Abstract
β-Secretase (BACE1, β-site APP cleaving enzyme 1) is an aspartic proteinase that has multiple functions in various physiological processes, such as cell differentiation, immunoregulation, and cell death. There is increasing evidence that changes in BACE1 activity are involved in many diseases, such as Alzheimer’s disease (AD), schizophrenia, epileptic behavior, and others. However, a deeper understanding of the molecular biology of BACE1 is necessary for further exploration of cell development, immunological regulation, and disease pathogenesis. Here, we review the molecular and cellular biology of BACE1, including its enzymatic properties, structure, biosynthesis, and physiological functions to provide a new perspective and rational assessment of drugability. Lastly, we discuss proposed strategies to control BACE1 activity for possible therapeutic application.
Introduction
In 1999, BACE1 was simultaneously discovered by five groups to be an aspartic proteinase [1–5]. The various research teams used very different experimental approaches to identify exactly the same protein. Since then, many clinicians, basic scientists, and industrial pharmacologists have made significant increases in the understanding of BACE1, and this progress has elucidated the role of BACE1 in multiple diseases, such as AD, schizophrenia, and epileptic behavior. However, most studies on BACE1 have focused on disease-related mechanisms or its pathological effects rather than its normal physiological properties. Thus, a deeper understanding of the basic pharmacological, biochemical, and molecular biological properties of BACE1 is necessary not only for elucidation of disease pathogenesis but also for drug discovery. Therefore, in this review, we provide an update on several important recent discoveries, including:
the primary structure of BACE1 and its basic posttranslational modifications, such as glycosylation, phosphorylation, palmitoylation, ubiquitination, and acetylation;
the enzymatic activity, substrates, and physiological functions of BACE1;
BACE1-binding partners; and
recombinant BACE1 in different expression systems, from Escherichia coli to mammals.
We will also discuss additional biological and physiological features of BACE1, particularly its biosynthesis, subcellular location, and degradation.
Understanding the biology of BACE1 will be crucial for development of safe and effective inhibitors or modulators for neurological disorder therapies.
Nomenclature and findings
There are two homologs of BACE1 [β-site amyloid precursor protein (APP) cleaving enzyme or β-secretase]. BACE1 and BACE2 comprise a new subfamily of membrane-anchored aspartyl proteases [6]. BACE1 [3.4.23.46] (Asp-2, membrane-bound aspartic proteinase or Memapsin 2) exists in neurons and cleaves APP at the Asp+1 site. BACE2 [3.4.23.45] (Asp1, Memapsin 1, or DRAP) shares ~60% homology with BACE1 but exists in peripheral tissues and cleaves APP at the Phe+19 or Phe+20 site. Both homologs of BACE (BACE1 and BACE2) share similar structure. They share 51% similarity in amino acid sequences, and also both proteins have a catalytic domain formed via DTG and DSG active site motifs, a single transmembrane domain, and a short C-terminal tail. BACE1 has been identified as the Alzheimer’s β-secretase, whereas BACE2 was mapped to the region of human chromosome 21 that is involved in Down syndrome [6], and is also related to diabetes [7,8]. The expression of BACE2 in the brain is significantly lower compared with BACE1 [6,9]. Thus, it can be distinguished from BACE1 by pro-segment autoprocessing, APP processing, and subcellular localization, as well as the distinct expression patterns in the brain [9].
BACE1 structure
Primary sequence
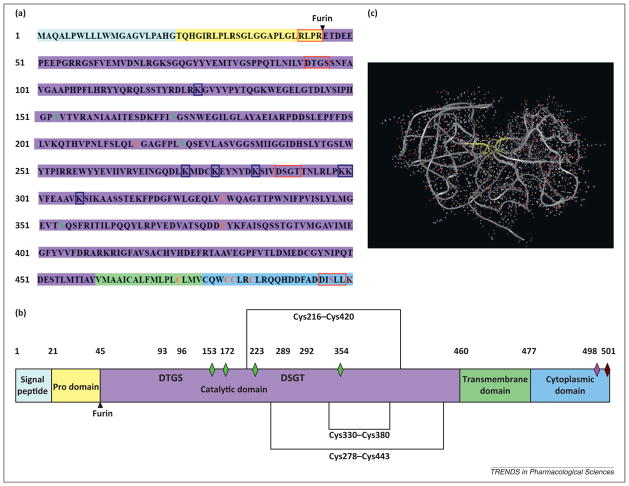
BACE1, a ~75-kD protein that has ~30% sequence homology with its pepsin family members, has two conserved active sites [DTGS (93–96) and DSGT (289–292)] and three pairs of disulfide bonds (216/420, 278/443, 330/380) (Figure 1a). Mutations in these disulfide bonds result in incomplete glycosylation and pro-peptide processing. Among them, the disulfide bond (330/380) in the C-terminal lobe is of significant importance in maintaining catalytic activity and proper structure, and the other two seem responsible for correct catalytic orientation.
Figure 1.
The human amino acid sequence of β-secretase (BACE1, β-site APP cleaving enzyme 1) and its domain structure. (a) BACE1 (human) sequence. The background color of amino acids matches their corresponding domain. Numbers on the left denote amino acids position. Boxes: (in red) structure conserved motif, (in blue) the acetylated sites of Lys residues. Capital letters in green, orange, pink, and brown indicate the sites of N-linked glycosylation, disulfide bonds formation, phosphorylation, and ubiquitination, respectively. (b) BACE1 domain structure. The diamonds with the same color as capital letters in the protein sequence indicate the same modification site in BACE1 domains. (c) BACE1 (monomer) 3D structure (53–447 amino acids of BACE1 were calculated for molecular modeling). Molecular modeling tools from Swiss-Prot are used and the yellow ribbons denote two aspartic activity motifs [24].
Post-translational modifications, such as N-glycosylation (Asn153, Asn172, Asn223, and Asn354) [10,11], phosphorylation (Ser498) [12], palmitoylation (Cys474, Cys478, Cys482, and Cys485) [13], ubiquitination (K501) [14], and acetylation (K126, K300, K307, K275, K279, K285, and K299) [15], also play roles in maintaining proper BACE1 function. N-Glycosylation and its sulfation facilitate propeptide processing, BACE1 maturation, and transportation. When phosphorylation occurs by casein kinase I, BACE1 is immediately recognized and carried by GGAs (Golgi-localized γ-ear-containing ARF-binding) to late endosomes or the cell surface.
Palmitoylation in the BACE1 C-terminal extension facilitates BACE1 dimerization and direction to lipid rafts. It increases BACE1 activity and suppresses its ectodomain shedding. GGAs carry ubiquitinated BACE1 to lysosomes for degradation. In the endoplasmic reticulum (ER), transient acetylation occurs with the help of ATase1 and ATase2 (lysine acetyltransferases). Then, acetylated BACE1 can be transported to the Golgi for further modification and maturation, and unacetylated BACE1 is hydrolyzed into fragments [16], suggesting that the high levels of BACE1 seen in brains with AD may be due to acetylated BACE1 that is unable to be degraded.
BACE1 domains
BACE1 contains five functional domains (Figure 1b). The signal domain directs BACE1 to the ER and releases pro-BACE into the luminal ER. The pro-domain, the shortest of human aspartic proteinases, exhibits no inhibitory activity to its mature enzyme, and pro-BACE1 itself possesses 30% enzymatic activity [17]. Then, in the late Golgi apparatus, furin identifies and cleaves pro-BACE1 at the RLPR↓ motif to form the mature enzyme. In addition, the pro-domain also helps to mediate correct folding of BACE1 for efficient exit from the ER [18,19].
In the catalytic domain, mutation of either aspartic residue renders the protein inactive. However, enzymatic activity is partially restored when vectors of either of the two active site (Asp+93, Asp+289) are coexpressed in one cell [20], suggesting that BACE1 homodimers exist.
The C-terminal extension is a distinctive feature of BACE and includes transmembrane and cytoplasmic domains. Without the C terminus, mutants mature faster as monomers [21]; by contrast, without the cytosolic domain or with the transmembrane domain substitution, BACE1 dimerizes normally, suggesting that the transmembrane region itself or the biological membrane is helpful for dimerization [21]. The transmembrane domain also dictates BACE1 retention in the ER and determines its enzymatic activity to substrates [22]. An ACDL sorting motif, DISLL, in cytosolic tail dictates BACE1 trafficking and recycling [10].
