Abstract
Cells of the dorsomedial/lateral hypothalamus (DMH/LH) that produce hypocretin (HCRT) promote arousal in part by activation of cells of the locus coeruleus (LC) which express tyrosine hydroxylase (TH). The suprachiasmatic nucleus (SCN) drives endogenous daily rhythms, including those of sleep and wakefulness. These circadian oscillations are generated by a transcriptional-translational feedback loop in which the Period (Per) genes constitute critical components. This cell-autonomous molecular clock operates not only within the SCN but also in neurons of other brain regions. However, the phenotype of such neurons and the nature of the phase controlling signal from the pacemaker are largely unknown. We used dual fluorescent in situ hybridization to assess clock function in vasopressin, HCRT and TH cells of the SCN, DMH/LH and LC, respectively, of male Syrian hamsters. In the first experiment, we found that Per1 expression in HCRT and TH oscillated in animals held in constant darkness with a peak phase that lagged that in AVP cells of the SCN by several hours. In the second experiment, hamsters induced to split their locomotor rhythms by exposure to constant light had asymmetric Per1 expression within cells of the middle SCN at 6 h before activity onset (AO) and in HCRT cells 9 h before and at AO. We did not observe evidence of lateralization of Per1 expression in the LC. We conclude that the SCN communicates circadian phase to HCRT cells via lateralized neural projections, and suggests that Per1 expression in the LC may be regulated by signals of a global or bilateral nature.
Introduction
Temporal coordination of mammalian physiology and behavior is maintained by the suprachiasmatic nucleus of the hypothalamus (SCN; [1]). Neurons of the SCN contain a molecular clock whose intrinsic transcriptional –translational loop operates with a ∼24-hour period. Circadian phase is marked by the expression of canonical clock genes whose transcription is sensitive to environmental cues, among which light is the principal zeitgeber. The endogenous pacemaker is thus entrained by the periodic environment and in turn regulates the phase of subordinate oscillators elsewhere in the brain and in the peripheral organs 2–13.
Among functions regulated by the SCN, the sleep-wake cycle is particularly prominent and biologically significant [14]. Hypocretins produced exclusively in the perifornical dorsomedial hypothalamus act to promote wakefulness not only through ascending projections, but also by exciting arousal-promoting midbrain regions, including the locus coeruleus [15]–[20]. Hypocretin signaling 21,22 as well as an intact SCN [14] are necessary for normal consolidation and temporal patterning of sleep. Activation of HCRT-expressing cells is modulated by the SCN [23]–[25]. Although the phenotype of some extra-SCN neuronal oscillators has been established [26]–[32], the expression of clock genes in neurons that regulate sleep and wakefulness is not established and their dependence upon the SCN pacemaker is unclear. Our first experiment was designed to determine whether clock genes are expressed in a circadian pattern within two arousal-promoting cell types, the hypocretin cells of the lateral/dorsomedial hypothalamus (LH/DMH) and the tyrosine hydroxylase (TH)-expressing cells of the locus coeruleus (LC).
Our results from free running hamsters established rhythmicity of clock gene expression in HCRT and TH expressing cells of the LH/DMH and LC, respectively. These findings raised the question of whether rhythmicity in these neurons was regulated by the SCN, and if so whether lateralized or diffusible signals might contribute. In order to address this question, we took advantage of the finding that upon prolonged exposure to constant light, most Syrian hamsters adopt a bimodal activity pattern [33]. Remarkably, clock gene transcription and neuronal activation of the left and right SCN enter an antiphase relationship in these circumstances [34]. This split preparation has proven useful in establishing the role of lateralized pacemaker projections in physiologically relevant circuits as opposed to diffusible humoral signals [35], [36]. We used behaviorally split Syrian hamsters to determine whether lateralized neuronal timing signals from the SCN set the phase of clock gene expression within HCRT neurons of the LH/DMH and TH-expressing neurons of the LC. If descending projections of the SCN control the function of wake-promoting regions, we expect to find asymmetry of Per1 expression in hypocretin or TH cells in the LH/DMH or LC respectively.
Methods
Animals and their Treatment
For experiment 1, adult male Syrian hamsters (Mesocricetus auratus, LVG strain) born and raised in 14 L: 10D were allowed ad libitum access to food and water throughout the experiment. All procedures were approved by the University of Massachusetts Institutional Animal Care and Use Committee (Assurance Number: A3551-01). As young adults, hamsters were transferred to constant darkness for 10 days. Activity in a running wheel (16.5 cm diameter) was monitored by computer (ClockLab software, Actimetrics, Evanston, IL). A least-squares regression line was fit to the activity onsets in order to estimate period and establish the time of onset of subjective night (CT12). Hamsters were killed at CT 3, 9, 12.5 or 22 (n = 5 per phase). Figure S1A depicts a representative actogram of an animal killed at CT12.5. The brains of these animals were removed immediately after decapitation under dim red light (<1 lux), and were rapidly frozen and stored at −80°C.
For experiment 2, Syrian hamsters were transferred as young adults (approximately 10 weeks of age) to a cage with access to a running wheel and maintained in LL (white fluorescent light; ∼200 lux at cage level) for 15±4 weeks. In addition, Chi square periodograms were analyzed to verify split locomotor patterns. Hamsters were classified as split if two activity bouts occurred each 24-hour cycle at intervals of approximately 12 h. A stable split was achieved approximately 8 weeks from transition into LL (range of 4 to 13 weeks; see figure S1B and [37]). The morning and evening activity bouts were designated as described by Pittendrigh and Daan [33]. Briefly, it is possible to identify which of the bouts can be extrapolated to the phase of activity onset (AO) before the split occurs, and this evening activity bout was used to predict the kill times. Twenty-six animals were killed 7 weeks after a stable split was established. Groups of 5 hamsters were rapidly decapitated at 0, 3, 6 (n = 5 each), or 9 (n = 6) hours before AO. Another five hamsters that displayed a single activity bout per 24 hours (non-split controls) were killed at 3 h before AO. The brains of these animals were collected and stored as described above. Two sets of DMH sections were not used in quantification due to loss during assay (n = 5 for 9 h, 6 h, 0 h before AO and n = 4 for 3 h before AO).
In situ Hybridization
Brains collected in both experiments were equilibrated to −20°C, notched on one side and sectioned in the coronal plane on a cryostat at a thickness of 20 µm before processing for in situ hybridization in order to determine levels of Per1 expression within vasopressin (AVP) cells of the SCN, HCRT cells of the LH/DMH, and tyrosine hydroxylase (TH) cells of the LC.
Probe Preparation. In order to prepare template for HCRT probes, the following primers were used to amplify a 0.29 kb RNA transcript isolated from Syrian hamster whole brain: FWD-5′-CAGCCTCTGCCCGACTGCTGTCGCCAGAAG-3′ and REV-5′-GACTCCGGAGCCTCCCCGGGGTGCTAAAGC-3′. The PCR product was gel purified (QiaxeII Agarose Gel Extraction kit), ligated into pGEM-T Easy Vector (Promega, Madison, WI) and transformed into XL 10-Gold Ultracompetent Cells (Stratagene). Transformed colonies were grown and plasmids were isolated and sequenced (Genewiz, Inc, South Plainfield, NJ) to confirm template sequence for production of riboprobes. In situ hybridization performed using 35S-labeled probe confirmed the expected anatomical distribution of HCRT expression in Syrian hamster brain (figure S2).
pBluescript II SK+ plasmid containing the hamster period 1 (haper1) cDNA sequence (AF249882 nt 215–1336, homologous to AF022992 nt 337–1120, ∼730 bp), was linearized with HindIII, for antisense, or SmaI, for sense probes. The antisense probe was transcribed with T3 polymerase and sense probe with T7 polymerase in presence of digoxigenin-11-UTP (Roche Diagnostics). Probes were isolated using ProbeQuant-50 spin column (GEHealthCare Bio-Sciences Corp) and checked on a 1% agarose gel prior to hybridization.
In order to permit colocalization with Per1, transcription of the remaining probes was carried out in the presence of fluorescein-12-UTP (Roche Diagnostics). For AVP, pGEM3 plasmid containing exon C of rat vasopressin cDNA (rAVP; [38] originally from Sherman Thomas G. University of Pittsburgh) was linearized with EcoR1 or HindIII for antisense and sense probe transcription, respectively. SP6 and T7 polymerase were used to transcribe antisense and sense probes (∼293 nt), respectively. For hypocretin, pGEM-T Easy plasmid containing the hamster hypocretin (HCRT) sequence (homologous to Mus musculus NM_010410.2, nt 187-480) was linearized with Apa1 for antisense or Spe-1 for sense probe preparation. SP6 and T7 polymerase were used to transcribe antisense and sense probes, respectively. For tyrosine hydroxylase, rat tyrosine hydroxylase (rTH; NM_012740; nt 14-1165; [39], [40] originally obtained from Donna M. Chikaraishi, Duke University School of Medicine) cDNA in pBS(-) was linearized with Pvu1 or HindIII for antisense and sense probe transcription, respectively. T3 polymerase or T7 polymerase was used to transcribe antisense and sense probes, respectively.
