Abstract
The complete sequences of three mitochondrial genomes from the land snail Cornu aspersum were determined. The mitogenome has a length of 14050 bp, and it encodes 13 protein-coding genes, 22 transfer RNA genes and two ribosomal RNA genes. It also includes nine small intergene spacers, and a large AT-rich intergenic spacer. The intra-specific divergence analysis revealed that COX1 has the lower genetic differentiation, while the most divergent genes were NADH1, NADH3 and NADH4. With the exception of Euhadra herklotsi, the structural comparisons showed the same gene order within the family Helicidae, and nearly identical gene organization to that found in order Pulmonata. Phylogenetic reconstruction recovered Basommatophora as polyphyletic group, whereas Eupulmonata and Pulmonata as paraphyletic groups. Bayesian and Maximum Likelihood analyses showed that C. aspersum is a close relative of Cepaea nemoralis, and with the other Helicidae species form a sister group of Albinaria caerulea, supporting the monophyly of the Stylommatophora clade.
Introduction
Mitochondria, powerhouses of the cell, are in charge of producing energy in the form of Adenosine triphosphate (ATP) that is usable by the cell in eukaryotic organisms. The metabolic pathway where this occurs is the oxidative phosphorylation (OXPHOS), and it requires a whole system of protein complexes, the electron transport chain (ETC), that are anchored to the inner membrane of the mitochondria. The enzymes that belong to the ETC are encoded by both mitochondrial (mtDNA) and nuclear (nDNA) genomes [1], where the nDNA-encoded peptides have mostly structural functions, whereas mtDNA-encoded peptides constitute the main catalytic centres [2].
The mitochondrial genome of metazoans is typically a circular double stranded DNA molecule of about 12–20 kb length, which contains 37 genes including 13 protein-coding genes, 2 ribosomal RNAs (12S rRNA and 16S rRNA) genes, and 22 transfer RNAs (tRNAs) [3]. Additionally it has an AT-rich non-coding region that contains the potential origin for mitochondrial DNA replication (POR) [4] and RNA transcription (i.e., the mitochondrial control region) [3], [5]. Over the last decades, mitochondrial genomes have been used for a wide range of comparative studies as phylogenetic markers to resolve evolutionary relationships [6]. Particularly the phylum mollusca, especially the Euthyneura clade, has been subject of intense debate regarding their phylogenetic relationships [4], [6]–[13]. The lineages associated with this group (i.e., Opistobranchia and Pulmonata) have been the subject of long controversy due to the different phylogenetic results obtained with morphological [14], [15] and molecular data [4], [11], [16]. In particular, within Pulmonata some authors had found contradictory results from molecular phylogenetic reconstructions. For example, the monophyly of Eupulmonata (Fig. 1A) has been documented based on the combination of mitochondrial and nuclear genes [13], [17], [18], whereas the paraphyly of this group (Fig. 1B) has been recovered using complete mitochondrial genomes [4], [16].
Figure 1. Competing hypotheses regarding the phylogenetic relationships among Euthyneuran species.
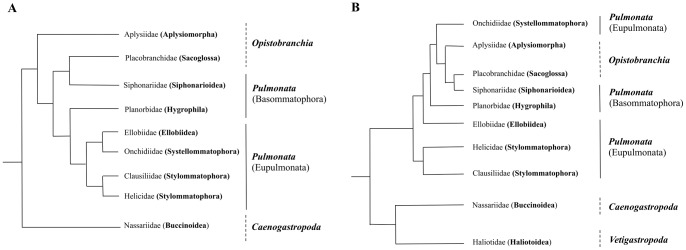
A) The monophyly of Eupulmonata has been documented based on the combination of mitochondrial and nuclear genes [13], [17], [18], and B) The paraphyly of Eupulmonata has been recovered using complete mitochondrial genomes [4], [16].
