1). Another set is the y-ions to the C-terminal side of the crosslink site (group 2 in Table S1), which contain two 18O with a mass shift of 4 Da. Essentially, these linear fragments are searchable in standard database, i.e., the mass can be matched with the corresponding peptide fragments. The freely available FindPept (web.expasy.org/findpept/) was used for the search in this study. Each observed mass value of these linear fragment ions should match to a partial sequence of the crosslinked peptides, but also unrelated sequences (false hits). High mass accuracy (typical 10 ppm in our FT MS/MS experiments) greatly limits false positives. Furthermore, multiple fragmentation ions collectively—and in combination with de novo sequencing as described below—narrow the hits to a selected few, if not one, candidate peptides.
Isotope pattern (8 Da mass shift with 18O-labeling) can also be readily used to isolate a set of fragmentation ions that contain two C-termini (y-ions containing the crosslink site, see group 4 in Table S1). First, these ions were excluded from database search (which is for linear peptides), reducing false hits. Second and more importantly, this markedly simplified set of tandem spectra could be used for de novo sequencing to yield sequence tags, as it was indeed the case in our study.
Partial Sequence Search via FindPept
The neutral peptide monoisotopic mass (obtained from the fragment ion bins of the mass shift of 0 and 4 Da, linear peptides, as shown in Scheme 1) were searched against the known IgG2 sequence using FindPept with user-defined mass error (10 ppm for the resolution of 7500 in FT-MS/MS in our experiments). FindPept also allows users to define the residue modifications, for example, alkylation at all cysteine residues (+58.005 Da for reaction with iodoacetic acid in our experiments). FindPept outputs a list of peptides that match the neutral peptide masses, and naturally, some are false hits. As such, several complimentary steps (constraints) were taken to confirm the actual sequences (higher probability and confidence level) and rule out false hits. It is worth noting that this is an iterative process, so the steps can be taken in a different order based on individual situation.
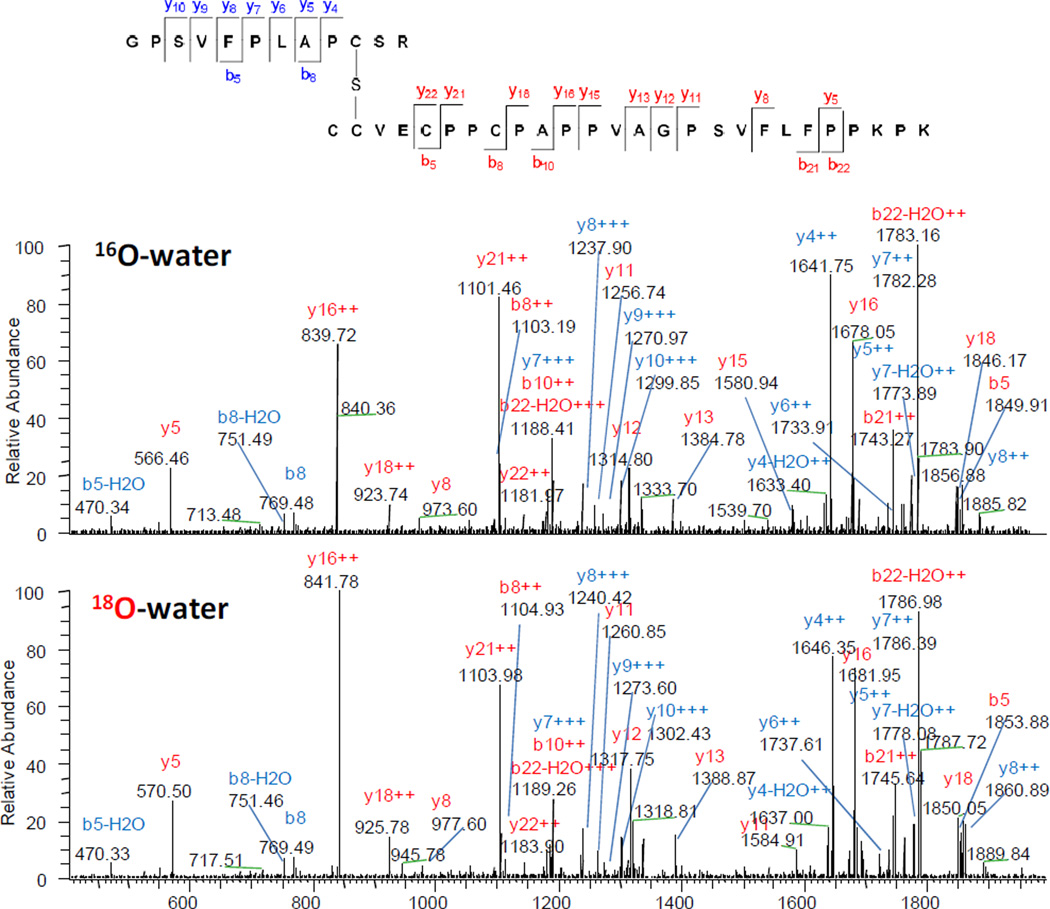
As an example, the process is demonstrated using a triply charged crosslinked peptide m/z 1351.33 (retention time at 91.17 min, G118-R129/C215-K240). The corresponding neutral monoisotopic mass of its fragment ions were searched against the IgG2 sequence. The full list of fragment ion peptides is shown in Table S3 and some are highlighted in Table 1.