Hybridization
The non-isotopic dual label in situ hybridization method used in this experiment was adapted from Watakabe et al. [41] and was carried out on slide mounted prehybridized brain sections. Slides were presoaked in hybridization buffer (final concentration 5xSSC, 50% Formamide, 5xDenhardt’s, 250ug/mL yeast tRNA, 500ug/mL salmon sperm DNA) for 2 hours at room temperature. Probes (400 ng/mL hybridization buffer) were heated to 80°C for 5 minutes and quenched on ice. Excess buffer was wicked off and probe was added (90uL) before the tissue was cover slipped (Hybri-slips, Sigma-Aldrich, St Louis, MO). Slides were incubated at 55°C for 36 hours.
Post hybridization
Coverslips were floated off in pre-warmed 5xSSC. Slides were washed in 0.2xSSC three times at 55°C, and washed in 1X Tris HCl (pH 7.5) at room temperature for 5 minutes. Tissue was incubated in 1% blocking buffer (PerkinElmer) for 1 hour at room temperature. Blocking reagent was blotted off and 230uL of anti-fluorescein-POD (1∶2000; Roche) overnight at 4°C. Slides were washed 3x in TNT (Tris-NaCl pH 7.5+0.05% Tween 20) for 10 minutes each. Tyramide Signal Amplification-plus DNP amplification (PerkinElmer) was carried out according to the suppliers’ instructions 1x 10 minutes, followed by 3 washes in TNT for 5 minutes each. Slides were incubated with 1∶2000 anti-DIG-AP (Roche) and 1∶2500 anti-DNP-Alexa488 (Roche) for 48 hours at 4°C. Slides were washed 3x in TNT for 10 minutes each at room temperature and washed once in TRIS pH 8.0 for 5 minutes at room temperature. HNPP/FR (1∶100) development was carried out 3 times at room temperature for 20 minutes each. The reaction was stopped in PBS-EDTA (2 washes at 5 minutes each). Tissue was cover slipped using CC mount (Sigma Aldrich).
Image Acquisition
Images were captured on a Zeiss 510 microscope using FITC/Rhodamine selection (Band Pass 505–530 nm and Long Pass 560 nm). The sections were excited with an Argon and Helium-neon laser using the excitation wavelengths of 488 nm (for the Alexafluor488 fluorophore) and 543 nm (for HNPP/FR). To prevent cross-talk of the two fluorophores, images were collected using multitrack optical sections. On average, ten-1 µm thick optical slices were taken. Images were saved as layered TIFF files (.lsm). The dispersed population of the HCRT field required, on average, three images (300×300um in dimension) to capture the medial, perifornical and lateral segments (similar to [42]). Standard fluorescent beads (Molecular Probes) were imaged during each imaging session. Quantification of cell number, Per1 intensity and colocalization was conducted using Image J (NIH software). The extent of colocalization was quantified using the Manders’ coefficient [43] within the region of interest. Colocalization within digital images is defined as the amount of overlap of two different channels within a multichannel image. To remove bias, many algorithms have been developed to quantify this overlap. The Manders’ coefficient describes the contribution of each channel to the overlap with minimal influence of fluorophore intensity [43]. A Manders’ coefficient of 0.5 or greater objectively confirms colocalization and represents 50% or more overlap of the two channels. Mean intensity within each region of interest was measured. Intensity values were normalized to the standard curve generated from the relative intensity levels of the standard fluorescent beads (Molecular Probes). In order to assess the distribution of intensities of Per1 expression within HCRT or TH cells in experiment one, the range of intensities was calculated for all sections and for each circadian phase the percent of cells falling in the highest quartile (values greater than quartile 3), moderate (values falling within quartile 2 and 3) and low (values less than quartile 2) Per1 intensity was determined. For experiment 2, within animal quartiles were determined to evaluate asymmetry in distribution of Per1 intensity between sides of the brain.
Statistical Analysis
For experiment 1, cell counts within each tissue quantified and mean Per1 intensity levels were evaluated across phases (n = 5 per phase except 3 h before AO n = 4) by the Kruskal-Wallis non-parametric ANOVA test. When overall statistical significance was observed (p<0.05), pairwise comparisons were carried out using the Mann-Whitney U-test. Similar statistical analysis was conducted by region (medial, perifornical and lateral) of the HCRT field [42]. The distribution of intensities of Per1 signal within these cell populations provides information regarding the pattern of clock gene expression within and between functionally interconnected but disparate nuclei and their subfields. To this end, the intensity of Per1 signal above threshold was assessed for all cells in each area for all animals in each experiment. Next, percentages of cells falling within the highest 25%, middle 50% or lowest 25% Per1 intensity bins were evaluated for AVP, HCRT, and TH cells of the SCN, DMH/LH and LC respectively at each circadian phase, and differences in this distribution between phases for each cell type were evaluated by nonparametric tests as described above. Within animal assessment of Per1 expression symmetry was carried out at each brain region of interest, including cell counts and quartile distribution (see next section for description). This provided a reference point for symmetry in DD housed hamsters, which aided in interpretation of the data from the next experiment.
For experiment 2, evaluation of Per1 expression was carried out in order to assess lateralization between the hemispheres of each brain region. Cell counts and intensity of Per1 expression were evaluated with Mann-Whitney U-test between the high Per1 expressing side (High side) versus the low Per1 expressing side (Low side). As a measure of asymmetry, the mean intensity of Per1 expression on the high side was divided by the mean Per1 intensity on the low side and compared to the control group using the Mann-Whitney U-test. The within animal distribution of Per1 intensity was assessed in two ways. First, the Kolmogorov-Smirnov test was used to determine whether the Per1 intensity distribution among HCRT cells differed between sides within each animal. Second, the percent of HCRT cells expressing a Per1 signal that fell in the high, moderate and low bin was determined within each animal and the group mean percentages were compared between the high and low sides using the Mann-Whitney U-test.
In order to determine whether lateralization of Per1 expression in the LH/DMH and LC was systematically related to asymmetry in the pacemaker, we assessed whether our quantification of the intensity of clock gene mRNA ipsilateral vs. that contralateral differed systematically from that in AVP cells of the SCN of the same animal. If a lateralized timing signal originating in the neurons of the SCN maintains the phase angle of Per1 expression within the arousal-promoting cells, then a consistent anatomical relationship is expected when assessing the ipsilateral versus contralateral sides of each brain region even though the circadian phase of peak asymmetry may differ between the SCN and the LH/DMH or LC. Spearman’s correlation was calculated in order to determine whether there was a phase-specific relationship between the level of asymmetry observed in the SCN and HCRT cells. Spearman’s correlations were also used to assess the relationship between characteristics of locomotor activity (volume, duration and intensity of the wheel revolutions) and the degree of asymmetry between HCRT fields.
Results
Experiment 1: Per1 Expression in Neuronal Cell types of free Running Hamsters
AVP cells of the SCN
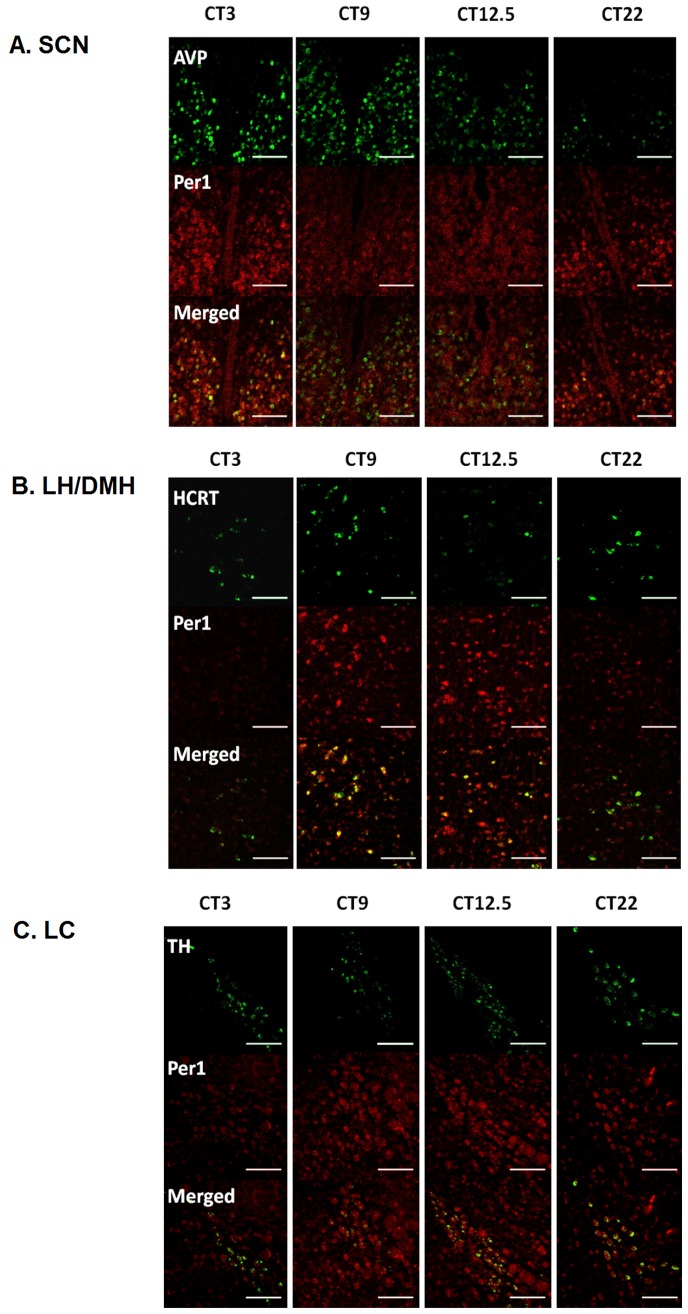
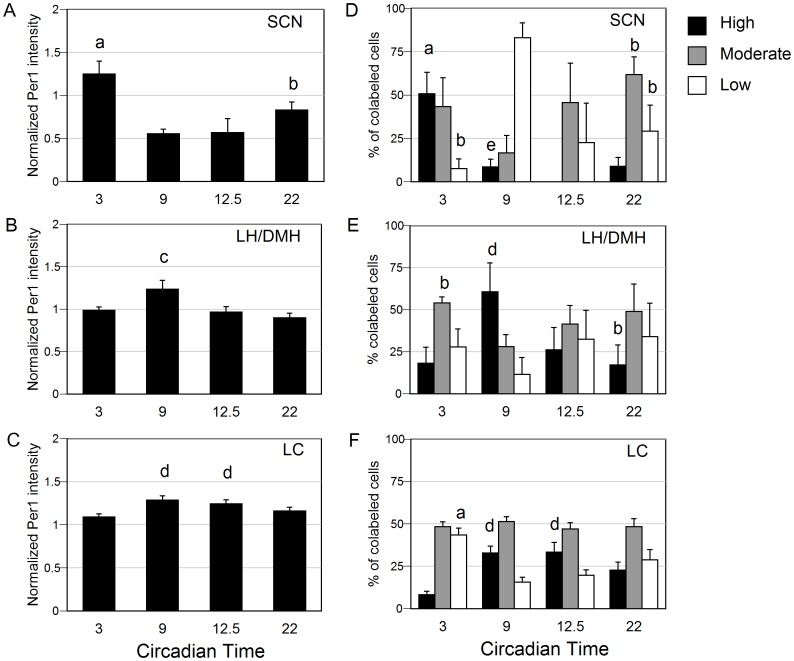
Representative images of the middle plane along the rostral-caudal axis of the SCN are presented in figure 1A. As expected, Per1 expression varied significantly with circadian time. The total number of AVP cells counted and the number of AVP cells that colabeled with Per1 were both two-fold higher at CT3 than at CT22 (p<0.05, table S1). The number of colabeled AVP and Per1 cells as well as the percentage of co-labeled AVP cells were higher at CT3 than at CT12 (p<0.05; table S1). Of the AVP cells colabeled with Per1, the normalized Per1 intensity level at CT3 was significantly greater than at all other phases (p<0.5; figure 2A). The range of Per1 signal intensities of all cells in the experiment was divided into three parts, and the distribution of high, moderate and low expressing cells was quantified for each circadian phase as a percentage of cells counted. The percentage of Per1/AVP colabeled cells that expressed high levels of Per1 was greater at CT3 than at each of the other phases (p<0.05; figure 2D).