Although a great number of mitochondrial genomes is reported in the Organelle Genome Resources of the NCBI for Euthyneura, there are some clades such as Stylommatophora that have few representatives, in which some of the mitochondrial genomes has been criticised because of the poor quality of its sequence [4]. In principle, the addition of new mitochondrial genomes from species of the family Helicidae will increase the taxonomic sampling in a clade that today has few representatives (Stylommatophora), and from a phylogenetic standpoint, will add valuable information to understand evolutionary relationships at different taxonomic levels. Moreover this information will also help to elucidate genomic features, such as gene order, that are unique to this group of snails, and those that are shared with other clades.
Here we report the mitochondrial genome of three individuals of the land snail Cornu aspersum, one of the most widespread invasive species in the world [19], [20]. This species is native from the West Mediterranean, and since the Holocene has successfully colonized all continents except Antarctica [21]. Currently, its distribution extends to vast areas in the temperate and subtropical regions of the world where it is found in agricultural areas, parks and domestic gardens within cities. At these locations, C. aspersum snails are quite abundant thus contributing to its consideration as an important pest [22]–[24]. The mitochondrial genomes of C. aspersum reported in this work correspond to three individuals from three populations in a latitudinal gradient from northern to southern Chile, characterized by contrasting climatic conditions [25]. These populations are genetically differentiated [26] and have been subject of at least three different events of introduction during the last century [27]. Such factors could underlie the physiological differences exhibited by these populations [25], [28], which are probably evidenced by the divergence of those genes related to the OXPHOS and ETC. Results from our comparative genomic analysis revealed that the mitochondrial genome of C. aspersum encodes for the 37 genes typical of most metazoans, and is characterized by the highly conserved gene order and gene content of pulmonate gastropods, with only few gene rearrangements that are shared within Helicidae. In accordance to previous studies, where complete mitochondrial genomes were used to infer phylogenetic relationships, we recovered Basommatophora as polyphyletic group, whereas Eupulmonata and Pulmonata as paraphyletic groups.
Materials and Methods
Specimen Collection and DNA Isolation
Adult individuals of Cornu aspersum were collected from three populations in Chile across a latitudinal range of approximately 1300 km: La Serena (29°54′ S, 71°15′ W), Constitución (35°20′ S, 72°25′ W) and Valdivia (39° 38′ S, 73° 5′ W). We selected these three localities based on their significant genetic [27] and climatic differentiations [25]. Snails were transported to the laboratory, and maintained at 20°C with a fixed photoperiod of 14L:10D. Additionally, snails were kept in humid litter soil and water was given ad libitum until DNA extractions were performed. DNA from a single snail of each population was obtained by the isolation of intact mitochondria from approximately 120 mg of fresh foot tissue, using the Mitochondrial Isolation Kit for Tissue (Thermo Scientific). The isolated mitochondrial pellet of each snail was used for the mtDNA extraction using the Mitochondrial DNA Isolation kit (BioVision).
Mitochondrial Genomes Sequencing and Assembly
The shotgun libraries of C. aspersum were sequenced using a combination of 454 (Roche Genome Sequencer GS FLX Titanium) and Sanger sequencing technologies on ABI 3730XL sequencers by Eurofins MWG Operon (Huntsville, USA). DNA samples were nebulized, individually bar-coded to perform emulsion-based clonal ampiflication (emPCR) and sequenced to approximately 20-fold coverage. After sequencing, raw sequence files were proof read, separated, and assembled, according to the bar-codes, into contigs in Celera Assembler v.6.1 [29]. Assembly data was evaluated with the statistical overview and quality scoring files of each single read. Sequences obtained in this work were deposited in GenBank under the accession numbers JQ417194 (La Serena), JQ417195 (Constitución) and JQ417196 (Valdivia).