Crystal structure
The crystal structure of an active form of BACE1 was solved in 2000, providing insight into regulation of its enzymatic activity [23]. BACE1 catalytic units are folded into the conserved N-terminal and C-terminal lobes (Figure 1c) [24]; the substrate-binding sites, enzymatic active sites with hydrogen bonds in the cleft center. Oligosaccharides limit the motion and orientation of the catalytic domain. The ‘flap’, a β-sheet structure (Tyr68–Glu77), provides access to the active site cleft and contributes to the substrate specificity by partially covering the cleft. Tyr71 and newly formed hydrogen bonds (such as Trp76–Tyr71, Ser35–Asp32, etc.) dictate the flap movement [25].
Activity and specificity of BACE1
The enzymatic reactions of BACE1
Pro-BACE1, monomeric BACE1, dimeric BACE1, and high molecular complex BACE1 are four species that exist naturally. Because monomeric BACE1 undergoes autoproteolysis in vitro, its activity fluctuates. In the endogenous environment, dimeric BACE1 and high molecular complex BACE1 have much higher kinetic activity, and the prodomain itself has little effect on kinetic parameters in a pro-BACE1 activity assay [21]. For recombinant BACE1, BACE1-FL (full-length BACE1) is more active than BACE1-NT (without C terminal) [21].
Substrates of BACE1
BACE1 accommodates 12 subsite residues (P4′–P8) in its substrate-binding cleft. The substrate residue preference is shown in Table 1. P1 is the most stringent site, and in P subsites, the inner ones are more stringent than the outer residues. Physiologically, BACE1 also cleaves substrates for various functions (Table 2).
Table 1.
Residue preference for BACE1 substrates
| Position | Amino acid residues preference |
|---|---|
| P4′ | Asp > Glu > Trp > Tyr > Phea |
| P3′ | Leu > Trp > Val > Ile > Thr > Asp > Glu |
| P2′ | Val > Ile > Ala > Glu > Leu |
| P1′ | Met > Glu > Gln > Ala > Asp |
| P1 | Leu > Phe > Met > Tyr > Thr |
| P2 | Asp > Asn > Met > Phe > Tyr > Leu > Glu > Ser > Ala > Gln > Lys |
| P3 | Ile > Val > Leu > Glu > His |
| P4 | Glu > Gln > Asp > Met > Asn |
| P5 | Asp > Phe > Tyr > Glu > Trp > Thr > Pro > Ser |
| P6 | Phe > Trp > Asp > Ile > Leu |
| P7 | Ile > Asp > Trp > Tyr > Phe > Glu |
| P8 | Arg > Tyr > Phe > Gln > Glu |
The residues found in native APP (labeled in blue) or Swedish mutant APP (labeled in red).
Table 2.
BACE1 substrates
| Name | Cleavage sites | Related phenomenon | |
|---|---|---|---|
| P1 | P1′ | ||
| APP | M | D | Aβ generation |
| APLP1 | Ma | Na | AICD generation |
| APLP2 | L | D | AICD generation |
| NRG1 | F | M | Axonal myelination |
| Voltage-gated sodium channels | L/F | E/Q | Reduced neuron cells excitability |
| PSGL-1 | L | S | Reduced leukocyte adhesion |
| ST6Gal1 | L | Q | Acute-phase hepatic reactions |
| LRP | Unknown | Unknown | LRP ectodomain releasing |
| mPGES-2 | La | Sa | Increased PGE-2 levels |
| IL-1R2 | Fa | Qa | Block NF-κB signal |
| Enterokinase | GYEb | Unknown | |
To be the possible cleavage site by analysis: relative kcat/Km [108].
Potential BACE1 cleavage site.
APP and its homologs
Generally, APP is cleaved by α-secretase, and this cleavage is considered as non-amyloidogenic. By contrast, APP can also be sequentially cleaved by BACE1 and γ-secretase, generating the Aβ peptide associated with AD (Box 1). Cholesterol is also associated with cleavage of APP by BACE1. If membrane cholesterol increases, APP relocalizes to lipid rafts already containing BACE1, possibly favoring the BACE1 pathway. In this way, abnormalities in cholesterol may be linked to an increase in APP processing by BACE1 and thus generation of Aβ [26].
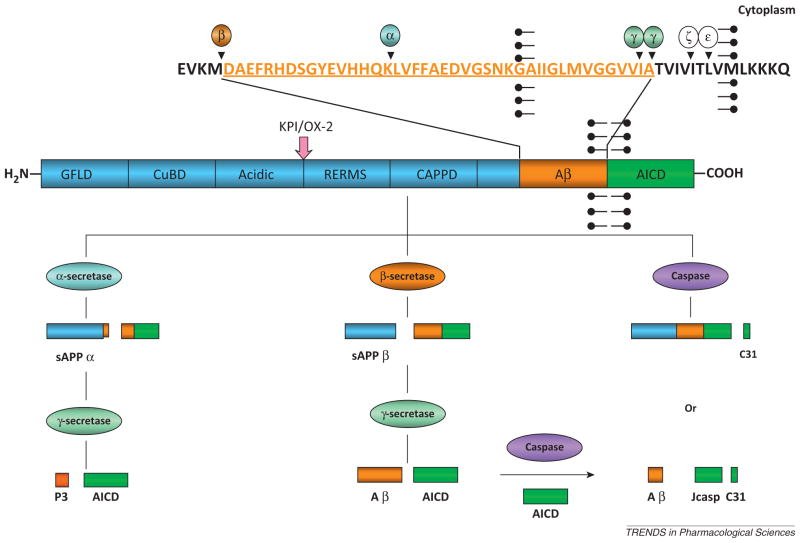
Box 1. APP processing.
APP is a key substrate for BACE1 in AD pathogenesis. After the first cleavage of APP by β-secretases, β-carboxyl-terminal fragments left on the membrane are also reported to be cytotoxic by disturbance of APP signals and neuronal ionic homeostasis (Figure I). Subsequently, γ-secretase cleaves β-CTFs (C-terminal fragments) and releases both Aβ and AICD. Depending on different physiological conditions, AICD is degraded by insulin-degrading enzyme or delivered to the nucleus for transcription. During cell apoptosis, caspases directly cleave APP at position Asp664 to release C31 (the last 31 amino acids of APP) or cleave AICD to generate both C31 and Jcasp fragments, which are neurotoxic. Further investigations reveal that caspase cleavage may be associated with regulation of AD abnormalities. With increased Aβ generation, APP695 expression is converted to Kunitz-type protease inhibitor (KPI) domain in the extracellular region of APP by prolonged activation of the extra-synaptic NMDA receptor in neurons.
To date, APP processing is found to include the following: α-processing, which precludes Aβ formation, β-processing, γ-processing, caspase processing, and other processing (ζ-processing and ε-processing).
Figure I.
Therapeutic targets in proteolysis of APP: non-amyloidogenic pathway and amyloidogenic pathways. Among the targets, β-secretase cleavage is believed to be a key step for Aβ generation, which is a major molecule that contributes to AD pathogenesis. All of the enzymes and their cleavages are validated to be involved in multiple physiological and pathological processes.