Figure 1. Representative confocal images taken at the level of the (A) SCN, (B) perifornical LH/DMH, (C) LC.
Within each brain region, top row shows cells detected in the Alexa488 channel, middle row shows the HNPP/FR signal indicating Per1 and bottom row is the merged image. A single ∼1micron thick section of a 10 section z-stack was selected as a representative image. Scale bar equals 100 microns. Within each brain region, a section taken from representative animals killed at CT3, CT9, CT12.5 or CT22 is shown.
Figure 2. Assessment of Per1 expression in experiment 1.
Left panels show mean colocalization of Per1, and right panels indicate distribution of Per1 expression intensities at each circadian phase, for AVP, HCRT, and TH cells in the brain areas examined. Normalized mean (± SEM) intensity values of Per1 expression is depicted within (A) AVP-expressing cells of the SCN, (B) HCRT-expressing cells of the LH/DMH and (C) TH-expressing cells of the LC. Percent of cells expressing highest 25% (black), moderate (grey), or lowest 25% (white) levels of Per1 within (D) AVP-expressing cells of the SCN, (E) HCRT-expressing cells of the LH/DMH and (F) TH-expressing cells of the LC. a- p<0.05 vs. CT9, 12.5, 22; b- p<0.05 vs. CT9; c- p<0.05 vs. CT3, 12.5; d- p<0.05 vs. CT3; e- p<0.05 vs. 12.5.
We assessed the symmetry of Per1 expression in the middle plane of the SCN of hamsters maintained in DD. Of the colabeled cells, the Per1 intensity level was similar between the sides of the SCN and the high to low ratio of Per1 expression did not differ between phases (figure S4A and S4D).
HCRT cells of the LH/DMH
Representative images of the perifornical HCRT field from animals killed at CT3, 9, 12.5 or 22 are included figure 1B. Of the phases sampled, Per1 expression was higher at CT9 than at CT3 and CT12.5 (P<0.05; figure 2B). A similar number and percentage of HCRT cells met the Manders’ coefficient (m≥0.5) of colocalization across the phases sampled (table S1). In order to assess regional distribution of Per1 expression within the hypocretin field the medial, perifornical and lateral regions were assessed separately. A similar expression pattern of Per1 was observed in the medial and perifornical regions. In contrast, Per1 intensity did not differ with phase in the lateral region (p>0.05; figure S3). Per1 intensity within HCRT fields was similar on both sides of the brain of DD hamsters, so that the high to low ratio was close to 1.0 at all phases analyzed (figure S4B and S4E). The percentage of Per1/HCRT colabeled cells that expressed high levels of Per1 was greater at CT9 than CT3 (P<0.05; figure 2E).
TH Cells of the LC
Representative images of the rostral LC are shown for each phase sampled (figure 1C). A similar number of TH-expressing cells and percentage of TH-expressing cells showed Per1 signal above background at each circadian time (table S1). Nevertheless, the mean level of Per1 expression in TH cells was higher at CT9 and CT12.5 than at CT3 (p<0.05, figure 2C). Per1 labeling in TH cells was bilaterally symmetrical at each phase (figure S4C and S4F). The percentage of Per1/TH double-labeled cells that expressed high levels of Per1 was greater at CT9 and CT12.5 than CT3 (P<0.05; figure 2F).
Experiment 2: Per1 Expression in Neuronal Cell types of Split Hamsters
AVP cells of the SCN
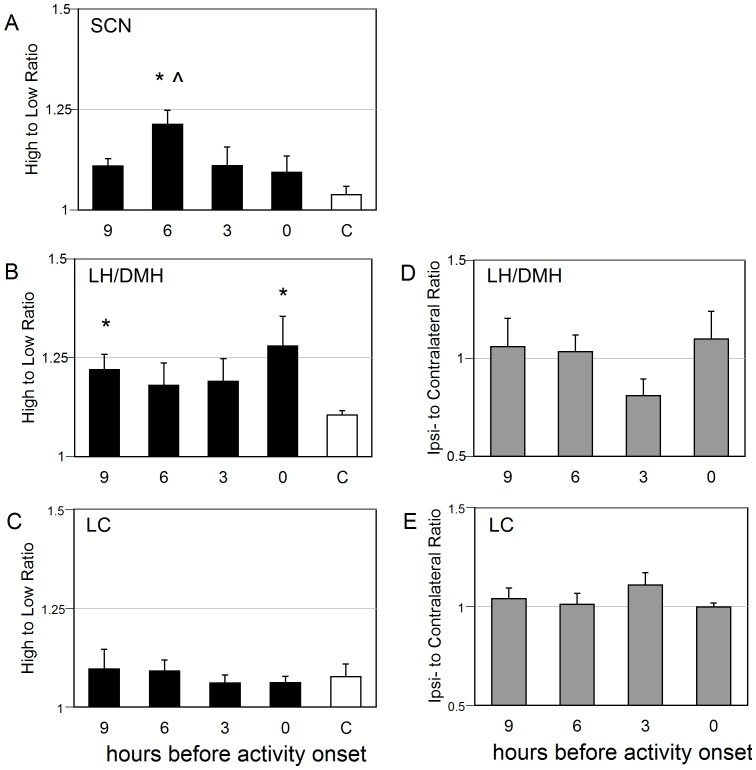
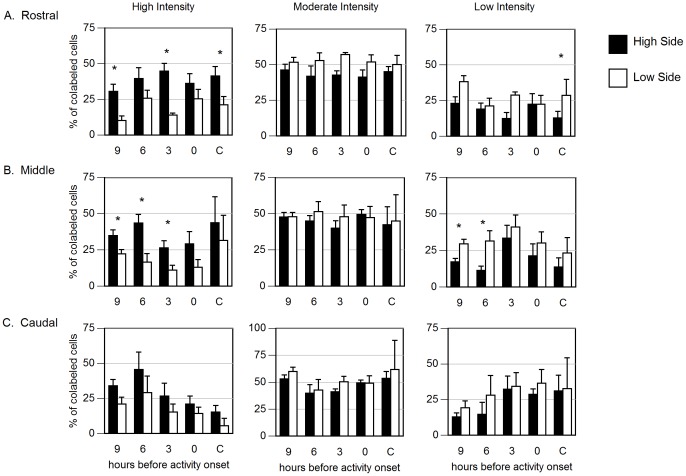
The number of cells expressing AVP on each side of the SCN, and the number of AVP cells that also expressed Per1, were similar across circadian phases in each plane (rostral, middle and caudal) of the SCN (see table S2). The number of Per1 cells was also similar between the sides of the SCN. Consistent with earlier reports, we found that LL exposure adequate to induce splitting of locomotor activity resulted in regionally and phenotypically specific asymmetry of Per1 expression in the SCN: within AVP cells colabeled with Per1 in the middle plane of the SCN, the ratio of high to low Per1 intensity was greater at 6 h before AO than at 9 h before AO (p<0.05; figure 3A) and greater than unsplit control (p<0.05). Of the AVP cells co-labeled with Per1, the percent in the highest quartile of Per1 expression differed between the coronal planes of the SCN. In the rostral SCN, significant asymmetry was evident at 9 h and 3 h before AO in (p<0.05 vs. low side; figure 4A). In the middle SCN, asymmetry occurred at 9 h, 6 h, and 3 h before AO (figure 4B). No asymmetry was seen at any phase in the caudal SCN (figure 4C).