Table 1.
Partial sequences determined from the mass of fragmentation ions for a triply charge precursor ion at the retention time of 91.17 min with m/z 1351.33 (molecular mass at 4048.963 Da). The longest b- and y-ions observed are underlined. All Cys are alkylated with IAA. The adjoining residues before cleavage are in parenthesis. 18O-labeling indicated that ions 1 and 2 are b-ions and ion 3 to 11 are y-ions. The crosslinked peptides were determined to be HC:G118-R129/HC:C215-K240
| # | m/z | Charge | User mass (Da) |
Theor.mass (Da) |
Δmass (ppm) |
peptide | Corresponding Tryptic Peptide |
Notes | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Sequence | Mass | |||||||||
| 1 | 470.2368 | 1 | 505.251 | 505.254 | 5.1 | (K) GPSVF(P) GPSVF(P) |
(K) 118GPSVFPLAPCSR129/(S) 118GPSVFPLAPCSR129/(S) |
1287.6282 | Chain 2 | |
| 2 | 769.4172 | 1 | 786.420 | 786.428 | 9.6 | (K)  GPSVFPLA(P) GPSVFPLA(P) |
||||
| 3 | 566.3647 | 1 | 565.357 | 565.359 | 3.1 | (F)PPKPK (D) (D) |
(K)  215CCVECPPCPAPPVAGPSVFLFPPKPK240 215CCVECPPCPAPPVAGPSVFLFPPKPK240 (D) (D) |
2911.3305 | Chain 1 | |
| 4 | 1313.7528 | 1 | 1312.745 | 1312.754 | 7.0 | (A)GPSVFLFPPKPK (D) (D) |
||||
| 5 | 1384.7951 | 1 | 1383.788 | 1383.791 | 2.4 | (V)AGPSVFLFPPKPK (D) (D) |
||||
| 6 | 839.4851 | 2 | 1676.956 | 1676.965 | 5.5 | (A)PPVAGPSVFLFPPKPK (D) (D) |
||||
| 7 | 1846.0661 | 1 | 1845.042 | 1845.055 | 7.1 | (C)PAPPVAGPSVFLFPPKPK (D) (D) |
||||
| 8 | 923.5283 | 2 | 1845.059 | 1845.055 | −2.0 | (C)PAPPVAGPSVFLFPPKPK (D) (D) |
||||
| 9 | 1101.0885 | 2 | 2200.162 | 2200.175 | 6.0 | (C)PPCPAPPVAGPSVFLFPPKPK (D) (D) |
||||
| 10 | 1181.5930 | 2 | 2361.171 | 2361.190 | 8.0 | (E)CPPCPAPPVAGPSVFLFPPKPK (D) (D) |
||||
| 11 | 1688.7689 | 1 | 1705.772 | 1705.788 | 9.4 | (V)HQDWLNGKEYKCK (V) (V) |
(R)  294VVSVLTVVHQDWLNGKEYKCK314 294VVSVLTVVHQDWLNGKEYKCK314 (V) (V) |
2502.2941 | ruled out for chain 2, see text |
|
A rewarding first step is to sort the peptides according to their positions in the full protein sequence. As illustrated in Table 1 and S3-2, typically, at least one sequence ladder could be readily identified. For example, the overwhelmingly large numbers of fragment ions (eight peptides #3–10 in Table 1) that share C-terminal sequences (CPPCPAPPVAGPSVFLFPPKPK) were found, essentially affirming this is part of the true sequence. An immediate implication is that the largest observed fragment ion (2361.190 Da) sets an upper limit for the mass of the other chain (1687 Da) by subtraction from the observed total crosslink peptide mass (4048.963 Da). Based on this criterion, fourteen peptides in Table S3-2 with a mass of significantly larger than 1687 Da were excluded.