Several alternatively spliced isoforms, APP695, APP751, and APP770, also exist. APP695 is expressed in neurons and is cleaved at Asp624 by BACE1 (Box 1). A recent study shows that this cleavage is essential for glial cell survival [27]. After cleavage by BACE1, sAPPβ is released. And the left truncated APP (C99) is further cleaved by γ-secretases into two parts (Aβ and AICD). All these three fragments play roles in multiple physiological processes. For example, Li and colleagues reported that recombinant brain soluble APPβ (sAPPβ) actually regulates the expression of transthyretin (TTR) and Klotho [28], both of which are APP-dependent genes and related to AD pathogenesis. In 2011, Chasseigneaux et al. reported that sAPPβ induces a decrease in cell adhesion and stimulates axon elongation [29]. Additionally, Baratchi and colleagues found that both sAPPs (sAPPα, sAPPβ) could independently promote proliferation and differentiation of hippocampal neural progenitor cells [30], suggesting that sAPPs play roles in early brain development. The reason why sAPPs are involved in the brain developmental process may be that sAPPs participate in changing ion levels in the nervous system, further modulating neuronal excitability [31], which might ultimately affect the developmental process. Using sAPPβ-treated human embryonic stem cell (hESC) clones, Freude and colleagues showed that sAPPβ spontaneously, robustly, and decisively induces hESC differentiation into neural type in a concentration-dependent manner [32]. It seems that sAPPα and sAPPβ may have differing importance in alternate stages of brain development in humans. sAPPβ could further be cleaved by an unknown enzyme to generate N-APP (~35 kD at N-terminal of sAPPβ) [33], which initiates neuronal cell death [33]. γ-Secretases subsequently cleave C99, generating APP intracellular domain (AICD) fragments whose transcriptional activity is preferentially produced by BACE1 cleavage on APP695 [34]. From the available evidence, APP cleavage by BACE1 has at least two opposing consequences: on the one hand, this processing is widely validated to generate Aβ, which triggers a feedback loop with APP, spreads to nearby healthy neurons, aggregates in the brain, and induces AD [35]; but on the other hand, Karen Ashe’s research team have shown that transgenic mice with non-oligomeric Aβ generation have decreased memory and synaptic plasticity, which may be due to the deficiency of BACE1 cleavage on APP when one or more copies of the BACE1 gene were deleted. Thus, the conclusion from this study is that BACE1 cleavage of APP may also be helpful for learning, memory, and synaptic plasticity [36].
However, recent research also shows that partial inhibition of BACE1 can improve memory deficits and synaptic plasticity [37], suggesting that BACE1 may reduce or impair neuroplasticity by precluding the production of sAPPα, which is important in neuronal plasticity, survival, and protection [38]. sAPPα was approximately 100-fold more potent than sAPPβ in protecting hippocampal neurons against excitotoxicity and Aβ toxicity, suggesting that reducing levels of sAPPα may contribute to neuronal degeneration in AD. [39]. More information is still needed to elucidate the function of BACE1 in neuroplasticity.
Neuregulins (Nrgs)
Nrg1 and Nrg3, both subtypes of neuronal growth factor, are additional BACE1 substrates [40]. NRG1 acts on the epidermal growth factor receptor (EGFR) family and is crucial for development. It exists in various isoforms generated by alternative splicing, which allows it to perform a wide variety of functions. Among these isoforms, Nrg1 type I and type III are validated BACE1 substrates in vivo [41]. In early postnatal stages, when myelination of axons is initiated, BACE1 is expressed at higher levels and cleaves Nrg1 type III between its EF and ME sites, releasing its EGF-like domain to bind with ErbB4 receptors, and thus activating downstream signals for myelination [41]. Indeed, compared with ADAM10, another signaling protein involved in similar processing, BACE1 is a more efficient regulator of initiation and maintenance of myelineation, as well as regulating the thickness of the myelin sheath during different developmental stages [42]. In BACE1−/− mice, axonal myelination is impaired and intact Nrg1s accumulate in the brains [43].
β Subunits of voltage-gated sodium channels (VGSCs)
VGSCs are large membrane-associated complexes, each comprising one α subunit and varying β subunits. The α subunits are functional on their own, but association with β subunits or other modulatory proteins permits altered voltage dependence and cellular localization. VGSCs with β subunits are found in the central nervous system (CNS), peripheral nervous system (PNS), and muscle tissues. They not only regulate the localization of Na+ channels, altering Na+ current, but are also involved in regulating neuron migration [44] and early CNS development. There are four subunits (β1, β2, β3, and β4) identified as BACE1 substrates in vitro, and two (β2 and β4) are verified substrates in vivo. β2 subunit is cleaved physiologically at 147L↓148 M by BACE1 [45]. After cleavage, the inactivated β2 subunit results in membrane channel disruption and, accordingly, decreases the excitability of the neuron. The β2-ICD fragment regulates α subunit expression, and decreased Nav1.1 mRNA levels are found in the brains of BACE1−/− mice [46,47]. BACE1 cleavage of β4 subunits increases neurite outgrowth and promotes Purkinje cell firing [48]. In 2009, Huth and coworkers [49] revealed that, in addition to cleaving the β2 or β4 subunit in sodium 1.2 channels, increased BACE1 might functionally substitute for β2. This non-enzymatic effect alters Nav1.2 current in murine neuroblastoma cells.
P-Selectin glycoprotein ligand 1 (PSGL-1) and β-galactoside α2,6-sialyltransferase (ST6Gal1)
BACE1 also cleaves immune-associated proteins, including PSGL-1 and ST6Gal1. PSGL-1, a homodimeric type I membrane protein, mediates leukocyte adhesion and the inflammatory response in the brain. BACE1 cleaves PSGL-1 at the Leu–Ser site near the membrane region [50]. ST6Gal I sialyltransferase, a type II membrane protein, plays roles in transferring sialic acid residues in vivo, and is secreted into the plasma after cleavage by BACE1 at its Leu37↓Gln38 or Lys40↓Glu41 site [51]. This soluble form of ST6Gal I is important for B cell differentiation and inflammation. Recently, Jones et al. found that soluble ST6Gal I released into circulation restricts myelopoiesis (formation of myeloid cells) [52]. In addition, Woodard-Grice et al. reported that BACE1 processing on ST6Gal I can regulate differentiation of monocytes into macrophages [53], suggesting that BACE1 may be crucial for macrophage function.
Lipoprotein receptor-related protein (LRP)
LRPs are type I membrane proteins that not only play roles in cholesterol transport in neurons but also mediate cell growth and differentiation. BACE1 interacts with LRPs in the lipid raft and initiates LRP ectodomain shedding, which could affect the endocytosis of APP and Aβ generation [54,55].
Prostaglandin E2 synthase-2 (PGES-2)
MicrosomalPGES-2 (mPGES-2), a Golgi membrane protein, colocalizes with BACE1 in neurons that are treated with Aβ in the medium [56]. BACE1 cleaves mPGES-2 in its N terminus, activates mPGES-2, and facilitates mPGES-2 translocation to the perinuclear region for generation of higher levels of prostaglandin E2 (PGE2), which, in turn, elevates Aβ production and induces neuronal cell death [56].
Interleukin-1 receptor II (IL-1R2), enterokinase, and other substrates
Whether IL-1R2 is a physiological substrate for BACE1 is still in question. It is reported that BACE1 cleaves IL-1R2 but the total level of IL-IR2 is unchanged in BACE1−/− cells [57]. Recently, evidence also suggests that BACE1 cleaves enterokinase and the neural cell adhesion molecules (L1 and CHL1) [58,59]. The broader issue of how many substrates interact with BACE1 is being explored, but research suggests that there are dozens of possibilities. A proteomics study uncovered more than 100 BACE1 candidate substrates [60], which provide us with a wide range to validate in vitro and in vivo using classical biochemistry or cellular biology methods.
Binding proteins
To function properly, BACE1 must bind with other proteins (Table 3). These binding partners are briefly reviewed below.
Table 3.
BACE1-binding partners
| Name | Function | Binding domain | |
|---|---|---|---|
| BACE1 | Partner | ||
| GGA | BACE1 trafficking | ACDL | VHS |
| PLSCR1 | BACE1 distribution | Dileucine motif | Cytoplasmic domain |
| RTN3/4 | Inhibit BACE1 cleavage | Cytoplasmic domain | Cytoplasmic domain |
| LR-II (sorLA) | Switch BACE1 to α/β cleavage | Cytoplasmic domain | Cytoplasmic domain |
| PAR-4 | BACE1 activity positive effector | Cytoplasmic domain | Cytoplasmic domain |
| PS1 | BACE1 maturation | Unknown | Unknown |
| CCS | Reduce SOD1 activation | Cytoplasmic domain | N-terminal domain |
| BRI3 | Unknown | Cytoplasmic domain | Cytoplasmic domain |
| APP | BACE1 endocytosis | Ectodomain | Ectodomain |
| Nicastrin (in vitro) | BACE1 activation | Unknown | Unknown |
| Sortlin | BACE1 retrograde trafficking | Ectodomain | Unknown |
| PrPc | APP processing of BACE1 | Ectodomain | Unknown |
Golgi-localized γ-ear-containing ARF-binding proteins (GGAs) and phospholipid scramblase 1 (PLSCR1)
By binding with the VHS (Vps-27, Hrs, and STAM) domain in GGAs, BACE1 is transported to endosomes or lysosomes. During cell apoptosis, caspase-3 inactivates GGA3, which not only changes the location of BACE1 but also increases its stability and synthesis [61]. PLSCR1, a protein involved in membrane phospholipids remodeling and movement, interacts with dileucine residues in the BACE1 tail and directly affects BACE1 distribution and recruitment to lipid rafts [62].