Figure 3. Asymmetry in the mean (±SEM) intensity of Per1 expression among hamsters maintained in LL in experiment 2.
Per1 expression was lateralized in cases in which the high to low ratio of Per1 expression exceeded that in non-split controls. The high to low ratio of Per1 expression is illustrated in the (A) AVP-expressing cells of the SCN, (B) HCRT-expressing cells of the LH/DMH and (C) TH-expressing cells of the LC. Consistency of lateralization to the high Per1 expressing SCN was assessed by the ipsilateral to contralateral ratios of Per1 expression in (D) HCRT-expressing cells of the LH/DMH and (E) TH-expressing cells of the LC. * p<0.05 vs. C; χ p<0.05 vs. 9 h before activity onset.
Figure 4. Assessment of the distribution of Per1 expression intensities in the SCN of hamsters exposed to LL.
Distribution of cells expressing high (left), moderate (middle two quartiles; center) or low (right) intensity of Per1 hybridization in (A) Rostral, (B) Middle or (C) Caudal SCN. * p<0.05 vs. low side.
HCRT-cells in the LH/DMH
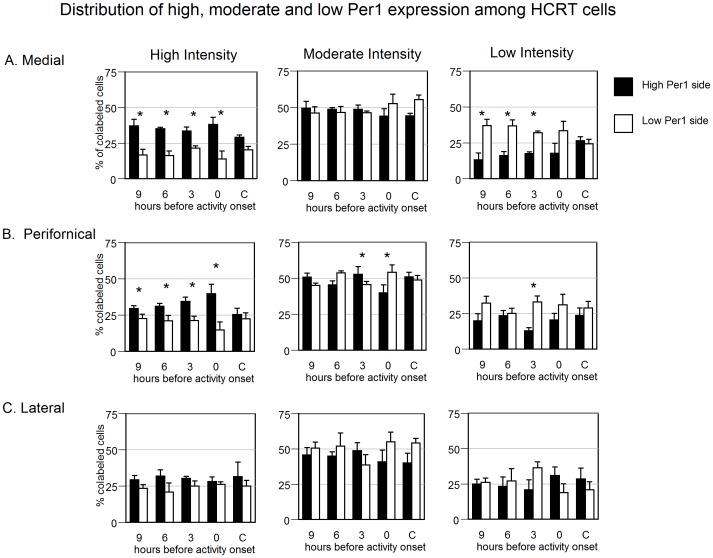
The total number of HCRT cells counted per side of the LH/DMH and the number of HCRT cells that expressed Per1 above the threshold was similar at all circadian phases (see table S2). In split hamsters, the asymmetry of Per1 labeling in HCRT cells exceeded that in unsplit controls at 9 h before AO and at activity onset (p<0.05; figure 3B). When evaluated by region, asymmetry of Per1 levels exceeded that in unsplit controls at 9 h before AO and at activity onset in the medial region (p<0.05; figure S5) and approached significance in the perifornical region at 9 h pAO versus unsplit controls (p = 0.07). The lateral region was symmetric at all phases. The percent of HCRT cells showing the highest quartile of Per1 expression was asymmetric at all phases assessed in split hamsters, but symmetric within unsplit controls. When assessed by region, the medial and perifornical HCRT field had significant lateralization in the percentage of high expressing Per1 cells (figure 5), but the lateral HCRT field did not (figure 5).
Figure 5. Per1 intensity quantified by category of intensity within HCRT cells.
Distribution of cells expressing high (left), moderate (center) or low (right) levels of Per1 in the (A) medial, (B) perifornical and (C) lateral regions of the HCRT-expressing cells of the LH/DMH. * p<0.05 vs. low side.
Per1 intensity levels within HCRT cells on the side ipsilateral vs. contralateral to the high Per1 expressing SCN (I:C ratio) differed at 9 h before AO in 4 of 5 animals (p<0.05, Kolmogorov-Smirnov test) whereas lateralization was inconsistent at the other phases sampled (occuring in only 1 of 5 animals at 3 h and 6 h before AO and in 2 of 5 hamsters sampled at AO). The I:C ratio of Per1 expression tended to be lower at 3 h vs. 0 h before activity onset in HCRT cells (p = 0.08; figure 3D). We sought to assess further the relationship of lateralization of Per1 expression in HCRT cells to asymmetry of circadian function in the SCN. Thus the relationship between the level of asymmetry in AVP- expressing cells of the middle SCN and the asymmetry of I:C or H:L ratios of Per1 expression in HCRT cells of the medial and perifornical regions were assessed using Spearman’s correlation. A significant positive correlation between the medial and perifornical HCRT I:C ratio with the H:L SCN ratio was observed at AO (figure S6).
No significant correlation between behavioral rhythms (locomotor activity bout volume, intensity or duration) and asymmetry of Per1 expression within HCRT cells was observed (Spearman correlation p>0.05 for each correlation analysis).
TH cells in the LC
The total number of TH cells counted per side of the LC and the number of TH cells that expressed Per1 was similar, as was the percentage of co-labeled TH cells (see table S2). There was no evidence of lateralization of Per1 expression in TH cells of split hamsters: the high to low ratio and the ipsilateral to contralateral ratios were close to 1.0 at all phases (figure 3C and 3E). As the ratios indicate a symmetric relationship between the sides of the LC, the data were collapsed and the distribution of Per1 expression across phases was assessed. Nevertheless, the percentage of cells in the highest quartile of Per1 expression was greater at 9 h than at 3 h before AO (p<0.05; figure 6).
Figure 6. Per1 intensity quantified by category of intensity within TH cells.
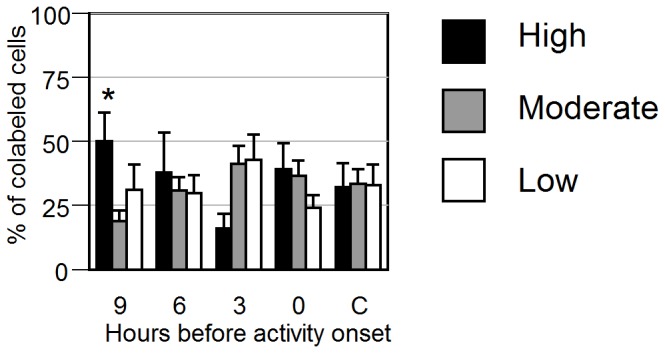
Distribution of cells expressing high (black), moderate (grey) or low (white) levels of Per1 in the TH-expressing cells of the LC. * P<0.05 vs. 3 hours before activity onset.
Discussion
Clock gene Expression within Arousal-Promoting Neurons
Core clock genes are expressed in multiple cell types in mice, rats and hamsters [44]–[49]. Within the brain, clock gene expression also appears to be widespread [2], [26], [28]–[30], [50]–[54]. The results of the current study indicate that the clock gene, per1, is expressed in HCRT cells of the LH/DMH and TH neurons of LC. Furthermore, the message levels of Per1 change across the day. As in the case of peripheral oscillators, the phase of the oscillation of Per1 expression in these arousal-regulating neurons lags that of the master pacemaker.
Our findings are consistent with earlier observations that the expression of Per1 within the SCN is high in the subjective day and low during the subjective night in hamsters [44], [54]–[59] as in other rodents [60]–[62], with peak expression around CT4 and a nadir at approximately CT18. We have expanded observations of regionally specific rhythmic Per1 expression in the SCN shell [57] to cellular co-expression in AVP neurons. The amplitude of the rhythms of clock gene expression detected through this dual label in situ hybridization method is lower than that found using isotopically labeled probes or immunostaining. This may reflect the focus on specific cell types that we were able to achieve using this method, or the fact that in the isotopic method the signal is generated from cells at multiple levels of the section so that stereological considerations contribute to the apparent amplitude. Additional technical issues, including the linearity of signal amplification of both the Per1 and the phenotypic signal, also make it difficult to compare directly the amplitude of the rhythm we examined with that inferred from other methods. Our main purpose in experiment 1 was to assess clock gene expression in particular cell types of two arousal-promoting nuclei that are likely under pacemaker control, and to determine its phase relationship to the SCN. The phase angle difference between clock gene expression in the SCN and other brain regions depends on the area and cell phenotypes investigated [29], [31], [63]; current results]. Peak clock gene expression in peripheral organs typically lags the master pacemaker by several hours [48], [64]. Similarly, peak reporter luminescence in explants of Per1-luc rats and PER2::LUC mice occurs around subjective dusk in the SCN region and in the mid-subjective night in the arcuate area, the lateral hypothalamus and paraventricular hypothalamic region [63], [65]. Limitations of sampling frequency restrict our ability to define with precision the phase of peak clock gene expression within the arousal-promoting cells. Nevertheless, our findings demonstrate a lag in the phase of Per1 expression between the vasopressinergic SCN and both HCRT- and TH-expressing cells.