Genome Annotation
Fragments of the whole mitochondrial DNA sequence were analyzed in MacClade 4.08 [30] and MEGA v.5.1 [31]. To control for sequencing errors, each partial sequence was evaluated at least twice. Ambiguous base pairs were resolved manually according to Roche’s flowcharts and the corresponding quality score values for each base in the reads. Annotations and editing procedures of the mitochondrial genomes of C. aspersum were done in Geneious v 4.8.5 [32]. Mitochondrial genes were identified by sequence comparison using DOGMA [33] and BLAST searches at NCBI (with the BlastN and BlastX algorithms [34]) against other Eupulmonata sequences (Table 1). The limits of both protein coding and ribosomal RNA genes were adjusted manually based on location of adjacent genes, and the presence of start and stop codons. Transfer RNA genes were located using ARWEN v.1.2 [35], DOGMA [33] and tRNAscan-SE v.1.21 [36], by means of the generalized invertebrate mitochondrial tRNA settings, after which they were manually adjusted based on specific anticodons in regions between identified genes.
Table 1. List of the Euthyneuran species included in the present study.
| Taxon | Order | Family | Species name | GenBank |
| PULMONATA | ||||
| Eupulmonata | Stylommatophora | |||
| Helicidae | Cornu aspersum | JQ417194 | ||
| Cornu aspersum | JQ417195 | |||
| Cornu aspersum | JQ417196 | |||
| Cepaea nemoralis | CMU23045 | |||
| Cylindrus obtusus | JN107636 | |||
| Euhadra herklotsi | Z71693 - Z71701 | |||
| Clausiliidae | Albinaria caerulea | X83390 | ||
| Succineidae | Succinea putris | JN627206 | ||
| Systellommatophora | ||||
| Onchidiidae | Onchidella celtica | AY345048 | ||
| Onchidella borealis | DQ991936 | |||
| Platevindex mortoni | GU475132 | |||
| Peronia peronii | JN619346 | |||
| Trimusculoidea | ||||
| Trimusculidae | Trimusculus reticulatus | JN632509 | ||
| Amphibolidea | ||||
| Amphibolidae | Salinator rhamphidia | JN620539 | ||
| Ellobiidea | ||||
| Ellobiidae | Myosotella myosotis | AY345053 | ||
| Auriculinella bidentata | JN606066 | |||
| Ovatella vulcani | JN615139 | |||
| Pedipes pedipes | JN615140 | |||
| Basommatophora | Hygrophila | |||
| Planorbidae | Biomphalaria glabrata | AY380531 | ||
| Lymnaeidae | Radix balthica | HQ330989 | ||
| Galba pervia | JN564796 | |||
| Siphonarioidea | ||||
| Siphonariidae | Siphonaria pectinata | AY345049 | ||
| Siphonaria gigas | JN627205 | |||
| OPISTHOBRANCHIA | ||||
| Aplysiomorpha | ||||
| Aplysiidae | Aplysia californica | AY569552 | ||
| Aplysia dactylomela | DQ991927 | |||
| Sacoglossa | ||||
| Placobranchidae | Elysia chlorotica | EU599581 | ||
| Volvatellidae | Ascobulla fragilis | AY345022 | ||
| Cephalaspidea | ||||
| Hydatinidae | Hydatina physis | DQ991932 | ||
| Acteonidae | Pupa strigosa | AB028237 | ||
| Notaspidea | ||||
| Pleurobranchidae | Berthellina ilisima | DQ991929 | ||
| Nudibranchia | ||||
| Chromodorididae | Chromodoris magnifica | DQ991931 | ||
| Polyceridae | Roboastra europaea | AY083457 | ||
| BASAL HETEROBRANCHIA | ||||
| Pyramidelloidea | ||||
| Pyramidellidae | Pyramidella dolabrata | AY345054 | ||
| CAENOGASTROPODA | ||||
| Neogastropoda | Buccinoidea | |||
| Nassariidae | Ilyanassa obsoleta | DQ238598 | ||
| VETIGASTROPODA | ||||
| Haliotoidea | ||||
| Haliotidae | Haliotis rubra | AY588938 |
Alignment and Divergence
Nucleotide translated sequences for the protein-coding genes from the three mtDNA sequences of C. aspersum, and other Euthyneuran species (Table 1) were aligned using the L-INS-I strategy from MAFFT [37]. Nucleotide alignments were generated using the amino acid alignment as a template using the web-based program TranslatorX [38]. Our taxonomic sampling included representative species of Pulmonata, Opistobranchia, Caenogastropoda and Vetigastropoda clades (Table 1). The last two groups were used as outgroups. The nucleotide and amino acid composition were estimated by Geneious v 4.8.5 [32]. Individual alignments were concatenated prior to phylogenetic analysis. Additionally, intra-specific divergences for each protein-coding gene were calculated based on these alignments. A p-distance method was performed using 1000 bootstrap replications for variance estimation using the program MEGA v.5.1 [31].