SorLAs and sortilins
SorLA and sortilin are both members of the Vps10p domain receptor family. SorLA interacts with, possibly inhibits, both BACE1 and APP through the sorLA tail. SorLA significantly reduced secreted Aβ levels when BACE was overexpressed, suggesting that sorLA influences β cleavage, which mediates APP distribution, and dictates APP processing [63,64]. Sortilins bind BACE1 directly and play roles in regulating subcellular distribution and retrograde trafficking of BACE1 [65].
RTNs and prostate apoptosis response-4 (PAR-4)
Reticulon/Nogo proteins (RTN3, RTN4-B/C, or Nogo-B/C) are components of myelin that interact with the cytoplasmic domain of BACE1 via its C-terminal QID sequence [66]. Overexpression of reticulon will increase BACE1 retention in the ER and, in turn, inhibit APP cleavage. Reduced RTN3 in AD brains is associated with increased BACE1 activity [67]. The two hydrophobic transmembrane domains in RTNs are crucial not only for maintaining structure and stability of RTNs but also for modulating enzymatic activity of BACE1 [66]. PAR-4, a leucine zipper protein, is a positive effector of BACE1 activity. The C-terminal of PAR-4 binds with the BACE1 tail and increases APP cleavage in the amyloidogenic pathway [68].
Other partners
Presenilins, the catalytic part of γ-secretase, are crucial in generating Aβ. Mutations in presenilin genes were found to be associated with inherited AD, and loss of presenilin function also induces sporadic AD [69]. Presenilin 1 (PS1) binds with BACE1 directly in vivo, regulates its trafficking, and facilitates generation of mature BACE1 [70]. The interactions between the ectodomain of both APP and BACE1 facilitate BACE1 endocytosis [71]. Cellular prion protein (PrPC), another BACE1 partner, has been shown to bind the pro-domain of BACE1 directly and inhibit BACE1 cleavage of wild type APP [72]. BACE1 also binds brain-specific type II membrane protein through both cytosolic tails [73].
The cysteine residues in the cytoplasmic tail of BACE1 have been shown to bind Cu (I) atom, and interact with the N-terminal domain of CCS [the copper chaperone for superoxide dismutase-1 (SOD1)], which increases oxidative stress in peripheral nerves [74]. Thus, BACE1 appears to be involved in regulating metal homoeostasis and oxidative stress in AD.
Recombinant BACE1 in different expression systems
In E. coli, recombinant pro-BACE1 cannot be cleaved by furins. In insect cells, furin-like enzymes typically cannot cleave recombinant BACE1 at putative furin processing sites, and recombinant BACE1 is always a mixture of pro-and mature enzymes [17].
Glycosylation is important for BACE1 to maintain its proteolytic activity and to allow it to be properly identified by furins because glycosylation has direct effects on substrate binding and protein interactions. Thus, in E. coli, lack of glycosylation of recombinant BACE1 results in significantly lower activity [23,75]. In insect cells, different glycosylation (Mannose-rich glycans) of recombinant BACE1 results in ~50% activity of the enzyme [76]. In mammalian cells, BACE1 glycosylation includes biantennary and triantennary oligosaccharides of the ‘complex’ type, which results in full enzymatic activity in BACE1 [76].
BACE1 transgenic mice
BACE1+/+ mice are viable, fertile, and healthy. They show an increase in 5-hydroxytryptamine (5-HT) turnover, less anxiety and more exploratory behaviors [77]. By contrast, BACE1−/− mice have an increased risk for premature mortality but show no obvious alterations in tissue morphology, hematology, and gross behavioral and neuromuscular parameters [78]; however, they have impaired spatial, reference, and temporal memory [79]. Also, BACE1−/− mice display reduced axonal myelination [80], deficiencies in assays of emotion or schizophrenia-like behavioral symptoms, altered synaptic plasticity, and enhanced locomotion [80]. Zhao et al. compared 5XFAD mice and Tg2576 mice (5XFAD mice form amyloid plaques at younger ages and exhibit neuron loss; Tg2576 mice form plaques at older ages and do not show cell death) and revealed that BACE1 levels in both mice became elevated without any change in BACE1 mRNA level, which means that a post-transcriptional mechanism contributes to the elevation of BACE1 protein levels. The authors also pointed out that high BACE1 levels were observed around Aβ plaque cores and colocalized with neuronal proteins, suggesting that amyloid plaques induce BACE1 elevation at early stages of pathology before neuron death occurs [81].
Biological and physiological features of BACE1
Biosynthesis, subcellular location, and degradation
The BACE1 gene is 30.6 kb in length and shows no AD-related mutations. Alternative splicing in exons 3 and 4 generates four variants with 501, 476, 457, and 432 amino acids, respectively, and the shorter variants have no cleavage activity on APP [82].
In the ER, BACE1 exists as an immature, N-glycosylated pro-protein (~60 kDa). Subsequently, pro-BACE1 is transported to Golgi for further modification. Then, the pro-peptide is removed and the mature enzyme (>16 h half-life) is trafficked to endosomes or the cell surface by GGAs. BACE1 localizes largely within cholesterol-rich lipid rafts where it interacts with substrates and binding partners.
Degradation of BACE1 is mainly through the following three ways: (i) BACE1 catalytic domain endoproteolysis; (ii) degradation though the proteasomal pathway; or (iii) though the lysosomal pathway. The K203 and K382 residues in BACE1 are essential amino acids for degradation by the ubiquitin–proteasome pathway [83] and UCHL1 (ubiquitin carboxyl-terminal hydrolase L1) participates in this process [84].
Factors regulating BACE1 expression and activity
BACE1 expression and its activity are controlled at various levels. At the level of protein translation, the GC-rich region in 5′ untranslated region (5′UTR) of BACE1 suppresses BACE1 translation. In addition, energy deprivation will increase eukaryotic initiation factor-2a phosphorylation, elevate 5′UTR scan-through, and increase BACE1 protein levels [85]. Moreover, miRNA also mediates BACE1 translational control [86].
Various transcription factors also control the BACE1 gene promoter, such as NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) [87], PPAR-γ (proliferator-activated receptor γ), HIF-1 (hypoxia inducible factor-1), and others [88,89]. We found that the tumor necrosis factor type I receptor (TNFRI) can mediate BACE1 through NF-κB activation by binding to NF-κB sites in the BACE1 promoter [90]. Thus, genetic deletion of TNFRI suppresses BACE1 transcriptional activation [90]. PPAR-γ activation suppresses BACE1 promoter activity [91]. In astrocytes, HIF-1 binding with the hypoxia-responsive element in the BACE1 promoter increases BACE1 expression, activates genes associated with energy metabolism, and initiates cell death [89]. Recently, p25 was verified to affect BACE1 promoter activity through STAT3 hyperphosphorylation, and the ratio of p25 to p35, together with CDK5 levels, is also important for BACE1 mRNA and protein levels, and further affects BACE1 activity and neurodegeneration [92].
In addition, sAPPα and γ-secretase also modulate BACE1 expression. By directly associating with BACE1, sAPPα modulates APP processing and decreases Aβ generation [93]. Therefore, restoring sAPPα levels or increasing its association with BACE1 may ameliorate AD pathogenesis [93]. γ-Secretase mediates oxidative stress-induced expression of BACE1 [94]. Jo et al. found that γ-secretase inhibitors could also downregulate oxidative stress-induced β-secretase activity [94], which might be another strategy for AD therapy.
BACE1 activity is also modified by the lipid environment. Glycosphingolipids, glycerophospholipids, sterols, and sphingosine-1-phosphate (S1P) have been shown to stimulate BACE1, and isoprenylation is required for BACE1 dimerization [95–98].
Interestingly, recently hormones have become attractive biological reagents to regulate BACE1 activity. For example, we discovered that estrogen suppresses BACE1 at the transcriptional level [99] and knockout of estrogen synthesizing enzyme, aromatase, resulted in AD-like pathology through decreasing BACE1 transcription [100]. The hypothalamic–pituitary–adrenal axis controls circulating levels of glucocorticoid hormones. The abnormal regulation of hormones in the hypothalamic–pituitary– adrenal axis, i.e., levels of glucocorticoid [101], is related to memory impairments and can be detected in AD brains, suggesting that this hormone may contribute to AD pathology as well as to the cognitive decline in AD. Moreover, using antidiabetic drug metformin (GlucophageR) increases biogenesis of Alzheimer’s amyloid peptides via upregulating BACE1 transcription [102].