Another two powerful constraints that can be applied to data analysis are based on the protease specificity (referred to as the tryptic rule) and mass shift conferred by 18O-labeling 18O/16O rule). For instance, the above mentioned eight peptides (peptides 3–10 in Table 1) are likely from a tryptic peptide as they all end with lysine, and indeed, a mass shift of 4 Da was observed for all the fragmentation ions from digestion in heavy and light water. Similarly, the other two overlapping partial sequences are likely the N-terminal fragments of a single tryptic peptide containing GPSVFPLA; and again, as expected, no mass shift was observed from 18O-labeling. Conversely, false hits can be ruled out; for example, the doubly charged fragment ion m/z 1101.0885 (Table S3-1) matches four peptide sequences W)GQGTLVTVSSASTKGPSVFPLAP(C), (T)APKLLIYGNSNRPSGVPDRF(S), Y)WGQGTLVTVSSASTKGPSVFPL, (C)PPCPAPPVAGPSVFLFPPKPK (D). Since a mass shift from 18O-labeling was observed, an internal fragment was ruled out, and therefore, this leaves only the last sequence with a C-terminal lysine as the only plausible choice.
(D). Since a mass shift from 18O-labeling was observed, an internal fragment was ruled out, and therefore, this leaves only the last sequence with a C-terminal lysine as the only plausible choice.
At this point, the fragment ion mass search data indicated the peptide at m/z 1351.33 highly likely contains CPPCPAPPVAGPSVFLFPPKPK. For the second chain, although mass search of two b-ions suggests the presence of GPSVFPLA, additional data were warranted for higher confidence level in the assignment as described next.
De novo sequencing
This compliments nicely with the database search and afford sequence tags25. As shown in Scheme 1, the identification of the sequence tags was conducted using the crosslinked y-ions (8 Da mass shift in 18O-digest, group 4 in Table S1), which obviously would not match any single chain peptides in the database. In Table S4, the observed m/z value is from the most abundant isotopic peak in each isotopic envelope because the monoisotopic peak is weak for large ions at low level. The mass difference between a pair of adjacent y-ions was calculated and compared manually to the mass of single amino acids and dipeptides within a mass error of 0.05 Da. Matching single amino acid residues or dipeptides are shown in red. The sequence tag SVFPLA was confirmed in the crosslink peptide chain G118- R129, lending a strong support to the existence of peptide chain G118-R129 as a component of the crosslinked peptide (Table S4).
In summary, the peptide chain C219-K240 (219CPPCPAPPVAGPSVFLFPPKPK240) and G118-A125 (118GPSVFPLA125) was identified at the end of Stage 2 as parts of the crosslinked peptide of m/z 1351.33.
Stage 3: Inference of full sequence for each chain
Extension to the Putative Full Sequences
Because the peptides were generated by trypsin digestion, the putative partial sequences of crosslinked peptide chains were extended to their corresponding full tryptic peptides (G118-R129, C215-K240, V294-K314) with mass of 1287.6282, 2911.3305, and 2502.2941 Da, respectively (Table 1). The mass difference between the observed intact crosslinked peptide (4048 Da) and the first tryptic peptide C215-K240 (2911 Da) is 1137 Da. This narrowed down the second crosslink chain to G118-R129 (1287.6282 Da) while a putative tryptic peptide V294-K314 (2502.2941 Da) is too large (combined mass) to be the second chain. This leaves peptide G118-R129 as an only plausible choice to pair with C215-K240. We were mindful that mis-cleavage might happen, which would be considered if the initial inference did not yield correct assignment.
Stage 4: Deduction of Crosslinking Chemistry and Site
Toward this end, two pieces of information are particularly useful: (1) elemental composition of the functional group involved and (2) peptide fragments not observed in tandem mass spectrometry.
Deducing Crosslinking Chemistry: Mass to Formula (Elemental Composition)
An example of the elemental composition calculation is illustrated in Table 2. Once the peptides G118-R129 and C215-K240 were established as the components of the cross-linked peptide, the difference between the observed mass of the crosslinked peptide (4048.9600 Da) and the combined mass of the two chains devoid of modifications (1287.6282+2911.3305=4198.9587 Da) was calculated to be 149.9987 Da. Four potential elemental compositions for the mass of 149.9987 Da were obtained via Thermo-Fisher Scientific Xcalibur (Table 2). The last two were eliminated based on their high delta ppm relative to the FT MS mass accuracy (typically ≤ 5 ppm). The high RDB (ring and double bond) value makes the second one unlikely too. This leaves the first one as the only plausible choice.
Table 2.