Multisynaptic relays between the pacemaker and subordinate oscillators may account at least in part for the latency with which Period gene expression peaks. The SCN may regulate DMH function by either direct [66]–[68] projections or by relays through the sub-paraventricular zone [69], [70]. In either case, there is clear anatomical evidence for SCN regulation of HCRT cell activation [23]. Moreover, the AVP-expressing cells of the SCN project more densely to the DMH than do the more ventrally situated VIP cells [66]. Yoshida et al. [68] observed that SCN neurons innervate the HCRT field medial to the fornix. The multiplicity of regional inputs to the HCRT field and heterogeneity of LH/DMH cell populations [71], [72] and activation patterns [73], [74] suggest that both direct and indirect SCN inputs may regulate Per1 expression in this region.
Although vasopressinergic outputs may serve as important relays [75], other SCN efferents whose peak phases may differ may also regulate subordinate oscillators including the DMH/LH and LC [65], [67], [76]. It is also not clear which components of the circadian mechanism are most directly responsive to SCN outputs. While the initial step in the entrainment of the SCN pacemaker by light and locomotor activity is transcriptional activation or suppression of Per1 [55], [60], [77], zeitgebers that shift the phase of subordinate oscillations may more directly regulate expression of other clock genes [78]–[80]. Further investigation of the entrainment mechanisms of subordinate oscillators in brain areas including the LH/DMH and LC is needed.
The amplitude of Per1 rhythms was higher in the AVP cells of the SCN than either HCRT neurons of the LH/DMH or TH neurons of the LC. We found HCRT cells that expressed Per1 at low levels at each phase sampled. We cannot discriminate between the possibility that Per1 expression oscillates only in a subpopulation of HCRT cells of the LH/DMH, or TH cells of the LC, or that most or all of these cells oscillate but peak at different phases. Such phase clusters occur within the SCN [81]–[84] and it is possible that SCN heterogeneity is conveyed to different cell populations in LH/DMH, LC, or other brain regions. We may have missed the peak or nadir of the Per1 oscillation in any or all of these areas due to the limitations of sampling frequency. On the other hand, we find a more pronounced oscillation than is suggested by the results of Salgado-Delgado et al. [31], who found no rhythm of PER1 immunoreactivity within the LH/DMH region of rats but reported that immunoreactive cell numbers increased when locomotor activity was induced between ZT2 and ZT10. Our examination of phenotype-specific Per1 expression may have enabled resolution of clock function that is not general to all cells in this region. Within the DMH/LH, HCRT and MCH cells have opposing functions that likely result in different phases of activation, but it remains to be determined whether clock gene expression is asynchronous [85]–[89]. A limitation of in situ hybridization or immuncytochemical studies is that they capture only one phase per animal. Use of a reporter gene driven by a clock gene promoter may allow for assessment of the HCRT population, in vivo or in vitro, in real time, provided that methodological hurdles to identification of cell phenotype along with clock gene expression can be surmounted [11], [90]. Use of tract-tracing approaches with phenotype labeling and phase markers may identify precisely which subpopulations of HCRT cells participate in a particular behavioral and/or physiological circuit and which possess a molecular clock.
Lateralized SCN Communication to Arousal-Promoting Neurons
Consistent with immunocytochemical studies, asymmetry of Per1 expression was evident in the rostral and middle regions of the SCN [62], [91], [92]. Yan et al. [91] proposed that the split state is marked by synchrony between the rostral and contralateral middle region of the SCN. In the present study, we observed one animal at each phase in which the side expressing high Per1 differed between the rostral and the middle SCN. Although a distributional analysis of Per1 expression provided a less stringent cutoff, the rostral and middle planes differed in less than half of the hamsters (6 out of 14 animals killed at 9 h, 6 h or 3 h before activity onset). These observations suggest that multiple phase relationships may occur among groups of SCN neurons in hamsters induced to split by exposure to constant light.
In some instances, diffusible signals may relay phase information from the SCN [36]. In others, blood-borne or temperature and/or food cues that mediate pacemaker influences on the phase of peripheral oscillators may regulate clock gene expression in the brain [4], [37], [46], [93] In each such case, clock function on the left and right sides of the brain would be expected to be symmetrical. Our intent was to use the split preparation as a tool to examine SCN control of efferent targets that might act as subordinate oscillators. Efforts to assess the relationship between the SCN and the LH/DMH and LC are complicated by the phase differences of peak Per1 expression between areas. Thus it is difficult to assess pacemaker control of clock gene expression in its efferent targets in a single animal, as transcription in the LH/DMH or LC at the time of sacrifice may have been controlled by events that occurred in the SCN many hours previously. The level of asymmetry within the HCRT field appeared to peak during the last few hours before activity onset, about 6 hours after the peak in asymmetry in the middle SCN. A similar lag was observed between peak Per1 in the SCN and HCRT cells of DD-housed hamsters. At 3 h before activity onset, the high Per1 expressing HCRT cells tended to be on the side contralateral to the high Per1 expressing SCN. At activity onset the opposite relationship exists. HCRT cells likely oscillate in antiphase in split hamsters, as is the case for AVP cells of the SCN. However, non-lateralized cues including diffusible SCN substances, humoral, temperature, or other activity-dependent cues available to both sides may diminish the extent of lateralization of clock gene expression [31], [36], [94]. The oscillation in free running hamsters of Per1 expression in TH cells of the LC, while statistically significant, was of lower amplitude than that in AVP cells of the SCN or HCRT cells of the LH/DMH. This may have made it difficult to detect lateralization in LL-exposed hamsters that split in experiment 2. Sampling at other phases relative to activity onset may be necessary to detect lateralization of clock gene expression in the LC.
Recently, Butler et al. [95] observed bimodal co-expression of FOS-ir within HCRT cells of hamsters induced to split by exposure to LL. They quantified co-expression in a region centered on the portion of lateral hypothalamus in which HCRT cells were most highly concentrated. Thus their analysis included a region lateral to that in which we found the greatest evidence of rhythmic Per1 expression in experiment 1. Butler et al. [95] observed a high amplitude fluctuation between the percentage of activated HCRT cells in split animals, but found no significant lateralization of HCRT/FOS co-labeling. Although they interpreted FOS expression to reflect entrainment of an efferent target of the SCN, they did not observe a measure of the phase of a circadian oscillator within the HCRT cells. In contrast, we assessed mRNA encoding a clock gene whose expression may not always directly correspond to cell activation [96], [97]. Different patterns of SCN input, or co-release of signals, may control activation vs. circadian phase, and the pattern of FOS/PER1 co-expression characteristic of retinorecipient cells of the SCN may not also be characteristic of SCN targets. This could be the case if output signals of the SCN responsible for entrainment of subordinate oscillators do not lead to activation of common promoter elements in clock and immediate early genes. Furthermore, FOS expression in the DMH/LH may be more readily influenced by feedback cues (including temperature and feeding) associated with the split behavior than is the transcription of clock genes. Beyond the differences between our study and that of Butler et al. [95] in the genes whose expression was assessed and the examination of protein vs. mRNA, we used a distributional analysis of message level rather than an all-or-none categorization of whether cells are activated or not.
Butler et al. [95] estimated a 6-hour lag between the phase of peak cell activation in the SCN shell and the HCRT cells. Peak FOS-ir within the SCN shell of behaviorally split hamsters occurred at approximately 3 hours before activity onset, whereas peak FOS within HCRT cells occurred at 3 hours after activity onset. This 6-hour lag is consistent with the lag we observe in Per1 expression within the AVP cells of the SCN and the HCRT cells of the LH/DMH. We observed a rhythm of Per1 expression in the medial and perifornical but not the lateral population of HCRT cells. This is interesting given the observations that HCRT cells in these subregions differ in projections and presumptive functions [70], [85], [98], [99] The numerous interactions (both efferent and afferent) between the HCRT field and other regions suggest that splitting of HCRT function may have significant behavioral and physiological impact.
In contrast to Per1 expression in the LH/DMH, the LC of behaviorally split hamsters remained symmetric, although the limited phases sampled may have prevented detection of lateralization at the level of the LC. Furthermore, the nadir of Per1 expression within TH-expressing cells of the LC occurred at about 3 hours before activity onset, suggesting that the phase relationship between the LC and activity onset in behaviorally split hamsters differs from that of DD-housed hamsters. Projections from the hypocretin field to the LC may be bilateral, therefore reducing the lateralized signal from the SCN to distant regions and originate across the HCRT field. Diffusible outputs of the SCN or even the DMH may regulate the circadian phase of the LC [14], [69], [70], [99]-[107], and this may contribute to lack of lateralization of Per1 expression. The effects of systemic humoral signals including adrenocortical hormones may also account in part for symmetry of clock function in the LC [108] as elsewhere in the brain [2]. Locomotor feedback may also reach the LC, preventing lateralized top-down signals from entraining clock gene expression in these cells [109], [110]. It remains an open question whether completely crossed or uncrossed cues from the SCN might regulate Per1 expression in the LC, but global and/or bilateral cues cannot be ruled out.
Conclusion
We have documented time-dependent changes in the expression of the core clock gene Per1 in arousal-promoting cells, and found that the phase of peak Per1 expression in these cells lags that observed in the SCN. We also observed regional lateralization of clock gene expression at specific phases between the sides of the medial HCRT population that is consistently related to that in AVP cells of the SCN. Lateralization of clock gene expression was not evident in TH-expressing cells of the LC. Identification of the transmitter(s) that control circadian phase will help elucidate integrative properties of timing cues. Investigation of the function of a molecular clock within specific neuronal populations will clarify the circadian modulation of behavior and physiology.