Because BACE1 is associated with many regulators and substrates, and its promotor is also validated to be similar to house-keeping genes, BACE1 might act as house-keeping protein to regulate multiple physiological processes, such as cell inflammation, differentiation, development, and cell death. Thus, in a therapeutic point of view, total inhibition of BACE1 activity would directly affect its substrates cleavage and, further, interrupt associated signal pathways, which could induce abnormal behavioral. Therefore, partial inhibition of BACE1 activity might be an alternative approach, although this may be a challenge to achieve.
BACE1 and diseases
AD is characterized by memory impairment accompanied by brain lesions in cognitive areas. One pathological hallmark of AD is the formation of amyloid plaques, generated from APP cleavage (Box 1). As we described earlier, APP is one of the major substrates of BACE1, and its Swedish mutation (KM change to NL at BACE1 cleavage site) [103] makes BACE1 cleavage more efficient. Moreover, BACE1 elevation in sprouting dystrophic axonal terminals coincides with accumulation of amyloid deposition, which then aggravates AD progression [104]. Additional multiple factors contribute to AD pathogenesis, including abnormal gene regulation associated with increased Aβ levels in AD. Compounds such as nonsteroidal anti-inflammatory drugs (NSAIDs) and 2,4-bis (p-hydroxyphenyl)-2-butenal that can target genetic regulators, such as PPAR-γ, NF-κB, and STAT3, may ultimately decrease BACE1 transcription or Aβ production [91,105].
In addition to AD, elevated BACE1 levels are also closely associated with traumatic brain injury (TBI), cerebral hypometabolism, hypoxia, ischemia, cellular stress, and disruptions in energy metabolism [106–108].
Recent investigations also suggest a link between BACE1 activity and schizophrenia, seizure, and epileptic behavior [80,109].
BACE1 as a pharmacological target
As BACE1 initiates Aβ generation, more efforts have been made to develop BACE1 inhibitors for clinical use in AD. Early BACE1 inhibitors were mainly designed to aid in BACE1 purification and structural analysis. After the crystal structure of BACE1 was elucidated, more drug-like compounds were designed, and these achieved better inhibition of Aβ in vivo [110]. Generally, these inhibitors are categorized into peptidic and non-peptide inhibitors. Some of them have demonstrated efficacy in lowering Aβ in AD transgenic mice, but when they were applied in clinical trials, AD patients showed no rescue of cognition [111]. One of the difficulties in developing BACE1 drug-like inhibitors is that, compared with humans, AD transgenic mice have a much faster Aβ plague development and a shorter life span, which results in different responses to these compounds. Because BACE1 has many physiological roles in cleaving substrates in vivo, some other difficulties in developing pharmaceutical inhibitors include their specificity and toxicity. It is critical to consider potential side effects because complete inhibition of BACE1 was reported to result in possible demyelination [40,43] as well as schizophrenia-like symptoms [112].
Several companies are involved in clinical trials of new BACE1 inhibitors, such as Roche (inhibitor RG7129), High Point Pharmaceuticals (inhibitor HPP854), Genetech, Eli Lilly (inhibitors LY2886721 and LY2811376) [113], Eisai (inhibitor E2609), and Merck (MK8931). Although LY2811376, an oral compound, was stopped in its Phase I clinical trial for possible linkage with retinal pigment epithelial defects in rat studies [113,114], Lilly LY2886721 has successfully entered a Phase II clinical trial and has been shown to lower Aβ levels in the brain as well as the plasma [115]. E2609, also an oral compound, has completed the drug safety assessment in Phase I, and will move to Phase II [116]. The compound MK8931 from Merck can lower cerebrospinal fluid (CSF) Aβ40 and Aβ42 down ~90% in a single dosage and 50–80% in multiple dosages, but it is still in Phase I safety evaluations [117]. Not only are these promising chemical compounds, existing drugs for different implications may offer potential therapeutic opportunities. For example, thalidomide, a TNF inhibitor, was reported to ameliorate AD-like pathology through inhibition of BACE1 in a mouse model of AD [118] and this existing drug has been tested in a Phase II clinical trial in AD patients [119]. Another existing drug, nilvadipine, which has been used for antihypertension, was reported to lower Aβ via indirect inhibition of BACE1 [120] and currently it is also being tested in a clinical trial for AD [121]. Thus, we should be patient for several more years for clear and convincing results from these clinical trials of BACE1 inhibitors. Meanwhile, we must also keep our minds open. Herbal medicine may provide such opportunities. For example, an anti-stroke medicine, which has clinical benefits in patients with stroke, was recently demonstrated to reduce AD-like pathology by decreasing abnormal APP processing, including BACE1 reduction [122].
Concluding remarks
In conclusion, BACE1 activity is closely related to AD and other CNS disease pathogenesis. However, the use BACE1 as a potential early biomarker for early diagnosis or drug treatment monitoring requires complete understanding of its function and activities. Although a great deal of information about structure and functions of BACE1 is available, many questions remain to be answered, such as: what are the basic physiological functions of BACE1 in the brain versus the peripheral system? Is it cell-type dependent? There are many factors that can regulate BACE1 and, in return, BACE1 also affects many other gene regulations. What are the causes for sporadic AD and other similar disorders? We know that BACE1 activity is closely associated with AD progression and it plays a key role in the process, and BACE1 itself exhibits many forms in terms of phosphorylated, glycosylated, acetylated forms, etc. Which form(s) is more relevant to AD? And which form(s) is more related to multiple sclerosis (MS) or other brain disorders? Finding answers to these questions would be helpful to understand the basic biology of BACE1 and to identify more valuable therapeutic targets, but also to provide guidance for future clinical trials for BACE1 inhibitors by highlighting potential side effects.
Acknowledgments
This paper is supported by grants from the National Institute on Aging (NIHR01AG032441 and RO1AG025888), the Alzheimer’s Association (Zenith Award, IIRG-07-59510 and IIRG-09-61521), and the American Health Assistance Foundation (G2006-118).