Elemental formula with mass of 149.9987 Da (the mass difference between the sum of unmodified peptides and crosslinked peptide). RDB means ring and double bond; ppm is part-per-million.
| Elemental Composition |
Proposed Structure | |||
|---|---|---|---|---|
| Formula | Cal. Mass (Da) | Delta ppm | RDB | |
| C4H6O4S | 149.9987 | 0 | 2.0 | S+CH2COOH+CH2COOH |
| C10ON | 149.9980 | 5 | 11.5 | Excluded |
| C5H2N4S | 150.0000 | −9 | 7.0 | Excluded |
| C2H4O3N3S | 149.9973 | 9 | 2.5 | Excluded |
The elemental composition of C4H6O4S contains sulfur, which only presents in cysteine and methionine. Each of the putative crosslinked peptide pair G118-R129 and C215-K240, and particularly the fragments that were not observed by tandem mass spectrometry (those not underlined in Table 1), contain cysteine but not methionine. During sample preparation, cysteine residues were reduced and alkylated by iodoacetic acid (IAA), so the mass for all peptides were calculated assuming cysteines are alkylated. Hence, removal of two alkyl groups (two C2H3O2) and a sulfur atom exactly matches the determined elemental composition. The mass of observed crosslink peptide (4048.963 Da) and the theoretical thioether peptide (4048.960 Da) are practically identical (with mass error of 0.74 ppm, see Table 3). All together, we surmised that the crosslinking chemistry is a thioether originating from a pair of cysteine residues (Scheme 2).
Table 3.
Crosslinked peptides identified in IgG2. Thioether crosslink sites are labeled in red and bold-face. The exact site at either C215 or C216 is unable to be differentiated with available data. Unless noted, all cysteine side chains are alkylated with IAA. Alkylated Lys is shown in blue. The longest b- or y-ions observed are underlined. Peptides are shown with the adjoining amino acid residues before cleavage in parenthesis. The peptide #8 contains a thioether (in red) and dehydroalanine (in green and asterisk) at C215-C216, as shown in Figure S5.
| # | Name | m/z(charge) | RT (min) | Observed Mass (Da) | Theoretical Mass (Da) | Mass Error (ppm) | Sequence | Cross-linking Site in Heavy Chain |
|---|---|---|---|---|---|---|---|---|
| 1 | G118R129/ C215K240 | 1351.33(3+) | 91.17 | 4048.963 | 4048.960 | 0.74 | 127 | |
| 215 or 216 | ||||||||
| 2 | G118R129/ K214K240 | 1394.03(3+) | 87.91 | 4177.059 | 4177.055 | 0.96 | Same | |
| 3 | G118R129/ 214K240 214K240 |
1413.36(3+) | 88.56 | 4235.064 | 4235.060 19 | 0.94 | Same | |
| 4 | C215K240/ C215K240 | 1419.92(4+) | 101.59 | 5672.670 | 5672.662 | 1.41 | 215 or 216 | |
| 215 or 216 | ||||||||
| 5 | C215K240/ K214K240 | 1452.20(4+) | 99.76 | 5800.765 24 | 5800.757 | 1.38 | Same | |
| 6 | K214K240/ K214K240 | 1484.22(4+) | 97.88 | 5928.854 | 5928.852 | 0.34 | Same | |
| 7 | K214K240/ 214K240 214K240 |
1498.47(4+) | 98.99 29 | 5986.858 | 5986.858 | 0.00 | Same | |
| 8 | K214K240 214K240 214K240 |
1475.73(4+) 31 | 98.49 | 5894.860 | 5894.856 | 0.68 | Same | |
| 9 | T210K240/ K214K240 | 1605.54(4+) | 96.15 | 6414.117 | 6414.112 | 0.78 | Same | |
| 10 | T210K240/ 214K240 36 214K240 36 |
1620.04(4+) | 97.39 | 6472.120 | 6472.117 | 0.46 | Same | |
Scheme 2.
Establishment of crosslink chemistry based on formula C4H6O4S obtained from elemental composition analysis of 149.9987 Da.
Locating Crosslink Site
Typically, the crosslink site can be localized by the largest b- and y-ions observed. For the crosslinked peptide m/z 1351.33, the largest b- and y-ions are GPSVFPLA/(PCSR, not observed), (CCVE, not observed)/CPPCPAPPVAGPSVFLFPPKPK for the chain HC:G118-R129 and HC:C215-K240, respectively. This indicates the crosslink site is in the corresponding PCSR and CCVE region (Figure 1). Compared to the highly stable valine and proline, cysteine and glutamic acid are chemically reactive, so they are more likely candidates for crosslinking.
Figure 1.
CID MS/MS spectrum of the triply charged precursor ions at m/z 1351.33 (16O-labeled C-termini) and 1354.00 (18O-labeled C-termini). Singly and doubly charge fragment ions that contain the individual chain of the crosslinked peptides (G118-R129/C215-K240, RT at 91.17 min) are highlighted in blue and red, respectively. Characteristic mass shift imparted by the heavier isotope 18O was observed (e.g., the mass shift of 4 Da for y5 in 16O-vs18O-water).
The elemental analysis described in the previous section indicates that a sulfur atom was removed, suggesting that a cysteine is involved. In addition, functional groups with a combined composition of C4H6O4 are eliminated from the theoretical peptide pairs, in which cysteinyl residues were assumed to be alkylated with IAA (C2H3O2). Taken together, our data indicated that the crosslink site is highly likely at HC:Cys127-HC:Cys215 or HC:Cys127-HC:Cys216 as shown in Figure 1. Because the two cysteines (Cys215 and Cys216) in the heavy chain are adjacent to each other, the exact crosslink site was unable to be unambiguously determined here.