Phylogenetic Analyses
Best Partition Scheme (BPS) analyses for the concatenated alignments were conducted with the program PartitionFinder [39], using the Bayesian Information Criterion (BIC) and a heuristic search algorithm. A total of 39 data blocks were defined, following the criteria of one data block for each codon position in each gene. Maximum Likelihood (ML) inference was performed using the graphical interface version (RAxML-GUI, [40]) of RAxML v.7.2.6 software [41]. Maximum likelihood trees were estimate under the GTRGAMMA substitution model and node support was calculated via rapid bootstrapping analyses of 1000 pseudo-replicates. In addition, Bayesian analyses were conducted using MrBayes v.3.2 [42]. Two simultaneous independent runs were performed for 10,000,000 iterations of a Markov Chain Monte Carlo algorithm, with six simultaneous chains, sampling every 1000 generations. The rate parameter was allowed to vary, and the parameter estimation was “unlinked” for the following parameters: the shape of the gamma distribution, the substitution matrix, the proportion of invariable sites, and the estimation of state frequencies. The “temperature” parameter was set at 0.2. Support for the nodes and parameter estimates were derived from a majority rule consensus of the last 5,000 trees sampled after convergence. The average standard deviation of split frequency remained <0.01 after the burn-in threshold.
Ethics Statement
This study did not involve endangered or protected species and was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Comisión Nacional de Investigación Científica y Tecnológica de Chile (CONICYT). All experiments were conducted according to current Chilean law. The protocol was approved by the Committee on the Ethics of Animal Experiments of the Universidad Austral de Chile (Permit Number: 02-2011). Because snails were obtained from public parks and gardens, no specific permissions were required for any of the three locations involved in this study (La Serena, Constitución and Valdivia).
Results
Genome Structural Features
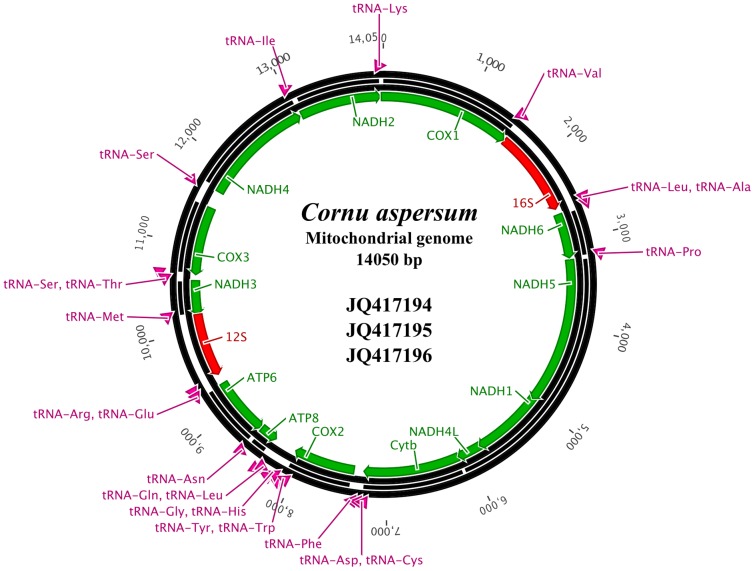
The size of the mitochondrial genome of C. aspersum is 14050 bp (Fig. 2), and contains 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes (Fig. 2). From these 37 genes, 13 are coded on the minus strand: tRNA-Q, tRNA-L2, ATP8, tRNA-N, ATP6, tRNA-R, tRNA-E, 12S-RNA, tRNA-M, NADH3, tRNA-S1, tRNA-T, and COX3 (Fig. 2). The mitochondrial genome of C. aspersum also includes nine small size-variable intergene spacer regions ranging from 2 to 47 bp, and a large AT-rich (81.2%) intergenic region (186 bp) located between COX3 and tRNA-S2 (i.e., Second Serine). This large intergene spacer contains three tandem repeated sequences (ATTATTA, ATTAGTG and TATAAATAT) of 7 bp length, distributed across the non-coding region. There are small gene overlaps at 14 gene borders, the largest has a length of 10 nucleotides and is located between NADH3 and tRNA-S1 (i.e., First Serine). The overall base composition of these mitochondrial genomes shows a high AT content (69.9%; Table 2).