References
- 1.Hussain I, et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 2.Sinha S, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 3.Vassar R, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 4.Yan R, et al. Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 5.Lin X, et al. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci USA. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan R, et al. BACE2 functions as an alternative alpha-secretase in cells. J Biol Chem. 2001;276:34019–34027. doi: 10.1074/jbc.M105583200. [DOI] [PubMed] [Google Scholar]
- 7.Casas S, et al. BACE2 plays a role in the insulin receptor trafficking in pancreatic β-cells. Am J Physiol Endocrinol Metab. 2010;299:E1087–E1095. doi: 10.1152/ajpendo.00420.2010. [DOI] [PubMed] [Google Scholar]
- 8.Holler CJ, et al. BACE2 expression increases in human neurodegenerative disease. Am J Pathol. 2012;180:337–350. doi: 10.1016/j.ajpath.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett BD, et al. Expression analysis of BACE2 in brain and peripheral tissues. J Biol Chem. 2000;275:20647–20651. doi: 10.1074/jbc.M002688200. [DOI] [PubMed] [Google Scholar]
- 10.Huse JT, et al. Maturation and endosomal targeting of β-site amyloid precursor protein-cleaving enzyme. The Alzheimer’s disease β-secretase. J Biol Chem. 2000;275:33729–33737. doi: 10.1074/jbc.M004175200. [DOI] [PubMed] [Google Scholar]
- 11.Haniu M, et al. Characterization of Alzheimer’s β-secretase protein BACE. A pepsin family member with unusual properties. J Biol Chem. 2000;275:21099–21106. doi: 10.1074/jbc.M002095200. [DOI] [PubMed] [Google Scholar]
- 12.Shiba T, et al. Insights into the phosphoregulation of β-secretase sorting signal by the VHS domain of GGA1. Traffic. 2004;5:437–448. doi: 10.1111/j.1600-0854.2004.00188.x. [DOI] [PubMed] [Google Scholar]
- 13.Vetrivel KS, et al. Alzheimer disease Aβ production in the absence of S-palmitoylation-dependent targeting of BACE1 to lipid rafts. J Biol Chem. 2009;284:3793–3803. doi: 10.1074/jbc.M808920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang EL, et al. Ubiquitin regulates GGA3-mediated degradation of BACE1. J Biol Chem. 2010;285:24108–24119. doi: 10.1074/jbc.M109.092742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costantini C, et al. A reversible form of lysine acetylation in the ER and Golgi lumen controls the molecular stabilization of BACE1. Biochem J. 2007;407:383–395. doi: 10.1042/BJ20070040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonas MC, et al. PCSK9 is required for the disposal of non-acetylated intermediates of the nascent membrane protein BACE1. EMBO Rep. 2008;9:916–922. doi: 10.1038/embor.2008.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi XP, et al. The pro domain of beta-secretase does not confer strict zymogen-like properties but does assist proper folding of the protease domain. J Biol Chem. 2001;276:10366–10373. doi: 10.1074/jbc.m009200200. [DOI] [PubMed] [Google Scholar]
- 18.Benjannet S, et al. Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J Biol Chem. 2001;276:10879–10887. doi: 10.1074/jbc.M009899200. [DOI] [PubMed] [Google Scholar]
- 19.Capell A, et al. Maturation and pro-peptide cleavage of beta-9secretase. J Biol Chem. 2000;275:30849–30854. doi: 10.1074/jbc.M003202200. [DOI] [PubMed] [Google Scholar]
- 20.Jin S, et al. Evidence for dimeric BACE-mediated APP processing. Biochem Biophys Res Commun. 2010;393:21–27. doi: 10.1016/j.bbrc.2010.01.064. [DOI] [PubMed] [Google Scholar]
- 21.Westmeyer GG, et al. Dimerization of beta-site beta-amyloid precursor protein-cleaving enzyme. J Biol Chem. 2004;279:53205–53212. doi: 10.1074/jbc.M410378200. [DOI] [PubMed] [Google Scholar]
- 22.Yan R, et al. The transmembrane domain of the Alzheimer’s beta-secretase (BACE1) determines its late Golgi localization and access to beta-amyloid precursor protein (APP) substrate. J Biol Chem. 2001;276:36788–36796. doi: 10.1074/jbc.M104350200. [DOI] [PubMed] [Google Scholar]
- 23.Hong L, et al. Structure of the protease domain of memapsin 2 (beta-secretase) complexed with inhibitor. Science. 2000;290:150–153. doi: 10.1126/science.290.5489.150. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu H, et al. Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production. Mol Cell Biol. 2008;28:3663–3671. doi: 10.1128/MCB.02185-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spronk SA, Carlson HA. The role of tyrosine 71 in modulating the flap conformations of BACE1. Proteins. 2011;79:2247–2259. doi: 10.1002/prot.23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquer C, et al. Local cholesterol increase triggers amyloid precursor protein-Bace1 clustering in lipid rafts and rapid endocytosis. FASEB J. 2011;25:1295–1305. doi: 10.1096/fj.10-168633. [DOI] [PubMed] [Google Scholar]
- 27.Bolkan BJ, et al. β-Secretase cleavage of the fly amyloid precursor protein is required for glial survival. J Neurosci. 2012;32:16181–16192. doi: 10.1523/JNEUROSCI.0228-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, et al. Soluble amyloid precursor protein (APP) regulates transthyretin and Klotho gene expression without rescuing the essential function of APP. Proc Natl Acad Sci USA. 2010;107:17362–17367. doi: 10.1073/pnas.1012568107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chasseigneaux S, et al. Secreted amyloid precursor protein beta and secreted amyloid precursor protein alpha induce axon outgrowth in vitro through Egr1 signaling pathway. PLoS ONE. 2011;6:e16301. doi: 10.1371/journal.pone.0016301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baratchi S, et al. Secreted amyloid precursor proteins promote proliferation and glial differentiation of adult hippocampal neural progenitor cells. Hippocampus. 2011;22:1517–1527. doi: 10.1002/hipo.20988. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa K, et al. Activation of K+ channels and suppression of neuronal activity by secreted beta-amyloid-precursor protein. Nature. 1996;379:74–78. doi: 10.1038/379074a0. [DOI] [PubMed] [Google Scholar]
- 32.Freude KK, et al. Soluble amyloid precursor protein induces rapid neural differentiation of human embryonic stem cells. J Biol Chem. 2011;86:24264–24274. doi: 10.1074/jbc.M111.227421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolaev A, et al. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Belyaev ND, et al. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a β-secretase-dependent pathway. J Biol Chem. 2010;285:41443–41454. doi: 10.1074/jbc.M110.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefort R, et al. Cross-linking of cell surface amyloid precursor protein leads to increased beta-amyloid peptide production in hippocampal neurons: implications for Alzheimer’s disease. J Neurosci. 2012;32:10674–10685. doi: 10.1523/JNEUROSCI.6473-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma H, et al. Involvement of beta-site APP cleaving enzyme 1 (BACE1) in amyloid precursor protein-mediated enhancement of memory and activity-dependent synaptic plasticity. Proc Natl Acad Sci USA. 2007;104:8167–8172. doi: 10.1073/pnas.0609521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura R, et al. Partial reduction of BACE1 improves synaptic plasticity, recent and remote memories in Alzheimer’s disease transgenic mice. J Neurochem. 2010;113:248–261. doi: 10.1111/j.1471-4159.2010.06608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishida A, et al. Secreted form of beta-amyloid precursor protein shifts the frequency dependency for induction of LTD, and enhances LTP in hippocampal slices. Neuroreport. 1997;8:2133–2137. doi: 10.1097/00001756-199707070-00009. [DOI] [PubMed] [Google Scholar]
- 39.Furukawa K, et al. Increased activity-regulating and neuroprotective efficacy of alpha-secretase-derived secreted amyloid precursor protein conferred by a C-terminal heparin-binding domain. J Neurochem. 1996;67:1882–1896. doi: 10.1046/j.1471-4159.1996.67051882.x. [DOI] [PubMed] [Google Scholar]