Supporting Information
Representative actograms of hamsters used in these experiments. (A) Locomotor activity record of an animal placed in constant darkness (DD) for ten days. Asterisk indicates kill time at CT12.5 for Experiment 1. (B) Record of a hamster maintained in LL for Experiment 2. Although only the 33 days leading up to the kill time are shown here, the morning and evening components can be identified by tracing the time of activity onset to the record preceeding the split (see figure 1 in Mahoney et al., 2010). This animal showed a typical split, and was killed at AO (asterisk).
(TIF)
HCRT expression in the Syrian hamster brain. Autoradiogram illustrating regional distribution of HCRT in the Syrian hamster as determined using 35S-labeled probe. Scale bar is 1 mm.
(TIF)
Expression of Per1 in subregions of the HCRT field. Normalized mean (±SEM) intensity values of Per1 expression within the medial (black), perifornical (gray) and lateral (white) regions of the hypocretin field of DD-housed hamsters. a- p<0.05 vs. CT, 12.5, 22.
(TIF)
Symmetry of Per1 expression within DD housed hamsters. Left panels show normalized Per1 intensity for the high (black) and low (white) expressing (A) AVP cells of the SCN, (B) HCRT cells of the LH/DMH and (C) TH cells of the LC. The right panels show the high to low ratio of Per1 expressionin in (D) AVP cells of the SCN, (E) HCRT cells of the LH/DMH and (F) TH cells of the LC. Mann Whitney U analysis indicate a lack of asymmetry in each cell type (p>0.05 between sides in A, B and C) and no effect of phase (p>0.05 between CT points in D, E and F). This analysis allows assessment of effects of LL to induce asymmetry in split hamsters in experiment 2.
(TIF)
Asymmetry of Per1 expression within subregions of the HCRT field. The high to low ratio of normalized Per1 intensity in the medial (black), perifornical (gray) and lateral (white) regions of the HCRT field. * p<0.05 vs. unsplit Controls; # p<0.05 vs. 3 h before AO.
(TIF)
Relationship between asymmetry of Per1 expression within subregions of the HCRT field and within AVP cells of the SCN. Asymmetry of Per1 expression within the medial HCRT field (A) plotted as the H:L ratios (top) and the I:C ratios of Per1 expression (bottom) versus the HL ratios of Per1 expression in AVP cells in the SCN at 9 h, 6 h, 3 h or 0 h before AO. Similar plots of asymmetry of the intermediate HCRT field are presented in (B). The p and rho values are indicated within each plot.
(TIF)
Cell counts for Experiment 1.
(DOCX)
Cell counts for Experiment 2.
(DOCX)
Acknowledgments
The authors thank Dale Callaham of the University of Massachusetts microscopy center for his time and assistance with the confocal microscope and the University of Massachusetts animal care personnel for their dedication to the animals used in this study.
Funding Statement
This study was supported by National Institute of Mental Health Grant RO1-MH-070019, and CEM was supported by the institutional training grant 5 T32 MH020051. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Welsh DK, Takahashi JS, Kay SA (2010) Suprachiasmatic nucleus: Cell autonomy and network properties. Annu Rev Physiol 72: 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amir S, Lamont EW, Robinson B, Stewart J (2004) A circadian rhythm in the expression of PERIOD2 protein reveals a novel SCN-controlled oscillator in the oval nucleus of the bed nucleus of the stria terminalis. J Neurosci 24: 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown MH, Nunez AA (1986) Hypothalamic circuits and circadian rhythms: Effects of knife cuts vary with their placement within the suprachiasmatic area. Brain Res Bull 16: 705–711. [DOI] [PubMed] [Google Scholar]
- 4. Guo H, Brewer JM, Champhekar A, Harris RB, Bittman EL (2005) Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc Natl Acad Sci U S A 102: 3111–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Inouye ST (1983) Does the ventromedial hypothalamic nucleus contain a self-sustained circadian oscillator associated with periodic feedings? Brain Res 279: 53–63. [DOI] [PubMed] [Google Scholar]
- 6. Inouye ST, Kawamura H (1979) Persistence of circadian rhythmicity in a mammalian hypothalamic "island" containing the suprachiasmatic nucleus. Proc Natl Acad Sci U S A 76: 5962–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu J, Zhang YH, Chou TC, Gaus SE, Elmquist JK, et al. (2001) Contrasting effects of ibotenate lesions of the paraventricular nucleus and subparaventricular zone on sleep-wake cycle and temperature regulation. J Neurosci 21: 4864–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer-Bernstein EL, Jetton AE, Matsumoto SI, Markuns JF, Lehman MN, et al. (1999) Effects of suprachiasmatic transplants on circadian rhythms of neuroendocrine function in golden hamsters. Endocrinology 140: 207–218. [DOI] [PubMed] [Google Scholar]
- 9. Mohawk JA, Green CB, Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ralph MR, Foster RG, Davis FC, Menaker M (1990) Transplanted suprachiasmatic nucleus determines circadian period. Science 247: 975–978. [DOI] [PubMed] [Google Scholar]
- 11. Tahara Y, Kuroda H, Saito K, Nakajima Y, Kubo Y, et al. (2012) In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol 22: 1029–1034. [DOI] [PubMed] [Google Scholar]
- 12. Yamazaki S, Kerbeshian MC, Hocker CG, Block GD, Menaker M (1998) Rhythmic properties of the hamster suprachiasmatic nucleus in vivo. J Neurosci 18: 10709–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, et al. (2004) LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A 101: 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang S, Zeitzer JM, Yoshida Y, Wisor JP, Nishino S, et al. (2004) Lesions of the suprachiasmatic nucleus eliminate the daily rhythm of hypocretin-1 release. Sleep 27: 619–627. [DOI] [PubMed] [Google Scholar]
- 15. Aston-Jones G, Chen S, Zhu Y, Oshinsky ML (2001) A neural circuit for circadian regulation of arousal. Nat Neurosci 4: 732–738. [DOI] [PubMed] [Google Scholar]
- 16. Dantz B, Edgar DM, Dement WC (1994) Circadian rhythms in narcolepsy: Studies on a 90 minute day. Electroencephalogr Clin Neurophysiol 90: 24–35. [DOI] [PubMed] [Google Scholar]
- 17. Hagan JJ, Leslie RA, Patel S, Evans ML, Wattam TA, et al. (1999) Orexin A activates locus coeruleus cell firing and increases arousal in the rat. Proc Natl Acad Sci U S A 96: 10911–10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kilduff TS, de Lecea L (2001) Mapping of the mRNAs for the hypocretin/orexin and melanin-concentrating hormone receptors: Networks of overlapping peptide systems. J Comp Neurol 435: 1–5. [DOI] [PubMed] [Google Scholar]
- 19. van den Pol AN, Ghosh PK, Liu RJ, Li Y, Aghajanian GK, et al. (2002) Hypocretin (orexin) enhances neuron activity and cell synchrony in developing mouse GFP-expressing locus coeruleus. J Physiol 541: 169–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeitzer JM, Buckmaster CL, Parker KJ, Hauck CM, Lyons DM, et al. (2003) Circadian and homeostatic regulation of hypocretin in a primate model: Implications for the consolidation of wakefulness. J Neurosci 23: 3555–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beuckmann CT, Yanagisawa M (2002) Orexins: From neuropeptides to energy homeostasis and sleep/wake regulation. J Mol Med (Berl) 80: 329–342. [DOI] [PubMed] [Google Scholar]
- 22. Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, et al. (1999) Narcolepsy in orexin knockout mice: Molecular genetics of sleep regulation. Cell 98: 437–451. [DOI] [PubMed] [Google Scholar]
- 23. Marston OJ, Williams RH, Canal MM, Samuels RE, Upton N, et al. (2008) Circadian and dark-pulse activation of orexin/hypocretin neurons. Mol Brain 1: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakurai T (2007) The neural circuit of orexin (hypocretin): Maintaining sleep and wakefulness. Nat Rev Neurosci 8: 171–181. [DOI] [PubMed] [Google Scholar]
- 25. Sutcliffe JG, de Lecea L (2002) The hypocretins: Setting the arousal threshold. Nat Rev Neurosci 3: 339–349. [DOI] [PubMed] [Google Scholar]
- 26. Hamada T, Honma S, Honma K (2011) Light responsiveness of clock genes, Per1 and Per2, in the olfactory bulb of mice. Biochem Biophys Res Commun 409: 727–731. [DOI] [PubMed] [Google Scholar]
- 27. Hickok JR, Tischkau SA (2010) In vivo circadian rhythms in gonadotropin-releasing hormone neurons. Neuroendocrinology 91: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Granados-Fuentes D, Prolo LM, Abraham U, Herzog ED (2004) The suprachiasmatic nucleus entrains, but does not sustain, circadian rhythmicity in the olfactory bulb. J Neurosci 24: 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kriegsfeld LJ, Korets R, Silver R (2003) GFP transgene. Eur J Neurosci 17: 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramanathan C, Smale L, Nunez AA (2008) Rhythms in expression of PER1 protein in the amygdala and bed nucleus of the stria terminalis of the diurnal grass rat (arvicanthis niloticus). Neurosci Lett 441: 86–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salgado-Delgado R, Nadia S, Angeles-Castellanos M, Buijs RM, Escobar C (2010) In a rat model of night work, activity during the normal resting phase produces desynchrony in the hypothalamus. J Biol Rhythms 25: 421–431. [DOI] [PubMed] [Google Scholar]