Final Confirmation and Additional Support
Confirmation by Data Matching
Once the putative crosslinking chemistry and site have been proposed, theoretical fragmentation spectra were calculated and compared with the observed spectra. The assignment is shown in Figure 1 and is highly consistent with the deduced structure. A handful fragment peaks of a few crosslinks were not assigned initially, and hence were subjected to further analysis as described next.
MS3 Analysis
MS3 analysis may provide additional structural information, especially for fragment ions that are difficult to assign in the MS/MS. For example, in Figure S3-2, two high intensity fragment ions at m/z 1196.50 (singly charged) and 1520.90 (doubly charged) were observed for the triply charged crosslinked peptide m/z 1413.37 (G118-R129/ 214-K240), but could not be assigned to typical b- or y-ions. To ascertain, MS3 analysis of these two unassigned ions revealed the sequences to be 118GPSVFPLAP
214-K240), but could not be assigned to typical b- or y-ions. To ascertain, MS3 analysis of these two unassigned ions revealed the sequences to be 118GPSVFPLAP SR129 and 214
SR129 and 214 C
C VECPPCPAPPVAGPSVFLFPPKPK240, in which a dehydroalanine replaces C127 in peptide G118-R129 and a free cysteine replaces the thioether at 216 in peptide
VECPPCPAPPVAGPSVFLFPPKPK240, in which a dehydroalanine replaces C127 in peptide G118-R129 and a free cysteine replaces the thioether at 216 in peptide  214-K240, respectively (Figure S3-4 & -5). These data further supported the proposed sequence and crosslink sites. Alkylation at Lys and Met as an artifact from sample preparation in peptide mapping was reported49. The alkylation at K214 in the crosslink peptide G118-R129/
214-K240, respectively (Figure S3-4 & -5). These data further supported the proposed sequence and crosslink sites. Alkylation at Lys and Met as an artifact from sample preparation in peptide mapping was reported49. The alkylation at K214 in the crosslink peptide G118-R129/ 214- K240 is in agreement with the literature 49.
214- K240 is in agreement with the literature 49.
Additional Peptides
Following the same work flow, full sequences, crosslinking chemistry and sites have been established for all ten candidate crosslinked peptides shown in red in Table S2. The final results of all identified crosslinked peptides are summarized in Table 3.
To evaluate the sensitivity of our method, the peak intensity from LC-MS analysis for each crosslinked peptide and its related (not crosslinked) peptides was used to estimate the degree of crosslinking as described by Zhang16, ranging from 0.2% to 5.0% (half less than 1%; see Table S6 for details). Comparable data were observed based on reducing SDS-PAGE, which indicated about 8% of total crosslinked species (see Figure S1A). It is also worth noting that no enrichment or separation was performed on the IgG2 samples prior to tryptic digestion (the first step of our work flow); in other words, the crosslinked peptides were analyzed in the presence of large excess of native peptides. Of course, considerably higher sensitivity can be achieved if the crosslinked proteins are separated or enriched prior to analysis.
Targeted Search Based on the Newly Established Crosslinking Chemistry
After the thioether crosslink chemistry was established, a targeted search for this particular modification was performed following well-established protocols. First, a theoretical database was built for all combination of a thioether crosslinking between any two cysteinyl residues. Then, all observed precursor ions were searched against the database. When a hit was found in the targeted mass search, the corresponding MS/MS data from both the 18O-water and 16O-water digests were examined for further structural confirmation. By this approach, four additional thioether peptides were found (Table S5, Figure S6 & S7). All contain a light chain C-terminal peptide, so each has only one newly created C-terminus (two 18O-incorporation) and therefore was not discriminated from single chain peptides in the initial screening stage. Again, these results showcase the utility of our approach to identify crosslinks in macromolecules derived from previously unknown crosslinking chemistry.