Figure 2. Schematic representation of the mitochondrial genome of Cornu aspersum.
The molecule contains 13 protein-coding genes, two rRNAs and 22 tRNAs. Arrows indicate the transcription direction.
Table 2. Structural features of the Eupulmonata mitochondrial genomes.
| Helicidae | Clausiliidae | Succineidae | Onchidiidae | Trimusculidae | Amphibolidae | Ellobiidea | ||||||||||
| Cornu aspersum | Cepaea nemoralis | Cylindrus obtusus | Euhadra herklotsi | Albinaria caerulea | Succinea putris | Onchidella borealis | Onchidella celtica | Platevindex mortoni | Peronia peronii | Trimusculus reticulatus | Salinator rhamphidia | Myosotella myosotis | Auriculinella bidentata | Ovatella vulcani | Pedipes pedipes | |
| Total size | 14050 | 14100 | 14610 | 14500 | 14130 | 14092 | 14510 | 14150 | 13991 | 13968 | 14044 | 14007 | 14246 | 14135 | 14274 | 16708 |
| %A | 30.8 | 26.2 | 25.8 | 29.5 | 32.8 | 33.8 | 29.3 | 25.3 | 27.3 | 27.1 | 26.4 | 26.7 | 23.7 | 25.8 | 25 | 28.6 |
| %T | 39.1 | 33.6 | 35.8 | 39.9 | 37.9 | 42.9 | 37 | 34.1 | 35.7 | 37.3 | 34.7 | 35.6 | 31.3 | 30.9 | 29.7 | 33.7 |
| %C | 13.6 | 18.9 | 16.6 | 14.1 | 13.8 | 10.8 | 18 | 18.9 | 26.8 | 15.4 | 18.2 | 16.9 | 21.3 | 20.4 | 21.6 | 18.4 |
| %G | 16.5 | 21.3 | 21.9 | 16.4 | 15.5 | 12.1 | 15.7 | 21.8 | 20.2 | 20.3 | 20.6 | 20.8 | 23.6 | 22.6 | 23.7 | 19.3 |
| %A+T | 69.9 | 59.8 | 61.5 | 69.4 | 70.7 | 76.7 | 66.3 | 59.4 | 63 | 64.4 | 61.1 | 62.3 | 55 | 56.7 | 54.7 | 62.3 |
| %G+C | 30.1 | 40.2 | 38.5 | 30.6 | 29.3 | 23.3 | 33.7 | 40.7 | 37 | 35.6 | 38.9 | 37.7 | 45 | 43.3 | 45.3 | 37.7 |
| POR | 186 | 158 | 189 | 43 | 42 | 47 | 41 | 43 | 23 | 54 | 47 | 44 | 46 | 44 | 44 | 397 |
| 12S rRNA | 708 | 710 | 714 | 697 | 759 | 755 | 704 | 708 | 695 | 714 | 719 | 714 | 712 | 704 | 711 | 786 |
| 16S rRNA | 984 | 1215 | 938 | 1024 | 1035 | 1020 | 1289 | 1056 | 1042 | 1033 | 1057 | 1025 | 1089 | 1047 | 1057 | 1138 |