- 32. Sellix MT, Egli M, Poletini MO, McKee DT, Bosworth MD, et al. (2006) Anatomical and functional characterization of clock gene expression in neuroendocrine dopaminergic neurons. Am J Physiol Regul Integr Comp Physiol 290: R1309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pittendrigh C, Daan S (1976) A functional analysis of circadian pacemakers in nocturnal rodents: V. pacemaker structure: A clock for all seasons. Journal Comparative Physiology 106: 333. [Google Scholar]
- 34. de la Iglesia HO, Meyer J, Carpino A Jr, Schwartz WJ (2000) Antiphase oscillation of the left and right suprachiasmatic nuclei. Science 290: 799–801. [DOI] [PubMed] [Google Scholar]
- 35. de la Iglesia HO, Meyer J, Schwartz WJ (2003) Lateralization of circadian pacemaker output: Activation of left- and right-sided luteinizing hormone-releasing hormone neurons involves a neural rather than a humoral pathway. J Neurosci 23: 7412–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silver R, LeSauter J, Tresco PA, Lehman MN (1996) A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature 382: 810–813. [DOI] [PubMed] [Google Scholar]
- 37. Mahoney CE, Brewer D, Costello MK, Brewer JM, Bittman EL (2010) Lateralization of the central circadian pacemaker output: A test of neural control of peripheral oscillator phase. Am J Physiol Regul Integr Comp Physiol 299: R751–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sherman TG, Day R, Civelli O, Douglass J, Herbert E, et al. (1988) Regulation of hypothalamic magnocellular neuropeptides and their mRNAs in the brattleboro rat: Coordinate responses to further osmotic challenge. J Neurosci 8: 3785–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lewis EJ, Harrington CA, Chikaraishi DM (1987) Transcriptional regulation of the tyrosine hydroxylase gene by glucocorticoid and cyclic AMP. Proc Natl Acad Sci U S A 84: 3550–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grima B, Lamouroux A, Blanot F, Biguet NF, Mallet J (1985) Complete coding sequence of rat tyrosine hydroxylase mRNA. Proc Natl Acad Sci U S A 82: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watakabe A, Komatsu Y, Ohsawa S, Yamamori T (2010) Fluorescent in situ hybridization technique for cell type identification and characterization in the central nervous system. Methods 52: 367–374. [DOI] [PubMed] [Google Scholar]
- 42. Estabrooke IV, McCarthy MT, Ko E, Chou TC, Chemelli RM, et al. (2001) Fos expression in orexin neurons varies with behavioral state. J Neurosci 21: 1656–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manders E, Verbeek F, Aten J (1993) Measurement of co-localization of objects in dual-colour confocal images. Journal of Microscopy 169: 375. [DOI] [PubMed] [Google Scholar]
- 44. Carr AJ, Johnston JD, Semikhodskii AG, Nolan T, Cagampang FR, et al. (2003) Photoperiod differentially regulates circadian oscillators in central and peripheral tissues of the syrian hamster. Curr Biol 13: 1543–1548. [DOI] [PubMed] [Google Scholar]
- 45. Doherty CJ, Kay SA (2010) Circadian control of global gene expression patterns. Annu Rev Genet 44: 419–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo H, Brewer JM, Lehman MN, Bittman EL (2006) Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: Effects of transplanting the pacemaker. J Neurosci 26: 6406–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peirson SN, Butler JN, Duffield GE, Takher S, Sharma P, et al. (2006) Comparison of clock gene expression in SCN, retina, heart, and liver of mice. Biochem Biophys Res Commun 351: 800–807. [DOI] [PubMed] [Google Scholar]
- 48. Tong Y, Guo H, Brewer JM, Lee H, Lehman MN, et al. (2004) Expression of haPer1 and haBmal1 in syrian hamsters: Heterogeneity of transcripts and oscillations in the periphery. J Biol Rhythms 19: 113–125. [DOI] [PubMed] [Google Scholar]
- 49. Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, et al. (2000) Resetting central and peripheral circadian oscillators in transgenic rats. Science 288: 682–685. [DOI] [PubMed] [Google Scholar]
- 50. Abe H, Honma S, Namihira M, Masubuchi S, Ikeda M, et al. (2001) Clock gene expressions in the suprachiasmatic nucleus and other areas of the brain during rhythm splitting in CS mice. Brain Res Mol Brain Res 87: 92–99. [DOI] [PubMed] [Google Scholar]
- 51. Abraham U, Prior JL, Granados-Fuentes D, Piwnica-Worms DR, Herzog ED (2005) Independent circadian oscillations of Period1 in specific brain areas in vivo and in vitro. J Neurosci 25: 8620–8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guilding C, Hughes AT, Piggins HD (2010) Circadian oscillators in the epithalamus. Neuroscience 169: 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Masubuchi S, Honma S, Abe H, Ishizaki K, Namihira M, et al. (2000) Clock genes outside the suprachiasmatic nucleus involved in manifestation of locomotor activity rhythm in rats. Eur J Neurosci 12: 4206–4214. [PubMed] [Google Scholar]
- 54. Yamamoto S, Shigeyoshi Y, Ishida Y, Fukuyama T, Yamaguchi S, et al. (2001) Expression of the Per1 gene in the hamster: Brain atlas and circadian characteristics in the suprachiasmatic nucleus. J Comp Neurol 430: 518–532. [PubMed] [Google Scholar]
- 55. Maywood ES, Mrosovsky N, Field MD, Hastings MH (1999) Rapid down-regulation of mammalian period genes during behavioral resetting of the circadian clock. Proc Natl Acad Sci U S A 96: 15211–15216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. de la Iglesia HO, Meyer J, Schwartz WJ (2004) Using per gene expression to search for photoperiodic oscillators in the hamster suprachiasmatic nucleus. Brain Res Mol Brain Res 127: 121–127. [DOI] [PubMed] [Google Scholar]
- 57. Hamada T, Antle MC, Silver R (2004) Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci 19: 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Johnston JD, Ebling FJ, Hazlerigg DG (2005) Photoperiod regulates multiple gene expression in the suprachiasmatic nuclei and pars tuberalis of the siberian hamster (phodopus sungorus). Eur J Neurosci 21: 2967–2974. [DOI] [PubMed] [Google Scholar]
- 59. Tournier BB, Menet JS, Dardente H, Poirel VJ, Malan A, et al. (2003) Photoperiod differentially regulates clock genes' expression in the suprachiasmatic nucleus of syrian hamster. Neuroscience 118: 317–322. [DOI] [PubMed] [Google Scholar]
- 60. Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF Jr, Reppert SM (1997) Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19: 1261–1269. [DOI] [PubMed] [Google Scholar]
- 61. Caldelas I, Poirel VJ, Sicard B, Pevet P, Challet E (2003) Circadian profile and photic regulation of clock genes in the suprachiasmatic nucleus of a diurnal mammal arvicanthis ansorgei. Neuroscience 116: 583–591. [DOI] [PubMed] [Google Scholar]
- 62. Yan L, Okamura H (2002) Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci 15: 1153–1162. [DOI] [PubMed] [Google Scholar]
- 63. Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, et al. (2002) Circadian rhythms in isolated brain regions. J Neurosci 22: 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N (1998) Antiphase circadian expression between BMAL1 and period homologue mRNA in the suprachiasmatic nucleus and peripheral tissues of rats. Biochem Biophys Res Commun 253: 199–203. [DOI] [PubMed] [Google Scholar]
- 65. Hughes AT, Guilding C, Piggins HD (2011) Neuropeptide signaling differentially affects phase maintenance and rhythm generation in SCN and extra-SCN circadian oscillators. PLoS One 6: e18926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abrahamson EE, Leak RK, Moore RY (2001) The suprachiasmatic nucleus projects to posterior hypothalamic arousal systems. Neuroreport 12: 435–440. [DOI] [PubMed] [Google Scholar]
- 67. Kalsbeek A, Yi CX, Cailotto C, la Fleur SE, Fliers E, et al. (2011) Mammalian clock output mechanisms. Essays Biochem 49: 137–151. [DOI] [PubMed] [Google Scholar]
- 68. Yoshida K, McCormack S, Espana RA, Crocker A, Scammell TE (2006) Afferents to the orexin neurons of the rat brain. J Comp Neurol 494: 845–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Deurveilher S, Semba K (2005) Indirect projections from the suprachiasmatic nucleus to major arousal-promoting cell groups in rat: Implications for the circadian control of behavioural state. Neuroscience 130: 165–183. [DOI] [PubMed] [Google Scholar]
- 70. Gompf HS, Aston-Jones G (2008) Role of orexin input in the diurnal rhythm of locus coeruleus impulse activity. Brain Res 1224: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cvetkovic V, Brischoux F, Jacquemard C, Fellmann D, Griffond B, et al. (2004) Characterization of subpopulations of neurons producing melanin-concentrating hormone in the rat ventral diencephalon. J Neurochem 91: 911–919. [DOI] [PubMed] [Google Scholar]