Formation of thioether
Thioether is a known modification for proteins50–55. For IgG1, a thioether crosslink was located at the disulfide bond of the light chain C-termini and the heavy chain hinge region52, 55. A generally accepted mechanism involves a β-elimination of disulfide to generate dehydroalanine followed by Michael addition by another cysteinyl thiol52–54, 56, 57. Basic conditions and structural flexibility generally favor its formation50–55. In addition, radical intermediates have been postulated for desulfurization57, 58. The hinge region of IgG2 is highly flexible and solvent exposed, and therefore very susceptible to this transformation. Indeed, our results indicated it occurs more frequently at the light chain C-termini and in the hinge region of IgG2. It is very interesting that the disulfide bonds of heavy chain C127 –heavy chain C215 (or C216) in the IgG2 A/B form (or B form) are also reactive (Figure S8). These thioether crosslinks at HC:Cys127-HC:Cys215 (or HC:Cys127-HC:Cys216), HC:Cys215-HC:Cys215 (or HC:Cys216-HC:Cys216), LC:Cys217-HC:Cys127, and LC:Cys217-HC:Cys215(or LC:Cys217- HC:Cys216) originated from native disulfides as shown in red in Figure S8. Thioether linkage is in agreement with the previous reports on IgG2 disulfide bond pairing59–61. In Table 3, the crosslink peptide #8 (HC:K214-K240*/HC:  214-K240) contains a thioether and an dehydroalanine. The corresponding linear peptide K214-K240* (214KCC*VECPPCPAPPVAGPSVFLFPPKPK240, the dehydroalanine at C215 or C216 was denoted as asterisk) was also found. All together, these data are consistent with thioether formation via dehydroalanine intermediates.
214-K240) contains a thioether and an dehydroalanine. The corresponding linear peptide K214-K240* (214KCC*VECPPCPAPPVAGPSVFLFPPKPK240, the dehydroalanine at C215 or C216 was denoted as asterisk) was also found. All together, these data are consistent with thioether formation via dehydroalanine intermediates.
CONCLUSIONS
The utility of our XChem-Finder strategy for the characterization of protein crosslinking with undefined chemistry is exemplified by the discovery of fourteen thioether peptides in IgG2. Essential to our approach is 18O-isotope labeling; it allows the facile detection of crosslinked peptides, and most significantly, divides the complex tandem mass spectra to sub-sets that can be processed by standard database search (FindPept that matches fragment ions with partial peptide sequences) and de novo sequencing (sequence tags). High-resolution spectral data also dramatically improve the confidence of assignment, and moreover, reveal the chemical nature of the crosslinking. While the reported work was manually processed, most steps can be automated. Hence our XChem-Finder strategy should be generally applicable for the discovery of crosslinked proteins, without prior defined chemistry, in both biological systems and biopharmaceuticals.
Supplementary Material
ACKNOWLEDGEMENT
We are grateful to Duclos Richard, Peter Zhou, Aleks Swietlow, and Wanlu Qu for their critical review of the manuscript and helpful suggestions. We also thank Bin Ma, Dan Maloney, and Cassandra Wigmore at Bioinformatics Solutions for helpful discussion on de novo sequencing. This activity is partially supported by an educational donation provided by Amgen and a grant from NIH NIGMS (1R01GM101396 to ZSZ). This is contribution number 1033 from the Barnett Institute.
REFERENCES
- 1.Liu H, Gaza-Bulseco G, Faldu D, Chumsae C, Sun J. J Pharm Sci. 2008;97:2426–2447. doi: 10.1002/jps.21180. [DOI] [PubMed] [Google Scholar]
- 2.DiMarco T, Giulivi C. Mass Spectrom Rev. 2007;26:108–120. doi: 10.1002/mas.20109. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava OP, Kirk MC, Srivastava K. J Biol Chem. 2004;279:10901–10909. doi: 10.1074/jbc.M308884200. [DOI] [PubMed] [Google Scholar]
- 4.Wilhelmus MM, Grunberg SC, Bol JG, van Dam AM, Hoozemans JJ, Rozemuller AJ, Drukarch B. Brain Pathol. 2009;19:612–622. doi: 10.1111/j.1750-3639.2008.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez B, Gonzalez A, Hermida N, Valencia F, de Teresa E, Diez J. Am J Physiol Heart Circ Physiol. 2010;299:H1–H9. doi: 10.1152/ajpheart.00335.2010. [DOI] [PubMed] [Google Scholar]