- 40.Hu X, et al. Genetic deletion of BACE1 in mice affects remyelination of sciatic nerves. FASEB J. 2008;22:2970–2980. doi: 10.1096/fj.08-106666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu X, et al. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- 42.Luo X, et al. Cleavage of neuregulin-1 by BACE1 or ADAM10 protein produces differential effects on myelination. J Biol Chem. 2011;286:23967–23974. doi: 10.1074/jbc.M111.251538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willem M, et al. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- 44.Brackenbury WJ, et al. Functional reciprocity between Na+ channel Nav1.6 and β1 subunits in the coordinated regulation of excitability and neurite outgrowth. Proc Natl Acad Sci USA. 2010;107:2283–2288. doi: 10.1073/pnas.0909434107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gersbacher MT, et al. Identification of BACE1 cleavage sites in human voltage-gated sodium channel beta 2 subunit. Mol Neurodegener. 2010;5:61. doi: 10.1186/1750-1326-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DY, et al. Reduced sodium channel Na(v)1.1 levels in BACE1-null mice. J Biol Chem. 2011;286:8106–8116. doi: 10.1074/jbc.M110.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DY, et al. BACE1 regulates voltage-gated sodium channels and neuronal activity. Nat Cell Biol. 2007;9:755–764. doi: 10.1038/ncb1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huth T, et al. β-Site APP-cleaving enzyme 1 (BACE1) cleaves cerebellar Na+ channel β4-subunit and promotes Purkinje cell firing by slowing the decay of resurgent Na+ current. Pflugers Arch. 2011;461:355–371. doi: 10.1007/s00424-010-0913-2. [DOI] [PubMed] [Google Scholar]
- 49.Huth T, et al. Non-proteolytic effect of β-site APP-cleaving enzyme 1 (BACE1) on sodium channel function. Neurobiol Dis. 2009;33:282–289. doi: 10.1016/j.nbd.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 50.Lichtenthaler SF, et al. The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J Biol Chem. 2003;278:48713–48719. doi: 10.1074/jbc.M303861200. [DOI] [PubMed] [Google Scholar]
- 51.Kitazume S, et al. Alzheimer’s beta-secretase cleaves a glycosyltransferase as a physiological substrate. Glycoconj J. 2004;20:59–62. doi: 10.1023/B:GLYC.0000016743.25495.45. [DOI] [PubMed] [Google Scholar]
- 52.Jones MB, et al. Role for hepatic and circulatory ST6Gal-1 sialyltransferase in regulating myelopoiesis. J Biol Chem. 2010;285:25009–25017. doi: 10.1074/jbc.M110.104406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodard-Grice AV, et al. Proteolytic shedding of ST6Gal-I by BACE1 regulates the glycosylation and function of α4β1 integrins. J Biol Chem. 2008;283:26364–26373. doi: 10.1074/jbc.M800836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang DE, et al. Modulation of amyloid β-protein clearance and Alzheimer’s disease susceptibility by the LDL receptor-related protein pathway. J Clin Invest. 2000;106:1159–1166. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Arnim CA, et al. The low density lipoprotein receptor-related protein (LRP) is a novel β-secretase (BACE1) substrate. J Biol Chem. 2005;280:17777–17785. doi: 10.1074/jbc.M414248200. [DOI] [PubMed] [Google Scholar]
- 56.Kihara T, et al. Aβ-induced BACE-1 cleaves N-terminal sequence of mPGES-2. Biochem Biophys Res Commun. 2010;393:728–733. doi: 10.1016/j.bbrc.2010.02.069. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn PH, et al. Regulated intramembrane proteolysis of the interleukin-1 receptor II by alpha-, beta-, and gamma-secretase. J Biol Chem. 2007;282:11982–11995. doi: 10.1074/jbc.M700356200. [DOI] [PubMed] [Google Scholar]
- 58.Hoffmeister A, et al. BACE1 is a newly discovered protein secreted by the pancreas which cleaves enteropeptidase in vitro. JOP. 2009;10:501–506. [PubMed] [Google Scholar]
- 59.Zhou L, et al. The neural cell adhesion molecules L1 and CHL1 are cleaved by BACE1 in vivo. J Neurochem. 2012;287:25927–25940. doi: 10.1074/jbc.M112.377465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hemming ML, et al. Identification of beta-secretase (BACE1) substrates using quantitative proteomics. PLoS ONE. 2009;4:e8477. doi: 10.1371/journal.pone.0008477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tesco G, et al. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kametaka S, et al. Identification of phospholipid scramblase 1 as a novel interacting molecule with beta-secretase (beta-site amyloid precursor protein (APP) cleaving enzyme (BACE)) J Biol Chem. 2003;278:15239–15245. doi: 10.1074/jbc.M208611200. [DOI] [PubMed] [Google Scholar]
- 63.Marks N, Berg MJ. BACE and gamma-secretase characterization and their sorting as therapeutic targets to reduce amyloidogenesis. Neurochem Res. 2010;35:181–210. doi: 10.1007/s11064-009-0054-1. [DOI] [PubMed] [Google Scholar]
- 64.Spoelgen R, et al. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finan GM, et al. BACE1 retrograde trafficking is uniquely regulated by the cytoplasmic domain of sortilin. J Biol Chem. 2011;286:12602–12616. doi: 10.1074/jbc.M110.170217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He W, et al. Reticulon family members modulate BACE1 activity and amyloid-beta peptide generation. Nat Med. 2004;10:959–965. doi: 10.1038/nm1088. [DOI] [PubMed] [Google Scholar]
- 67.Hu X, et al. Transgenic mice overexpressing reticulon 3 develop neuritic abnormalities. EMBO J. 2007;26:2755–2767. doi: 10.1038/sj.emboj.7601707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie J, Guo Q. PAR-4 is involved in regulation of beta-secretase cleavage of the Alzheimer amyloid precursor protein. J Biol Chem. 2005;280:13824–13832. doi: 10.1074/jbc.M411933200. [DOI] [PubMed] [Google Scholar]
- 69.Supnet C, Bezprozvanny I. Presenilins function in ER calcium leak and Alzheimer’s disease pathogenesis. Cell Calcium. 2011;50:303–309. doi: 10.1016/j.ceca.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuzuya A, et al. Presenilin 1 is involved in the maturation of beta-site amyloid precursor protein-cleaving enzyme 1 (BACE1) J Neurosci Res. 2007;85:153–165. doi: 10.1002/jnr.21104. [DOI] [PubMed] [Google Scholar]
- 71.Huang XP, et al. Internalization of exogenously added memapsin 2 (beta-secretase) ectodomain by cells is mediated by amyloid precursor protein. J Biol Chem. 2004;279:37886–37894. doi: 10.1074/jbc.M402130200. [DOI] [PubMed] [Google Scholar]
- 72.Griffiths HH, et al. Prion protein interacts with BACE1 protein and differentially regulates its activity toward wild type and Swedish mutant amyloid precursor protein. J Biol Chem. 2011;286:33489–33500. doi: 10.1074/jbc.M111.278556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wickham L, et al. Beta-amyloid protein converting enzyme 1 and brain-specific type II membrane protein BRI3: binding partners processed by furin. J Neurochem. 2005;92:93–102. doi: 10.1111/j.1471-4159.2004.02840.x. [DOI] [PubMed] [Google Scholar]
- 74.Fischer LR, et al. Absence of SOD1 leads to oxidative stress in peripheral nerve and causes a progressive distal motor axonopathy. Exp Neurol. 2012;233:163–171. doi: 10.1016/j.expneurol.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ermolieff J, et al. Proteolytic activation of recombinant pro-memapsin 2 (pro-beta-secretase) studied with new fluorogenic substrates. Biochemistry. 2000;39:16263. doi: 10.1021/bi005122i. [DOI] [PubMed] [Google Scholar]
- 76.Charlwood J, et al. Characterization of the glycosylation profiles of Alzheimer’s beta-secretase protein Asp-2 expressed in a variety of cell lines. J Biol Chem. 2001;276:16739–16748. doi: 10.1074/jbc.M009361200. [DOI] [PubMed] [Google Scholar]
- 77.Harrison SM, et al. BACE1 (beta-secretase) transgenic and knockout mice: identification of neurochemical deficits and behavioral changes. Mol Cell Neurosci. 2003;24:646–655. doi: 10.1016/s1044-7431(03)00227-6. [DOI] [PubMed] [Google Scholar]
- 78.Luo Y, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- 79.Ohno M, et al. Temporal memory deficits in Alzheimer’s mouse models: rescue by genetic deletion of BACE1. Eur J Neurosci. 2006;23:251–260. doi: 10.1111/j.1460-9568.2005.04551.x. [DOI] [PubMed] [Google Scholar]
- 80.Hu X, et al. BACE1 deficiency causes altered neuronal activity and neurodegeneration. J Neurosci. 2010;30:8819–8829. doi: 10.1523/JNEUROSCI.1334-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao J, et al. Beta-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer’s disease pathogenesis. J Neurosci. 2007;27:3639–3649. doi: 10.1523/JNEUROSCI.4396-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mowrer KR, Wolfe MS. Promotion of BACE1 mRNA alternative splicing reduces amyloid beta-peptide production. J Biol Chem. 2008;283:18694–18701. doi: 10.1074/jbc.M801322200. [DOI] [PubMed] [Google Scholar]
- 83.Wang R, et al. Lys203 and Lys382 are essential for the proteasomal degradation of BACE1. Curr Alzheimer Res. 2012;9:606–615. doi: 10.2174/156720512800618026. [DOI] [PubMed] [Google Scholar]
- 84.Zhang M, et al. Control of BACE1 degradation and APP processing by ubiquitin carboxyl-terminal hydrolase L1. J Neurochem. 2012;120:1129–1138. doi: 10.1111/j.1471-4159.2011.07644.x. [DOI] [PubMed] [Google Scholar]
- 85.O’Connor T, et al. Phosphorylation of the translation initiation factor eIF2α increases BACE1 levels and promotes amyloidogenesis. Neuron. 2008;60:988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hebert SS, et al. MicroRNA regulation of Alzheimer’s amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 87.Sambamurti K, et al. Gene structure and organization of the human beta-secretase (BACE) promoter. FASEB J. 2004;18:1034–1036. doi: 10.1096/fj.03-1378fje. [DOI] [PubMed] [Google Scholar]