- 72. Lee S, Bookout AL, Lee CE, Gautron L, Harper MJ, et al. (2012) Laser-capture microdissection and transcriptional profiling of the dorsomedial nucleus of the hypothalamus. J Comp Neurol 520: 3617–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Alam MN, Gong H, Alam T, Jaganath R, McGinty D, et al. (2002) Sleep-waking discharge patterns of neurons recorded in the rat perifornical lateral hypothalamic area. J Physiol 538: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fadel J, Bubser M, Deutch AY (2002) Differential activation of orexin neurons by antipsychotic drugs associated with weight gain. J Neurosci 22: 6742–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tsunematsu T, Fu LY, Yamanaka A, Ichiki K, Tanoue A, et al. (2008) Vasopressin increases locomotion through a V1a receptor in orexin/hypocretin neurons: Implications for water homeostasis. J Neurosci 28: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang C, Truong KK, Zhou QY (2009) Efferent projections of prokineticin 2 expressing neurons in the mouse suprachiasmatic nucleus. PLoS One 4: e7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Akiyama M, Kouzu Y, Takahashi S, Wakamatsu H, Moriya T, et al. (1999) Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci 19: 1115–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, et al. (2012) The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A 109: 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meng QJ, McMaster A, Beesley S, Lu WQ, Gibbs J, et al. (2008) Ligand modulation of REV-ERBalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci 121: 3629–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jordan SD, Lamia KA (2012) AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cheng HY, Alvarez-Saavedra M, Dziema H, Choi YS, Li A, et al. (2009) Segregation of expression of mPeriod gene homologs in neurons and glia: Possible divergent roles of mPeriod1 and mPeriod2 in the brain. Hum Mol Genet 18: 3110–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ciarleglio CM, Gamble KL, Axley JC, Strauss BR, Cohen JY, et al. (2009) Population encoding by circadian clock neurons organizes circadian behavior. J Neurosci 29: 1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ (2011) Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS One 6: e15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K (2007) Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A 104: 7664–7669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. de Lecea L, Sutcliffe JG (2005) The hypocretins and sleep. FEBS J 272: 5675–5688. [DOI] [PubMed] [Google Scholar]
- 86. Willie JT, Chemelli RM, Sinton CM, Tokita S, Williams SC, et al. (2003) Distinct narcolepsy syndromes in orexin receptor-2 and orexin null mice: Molecular genetic dissection of non-REM and REM sleep regulatory processes. Neuron 38: 715–730. [DOI] [PubMed] [Google Scholar]
- 87. Hassani OK, Lee MG, Jones BE (2009) Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep-wake cycle. Proc Natl Acad Sci U S A 106: 2418–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Torterolo P, Sampogna S, Chase MH (2009) MCHergic projections to the nucleus pontis oralis participate in the control of active (REM) sleep. Brain Res 1268: 76–87. [DOI] [PubMed] [Google Scholar]
- 89. Torterolo P, Lagos P, Monti JM (2011) Melanin-concentrating hormone: A new sleep factor? Front Neurol 2: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wendt MK, Molter J, Flask CA, Schiemann WP (2011) In vivo dual substrate bioluminescent imaging. J Vis Exp (56). pii: 3245. doi: 10.3791/3245 [DOI] [PMC free article] [PubMed]
- 91. Yan L, Foley NC, Bobula JM, Kriegsfeld LJ, Silver R (2005) Two antiphase oscillations occur in each suprachiasmatic nucleus of behaviorally split hamsters. J Neurosci 25: 9017–9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yan L, Silver R (2008) Day-length encoding through tonic photic effects in the retinorecipient SCN region. Eur J Neurosci 28: 2108–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Saini C, Morf J, Stratmann M, Gos P, Schibler U (2012) Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev 26: 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Buhr ED, Yoo SH, Takahashi JS (2010) Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Butler MP, Rainbow MN, Rodriguez E, Lyon SM, Silver R (2012) Twelve-hour days in the brain and behavior of split hamsters. Eur J Neurosci 36: 2556–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mead S, Ebling FJ, Maywood ES, Humby T, Herbert J, et al. (1992) A nonphotic stimulus causes instantaneous phase advances of the light-entrainable circadian oscillator of the syrian hamster but does not induce the expression of c-fos in the suprachiasmatic nuclei. J Neurosci 12: 2516–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rea MA, Michel AM, Lutton LM (1993) Is fos expression necessary and sufficient to mediate light-induced phase advances of the suprachiasmatic circadian oscillator? J Biol Rhythms 8 Suppl: S59–64 [PubMed] [Google Scholar]
- 98. Gonzalez JA, Jensen LT, Fugger L, Burdakov D (2012) Convergent inputs from electrically and topographically distinct orexin cells to locus coeruleus and ventral tegmental area. Eur J Neurosci 35: 1426–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Peyron C, Tighe DK, van den Pol AN, de Lecea L, Heller HC, et al. (1998) Neurons containing hypocretin (orexin) project to multiple neuronal systems. J Neurosci 18: 9996–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kalsbeek A, Foppen E, Schalij I, Van Heijningen C, van der Vliet J, et al. (2008) Circadian control of the daily plasma glucose rhythm: An interplay of GABA and glutamate. PLoS One 3: e3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, et al. (2001) Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science 294: 2511–2515. [DOI] [PubMed] [Google Scholar]
- 102. Watanabe K, Vanecek J, Yamaoka S (2000) In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res 877: 361–366. [DOI] [PubMed] [Google Scholar]
- 103. Devilbiss DM, Waterhouse BD, Berridge CW, Valentino R (2012) Corticotropin-releasing factor acting at the locus coeruleus disrupts thalamic and cortical sensory-evoked responses. Neuropsychopharmacology 37: 2020–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Horvath TL, Peyron C, Diano S, Ivanov A, Aston-Jones G, et al. (1999) Hypocretin (orexin) activation and synaptic innervation of the locus coeruleus noradrenergic system. J Comp Neurol 415: 145–159. [PubMed] [Google Scholar]
- 105. Carter ME, Brill J, Bonnavion P, Huguenard JR, Huerta R, et al. (2012) Mechanism for hypocretin-mediated sleep-to-wake transitions. Proc Natl Acad Sci U S A 109: E2635–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Date Y, Ueta Y, Yamashita H, Yamaguchi H, Matsukura S, et al. (1999) Orexins, orexigenic hypothalamic peptides, interact with autonomic, neuroendocrine and neuroregulatory systems. Proc Natl Acad Sci U S A 96: 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Espana RA, Reis KM, Valentino RJ, Berridge CW (2005) Organization of hypocretin/orexin efferents to locus coeruleus and basal forebrain arousal-related structures. J Comp Neurol 481: 160–178. [DOI] [PubMed] [Google Scholar]
- 108. Li M, Han F, Shi Y (2011) Expression of locus coeruleus mineralocorticoid receptor and glucocorticoid receptor in rats under single-prolonged stress. Neurol Sci 32: 625–631. [DOI] [PubMed] [Google Scholar]
- 109. Aston-Jones G, Bloom FE (1981) Norepinephrine-containing locus coeruleus neurons in behaving rats exhibit pronounced responses to non-noxious environmental stimuli. J Neurosci 1: 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Berridge CW, Waterhouse BD (2003) The locus coeruleus-noradrenergic system: Modulation of behavioral state and state-dependent cognitive processes. Brain Res Brain Res Rev 42: 33–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative actograms of hamsters used in these experiments. (A) Locomotor activity record of an animal placed in constant darkness (DD) for ten days. Asterisk indicates kill time at CT12.5 for Experiment 1. (B) Record of a hamster maintained in LL for Experiment 2. Although only the 33 days leading up to the kill time are shown here, the morning and evening components can be identified by tracing the time of activity onset to the record preceeding the split (see figure 1 in Mahoney et al., 2010). This animal showed a typical split, and was killed at AO (asterisk).
(TIF)
HCRT expression in the Syrian hamster brain. Autoradiogram illustrating regional distribution of HCRT in the Syrian hamster as determined using 35S-labeled probe. Scale bar is 1 mm.
(TIF)
Expression of Per1 in subregions of the HCRT field. Normalized mean (±SEM) intensity values of Per1 expression within the medial (black), perifornical (gray) and lateral (white) regions of the hypocretin field of DD-housed hamsters. a- p<0.05 vs. CT, 12.5, 22.
(TIF)
Symmetry of Per1 expression within DD housed hamsters. Left panels show normalized Per1 intensity for the high (black) and low (white) expressing (A) AVP cells of the SCN, (B) HCRT cells of the LH/DMH and (C) TH cells of the LC. The right panels show the high to low ratio of Per1 expressionin in (D) AVP cells of the SCN, (E) HCRT cells of the LH/DMH and (F) TH cells of the LC. Mann Whitney U analysis indicate a lack of asymmetry in each cell type (p>0.05 between sides in A, B and C) and no effect of phase (p>0.05 between CT points in D, E and F). This analysis allows assessment of effects of LL to induce asymmetry in split hamsters in experiment 2.
(TIF)
Asymmetry of Per1 expression within subregions of the HCRT field. The high to low ratio of normalized Per1 intensity in the medial (black), perifornical (gray) and lateral (white) regions of the HCRT field. * p<0.05 vs. unsplit Controls; # p<0.05 vs. 3 h before AO.
(TIF)
Relationship between asymmetry of Per1 expression within subregions of the HCRT field and within AVP cells of the SCN. Asymmetry of Per1 expression within the medial HCRT field (A) plotted as the H:L ratios (top) and the I:C ratios of Per1 expression (bottom) versus the HL ratios of Per1 expression in AVP cells in the SCN at 9 h, 6 h, 3 h or 0 h before AO. Similar plots of asymmetry of the intermediate HCRT field are presented in (B). The p and rho values are indicated within each plot.
(TIF)
Cell counts for Experiment 1.
(DOCX)
Cell counts for Experiment 2.
(DOCX)