- 6.Nemes Z, Devreese B, Steinert PM, Van Beeumen J, Fesus L. FASEB J. 2004;18:1135–1137. doi: 10.1096/fj.04-1493fje. [DOI] [PubMed] [Google Scholar]
- 7.Friedman M. J Agric Food Chem. 1999;47:1295–1319. doi: 10.1021/jf981000+. [DOI] [PubMed] [Google Scholar]
- 8.Nashef AS, Osuga DT, Lee HS, Ahmed AI, Whitaker JR, Feeney RE. J Agric Food Chem. 1977;25:245–251. doi: 10.1021/jf60210a020. [DOI] [PubMed] [Google Scholar]
- 9.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh P, Panchaud A, Goodlett DR. Anal Chem. 2010;82:2636–2642. doi: 10.1021/ac1000724. [DOI] [PubMed] [Google Scholar]
- 11.Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. Proc Natl Acad Sci U S A. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee YJ, Lackner LL, Nunnari JM, Phinney BS. J Proteome Res. 2007;6:3908–3917. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- 13.Hoopmann MR, Weisbrod CR, Bruce JE. J Proteome Res. 2010;9:6323–6333. doi: 10.1021/pr100572u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du X, Chowdhury SM, Manes NP, Wu S, Mayer MU, Adkins JN, Anderson GA, Smith RD. J Proteome Res. 2011;10:923–931. doi: 10.1021/pr100848a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury SM, Du X, Tolic N, Wu S, Moore RJ, Mayer MU, Smith RD, Adkins JN. Anal Chem. 2009;81:5524–5532. doi: 10.1021/ac900853k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z. Anal Chem. 2009;81:8354–8364. doi: 10.1021/ac901193n. [DOI] [PubMed] [Google Scholar]
- 17.Zang T, Lee BW, Cannon LM, Ritter KA, Dai S, Ren D, Wood TK, Zhou ZS. Bioorg Med Chem Lett. 2009;19:6200–6204. doi: 10.1016/j.bmcl.2009.08.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan W, Zhao G, Al-Saad K, Siems WF, Zhou ZS. Rapid Commun Mass Spectrom. 2004;18:319–324. doi: 10.1002/rcm.1335. [DOI] [PubMed] [Google Scholar]
- 19.Gao Q, Xue S, Doneanu CE, Shaffer SA, Goodlett DR, Nelson SD. Anal Chem. 2006;78:2145–2149. doi: 10.1021/ac051339c. [DOI] [PubMed] [Google Scholar]
- 20.Gao Q, Xue S, Shaffer SA, Doneanu CE, Goodlett DR, Nelson SD. Eur J Mass Spectrom (Chichester, Eng) 2008;14:275–280. doi: 10.1255/ejms.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zelter A, Hoopmann MR, Vernon R, Baker D, MacCoss MJ, Davis TN. J Proteome Res. 2010;9:3583–3589. doi: 10.1021/pr1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, Mueller M, Aebersold R. Nat Methods. 2008;5:315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seebacher J, Mallick P, Zhang N, Eddes JS, Aebersold R, Gelb MH. J Proteome Res. 2006;5:2270–2282. doi: 10.1021/pr060154z. [DOI] [PubMed] [Google Scholar]
- 24.Seidler J, Zinn N, Boehm ME, Lehmann WD. Proteomics. 2010;10:634–649. doi: 10.1002/pmic.200900459. [DOI] [PubMed] [Google Scholar]
- 25.Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzner RA, Larson Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Anal Chem. 2008;80:8799–8806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroon DJ, Baldwin-Ferro A, Lalan P. Pharm Res. 1992;9:1386–1393. doi: 10.1023/a:1015894409623. [DOI] [PubMed] [Google Scholar]
- 27.Van Buren N, Rehder D, Gadgil H, Matsumura M, Jacob J. J Pharm Sci. 2009;98:3013–3030. doi: 10.1002/jps.21514. [DOI] [PubMed] [Google Scholar]
- 28.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Anal Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 29.Ren D, Pipes GD, Liu D, Shih LY, Nichols AC, Treuheit MJ, Brems DN, Bondarenko PV. Anal Biochem. 2009;392:12–21. doi: 10.1016/j.ab.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z. Anal Chem. 2005;77:6364–6373. doi: 10.1021/ac050857k. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z. Anal Chem. 2011;83:8642–8651. doi: 10.1021/ac2020917. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z. Anal Chem. 2004;76:3908–3922. doi: 10.1021/ac049951b. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z. J Am Soc Mass Spectrom. 2012;23:764–772. doi: 10.1007/s13361-011-0334-2. [DOI] [PubMed] [Google Scholar]
- 34.Schnolzer M, Jedrzejewski P, Lehmann WD. Electrophoresis. 1996;17:945–953. doi: 10.1002/elps.1150170517. [DOI] [PubMed] [Google Scholar]