- 88.Sun X, et al. Hypoxia facilitates Alzheimer’s disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci USA. 2006;103:18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X, et al. Hypoxia-inducible factor 1alpha (HIF-1alpha)-mediated hypoxia increases BACE1 expression and beta-amyloid generation. J Biol Chem. 2007;282:10873–10880. doi: 10.1074/jbc.M608856200. [DOI] [PubMed] [Google Scholar]
- 90.He P, et al. Deletion of tumor necrosis factor death receptor inhibits amyloid beta generation and prevents learning and memory deficits in Alzheimer’s mice. J Cell Biol. 2007;178:829–841. doi: 10.1083/jcb.200705042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sastre M, et al. Nonsteroidal anti-inflammatory drugs repress beta-secretase gene promoter activity by the activation of PPARγ. Proc Natl Acad Sci USA. 2006;103:443–448. doi: 10.1073/pnas.0503839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sadleir KR, Vassar R. Cdk5 protein inhibition and Aβ42 increase BACE1 protein level in primary neurons by a post-transcriptional mechanism: implications of CDK5 as a therapeutic target for Alzheimer disease. J Biol Chem. 2012;287:7224–7235. doi: 10.1074/jbc.M111.333914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Obregon D, et al. Soluble amyloid precursor protein-α modulates β-secretase activity and amyloid-β generation. Nat Commun. 2012;3:777. doi: 10.1038/ncomms1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jo DG, et al. Evidence that gamma-secretase mediates oxidative stress-induced beta-secretase expression in Alzheimer’s disease. Neurobiol Aging. 2010;31:917–925. doi: 10.1016/j.neurobiolaging.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takasugi N, et al. BACE1 activity is modulated by cell-associated sphingosine-1-phosphate. J Neurosci. 2011;31:6850–6857. doi: 10.1523/JNEUROSCI.6467-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kalvodova L, et al. Lipids as modulators of proteolytic activity of BACE: involvement of cholesterol, glycosphingolipids, and anionic phospholipids in vitro. J Biol Chem. 2005;280:36815–36823. doi: 10.1074/jbc.M504484200. [DOI] [PubMed] [Google Scholar]
- 97.Cole SL, et al. Statins cause intracellular accumulation of amyloid precursor protein, beta-secretase-cleaved fragments, and amyloid beta-peptide via an isoprenoid-dependent mechanism. J Biol Chem. 2005;280:18755–18770. doi: 10.1074/jbc.M413895200. [DOI] [PubMed] [Google Scholar]
- 98.Parsons RB, et al. Statins inhibit the dimerization of beta-secretase via both isoprenoid- and cholesterol-mediated mechanisms. Biochem J. 2006;399:205–214. doi: 10.1042/BJ20060655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McAllister C, et al. Genetic targeting aromatase in male amyloid precursor protein transgenic mice down-regulates beta-secretase (BACE1) and prevents Alzheimer-like pathology and cognitive impairment. J Neurosci. 2010;30:7326–7334. doi: 10.1523/JNEUROSCI.1180-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yue X, et al. Brain estrogen deficiency accelerates Aβ plaque formation in an Alzheimer’s disease animal model. Proc Natl Acad Sci USA. 2005;102:19198–19203. doi: 10.1073/pnas.0505203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Green KN, et al. Glucocorticoids increase amyloid-beta and tau pathology in a mouse model of Alzheimer’s disease. J Neurosci. 2006;26:9047–9056. doi: 10.1523/JNEUROSCI.2797-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Y, et al. Antidiabetic drug metformin (GlucophageR) increases biogenesis of Alzheimer’s amyloid peptides via up-regulating BACE1 transcription. Proc Natl Acad Sci USA. 2009;106:3907–3912. doi: 10.1073/pnas.0807991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mullan M, et al. Pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1:345–347. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- 104.Zhang XM, et al. Beta-secretase-1 elevation in transgenic mouse models of Alzheimer’s disease is associated with synaptic/axonal pathology and amyloidogenesis: implications for neuritic plaque development. Eur J Neurosci. 2009;30:2271–2283. doi: 10.1111/j.1460-9568.2009.07017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee YJ, et al. Inhibitory effect of a tyrosine-fructose Maillard reaction product, 2,4-bis(p-hydroxyphenyl)-2-butenal on amyloid-beta generation and inflammatory reactions via inhibition of NF-kappaB and STAT3 activation in cultured astrocytes and microglial BV-2 cells. J Neuroinflammation. 2011;8:132. doi: 10.1186/1742-2094-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Walker KR, et al. Depletion of GGA1 and GGA3 mediates postinjury elevation of BACE1. J Neurosci. 2012;32:10423–10437. doi: 10.1523/JNEUROSCI.5491-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guglielmotto M, et al. The up-regulation of BACE1 mediated by hypoxia and ischemic injury: role of oxidative stress and HIF1α. J Neurochem. 2009;108:1045–1056. doi: 10.1111/j.1471-4159.2008.05858.x. [DOI] [PubMed] [Google Scholar]
- 108.Kwak YD, et al. Differential regulation of BACE1 expression by oxidative and nitrosative signals. Mol Neurodegener. 2011;6:17. doi: 10.1186/1750-1326-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hitt BD, et al. BACE1−/− mice exhibit seizure activity that does not correlate with sodium channel level or axonal localization. Mol Neurodegener. 2010;5:31. doi: 10.1186/1750-1326-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mancini F, et al. Beta-secretase as a target for Alzheimer’s disease drug discovery: an overview of in vitro methods for characterization of inhibitors. Anal Bioanal Chem. 2011;400:1979–1996. doi: 10.1007/s00216-011-4963-x. [DOI] [PubMed] [Google Scholar]
- 111.Chang W, et al. β-Secretase inhibitor GRL-8234 rescues age-related cognitive decline in APP transgenic mice. FASEBJ. 2011;25:775–784. doi: 10.1096/fj.10-167213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Savonenko AV, et al. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc Natl Acad Sci USA. 2008;105:5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Citron MP, et al. Central inhibition of Alzheimer’s β-secretase in human: proof of concept with LY2811376. 10th AD/PD International Conference Barcelona, Program SYMPOSIUM33: BETA- ALPHA- SECRETASES; 2011. p. 43. [Google Scholar]
- 114.May PC, et al. Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor. J Neurosci. 2011;31:16507–16516. doi: 10.1523/JNEUROSCI.3647-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.May PC, et al. Preclinical to clinical translation of APP biomarker responses to LY2886721, a potent, oral BACE1 inhibitor in Phase II development for Alzheimer’s disease. 2012 Neuroscience Meeting, Nanosymposium, Society for Neuroscience, Poster 542.08/E34; 2012. p. 334. [Google Scholar]
- 116.Fukushima T, Lucas F. Novel BACE1 inhibitor, E2609 (P1-336), lowers Aβ levels in the CSF and plasma in nonhuman primates. Alzheimer’s Association International Conference.2012. [Google Scholar]
- 117.Forman MS, et al. The novel BACE inhibitor MK8931 dramatically lowers CSF Ab peptides in healthy subjects: results from a rising single dose study. 2012 Annual Meeting of the American Academy of Neurology, Clinical Trials Forum.2012. [Google Scholar]
- 118.He P, et al. Long-term treatment of thalidomide ameliorates Alzheimer-like pathology through inhibition of β-secretase in a mouse model of Alzheimer’s disease. PLoS ONE. 2013;8:e55091. doi: 10.1371/journal.pone.0055091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sabbagh MN, et al. Rationale and strategy for thalidomide as a BACE1 inhibitor for a Phase II randomized clinical trial in mild to moderate Alzheimer’s disease. J Alzheimers Dis. 2010;19:621–630. [Google Scholar]
- 120.Paris D, et al. Selective antihypertensive dihydropyridines lower Aβ accumulation by targeting both the production and the clearance of Aβ across the blood–brain barrier. Mol Med. 2011;17:149–162. doi: 10.2119/molmed.2010.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kennelly SP, et al. Demonstration of safety in Alzheimer’s patients for intervention with an anti-hypertensive drug Nilvadipine: results from a 6-week open label study. Int J Geriatr Psychiatry. 2011;6:1038–1045. doi: 10.1002/gps.2638. [DOI] [PubMed] [Google Scholar]
- 122.He P, et al. Chronic administration of anti-stroke herbal medicine TongLuoJiuNao reduces abnormal processing of amyloid precursor protein in a mouse model of Alzheimer’s disease. PLoS ONE. doi: 10.1371/journal.pone.0058181. http://dx.doi.org/10.1371/journal.pone.0058181. [DOI] [PMC free article] [PubMed]