- 35.Ye X, Luke B, Andresson T, Blonder J. Brief Funct Genomic Proteomic. 2009;8:136–144. doi: 10.1093/bfgp/eln055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fenselau C, Yao X. J Proteome Res. 2009;8:2140–2143. doi: 10.1021/pr8009879. [DOI] [PubMed] [Google Scholar]
- 37.Yao X, Afonso C, Fenselau C. J Proteome Res. 2003;2:147–152. doi: 10.1021/pr025572s. [DOI] [PubMed] [Google Scholar]
- 38.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 39.Bantscheff M, Dumpelfeld B, Kuster B. Rapid Commun Mass Spectrom. 2004;18:869–876. doi: 10.1002/rcm.1418. [DOI] [PubMed] [Google Scholar]
- 40.Stewart II, Thomson T, Figeys D. Rapid Commun Mass Spectrom. 2001;15:2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 41.Koehler CJ, Arntzen MO, de Souza GA, Thiede B. Anal Chem. 2013 doi: 10.1021/ac3035508. [DOI] [PubMed] [Google Scholar]
- 42.Koehler CJ, Arntzen MO, Strozynski M, Treumann A, Thiede B. Anal Chem. 2011;83:4775–4781. doi: 10.1021/ac200229w. [DOI] [PubMed] [Google Scholar]
- 43.Nakazawa T, Yamaguchi M, Okamura TA, Ando E, Nishimura O, Tsunasawa S. Proteomics. 2008;8:673–685. doi: 10.1002/pmic.200700084. [DOI] [PubMed] [Google Scholar]
- 44.Liu M, Cheetham J, Cauchon N, Ostovic J, Ni W, Ren D, Zhou ZS. Anal Chem. 2012;84:1056–1062. doi: 10.1021/ac202652z. [DOI] [PubMed] [Google Scholar]
- 45.Ni W, Dai S, Karger BL, Zhou ZS. Anal Chem. 2010;82:7485–7491. doi: 10.1021/ac101806e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alfaro JF, Gillies LA, Sun HG, Dai S, Zang T, Klaene JJ, Kim BJ, Lowenson JD, Clarke SG, Karger BL, Zhou ZS. Anal Chem. 2008;80:3882–3889. doi: 10.1021/ac800251q. [DOI] [PubMed] [Google Scholar]
- 47.Gururaja TL, Payan DG, Anderson DC. Biopolymers. 2007;88:55–63. doi: 10.1002/bip.20626. [DOI] [PubMed] [Google Scholar]
- 48.Banerjee S, Mazumdar S. J Mass Spectrom. 2010;45:1212–1219. doi: 10.1002/jms.1817. [DOI] [PubMed] [Google Scholar]
- 49.Gurd FR. Methods Enzymol. 1972;25:424–438. doi: 10.1016/S0076-6879(72)25038-8. [DOI] [PubMed] [Google Scholar]
- 50.Datola A, Richert S, Bierau H, Agugiaro D, Izzo A, Rossi M, Cregut D, Diemer H, Schaeffer C, Van Dorsselaer A, Giartosio CE, Jone C. ChemMedChem. 2007;2:1181–1189. doi: 10.1002/cmdc.200700042. [DOI] [PubMed] [Google Scholar]
- 51.Lispi M, Datola A, Bierau H, Ceccarelli D, Crisci C, Minari K, Mendola D, Regine A, Ciampolillo C, Rossi M, Giartosio CE, Pezzotti AR, Musto R, Jone C, Chiarelli F. J Pharm Sci. 2009;98:4511–4524. doi: 10.1002/jps.21774. [DOI] [PubMed] [Google Scholar]
- 52.Cohen SL, Price C, Vlasak J. J Am Chem Soc. 2007;129:6976–6977. doi: 10.1021/ja0705994. [DOI] [PubMed] [Google Scholar]
- 53.Florence TM. Biochem J. 1980;189:507–520. doi: 10.1042/bj1890507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galande AK, Trent JO, Spatola AF. Biopolymers. 2003;71:534–551. doi: 10.1002/bip.10532. [DOI] [PubMed] [Google Scholar]
- 55.Tous GI, Wei Z, Feng J, Bilbulian S, Bowen S, Smith J, Strouse R, McGeehan P, Casas-Finet J, Schenerman MA. Anal Chem. 2005;77:2675–2682. doi: 10.1021/ac0500582. [DOI] [PubMed] [Google Scholar]
- 56.Zhao G, Zhou ZS. Bioorg Med Chem Lett. 2001;11:2331–2335. doi: 10.1016/s0960-894x(01)00440-1. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Rejtar T, Zhou ZS, Karger BL. Rapid Commun Mass Spectrom. 2010;24:267–275. doi: 10.1002/rcm.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou ZS, Smith AE, Matthews RG. Bioorg Med Chem Lett. 2000;10:2471–2475. doi: 10.1016/s0960-894x(00)00498-4. [DOI] [PubMed] [Google Scholar]
- 59.Dillon TM, Ricci MS, Vezina C, Flynn GC, Liu YD, Rehder DS, Plant M, Henkle B, Li Y, Deechongkit S, Varnum B, Wypych J, Balland A, Bondarenko PV. J Biol Chem. 2008;283:16206–16215. doi: 10.1074/jbc.M709988200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wypych J, Li M, Guo A, Zhang Z, Martinez T, Allen MJ, Fodor S, Kelner DN, Flynn GC, Liu YD, Bondarenko PV, Ricci MS, Dillon TM, Balland A. J Biol Chem. 2008;283:16194–16205. doi: 10.1074/jbc.M709987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang B, Harder AG, Connelly HM, Maheu LL, Cockrill SL. Anal Chem. 2010;82:1090–1099. doi: 10.1021/ac902466z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.