| Cox1 | 1530 (TTG/TAA) | 1492 (TTG/TAA) | 1527 (TTG/TAA) | 1445 (TTG/T ) | 1529 (TTG/TA) | 1548 (TTG/TAG) | 1527 (TTG/TAA) | 1527 (TTG/TAG) | 1494 (ATT/TAA) | 1525 (TTG/T) | 1527 (TTG/TAG) | 1527 (TTG/TAA) | 1527 (ATG/TAA) | 1533 (TTG/TAA) | 1533 (TTG/TAA) | 1527 (TTG/TAA) |
| Cox2 | 664 (ATG/T) | 654 (ATT/TAG) | 687 (ATG/TAG) | 684 (TTG/TAG) | 685 (ATG/TAA) | 649 (ATG/T) | 672 (TTG/TAA) | 681 (TTG/TAA) | 666 (ATG/TAA) | 666 (TTG/TAA) | 666 (TTG/TAA) | 664 (GTG/T) | 669 (GTG/TAA) | 687 (TTG/TAG) | 666 (TTG/TAG) | 681 (GTG/TAA) |
| Cox3 | 780 (ATG/TAA) | 814 (ATA/T) | 813 (ATG/TAA) | 644 (ATG/T)* | 780 (ATG/TAA) | 783 (ATG/TAG) | 778 (ATG/T) | 778 (ATG/T) | 810 (ATA/TAA) | 804 (ATG/T) | 778 (ATG/T) | 781 (ATG/T) | 778 (ATG/T) | 778 (ATG/T) | 778 (ATG/T) | 780 (GTG/TAG) |
| Cytb | 1099 (ATA/T) | 1143 (ATA/T) | 1126 (ATG/T) | 1035 (GTA/T)* | 1103 (ATA/TA) | 1107 (TTG/TAG) | 1108 (TTG/T) | 1122 (ATT/TAA) | 1099 (ATT/T) | 1108 (TTG/T) | 1110 (TTG/TAA) | 1111 (TTG/T) | 1110 (TTG/TAG) | 1111 (TTG/T) | 1110 (TTG/TAA) | 1108 (TTG/T) |
| Nadh1 | 873 (ATG/TAG) | 883 (ATA/T) | 873 (ATA/TAG) | 891 (ATT/TAA) | 900 (ATG/TAA) | 916 (TTG/T) | 906 (TTG/TAA) | 906 (TTG/TAA) | 882 (ATT/TAA) | 906 (TTG/TAA) | 906 (TTG/TAA) | 958 (TTG/T) | 882 (ATT/TAG) | 906 (GTG/TAA) | 906 (TTG/TAA) | 903 (ATG/TAG) |
| Nadh2 | 927 (ATA/TAG) | 946 (ATG/T) | 979 (ATA/T) | 928 (ATG/T) | 924 (ATG/TAA) | 975 (TTG/TAA) | 922 (GTG/TAA) | 922 (ATG/T) | 922 (GTG/T) | 939 (GTG/TAG) | 916 (TTG/T) | 925 (TTG/T) | 948 (ATG/TAG) | 945 (ATG/TAG) | 942 (CTG/T) | 927 (ATG/TAA) |
| Nadh3 | 343 (ATG/T) | 405 (ATG/T) | 357 (ATA/TAA) | 184 (TTA/T)* | 352 (ATA/T) | 352 (ATG/T) | 352 (ATG/TAA) | 352 (ATG/T) | 279 (TTG/TAA) | 327 (ATT/TAA) | 357 (ATG/TAG) | 349 (TTG/T) | 334 (ATA/T) | 354 (TTG/TAA) | 354 (TTG/TAA) | 372 (ATG/TAA) |
| Nadh4 | 1293 (ATG/TAA) | 1252 (ATA/T) | 1329 (ATC/TAA) | 975 (TTG/T)* | 1314 (ATG/TAA) | 1326 (ATG/TAA) | 1305 (ATA/TAA) | 1308 (GTG/TAA) | 1326 (ATG/TAA) | 1318 (TTG/T) | 1306 (TTG/T) | 1311 (TTG/TAG) | 1305 (TTG/TAA) | 1311 (TTG/TAG) | 1308 (TTG/TAG) | 1302 (ATG/TAG) |
| Nadh4L | 264 (ATA/TAG) | 238 (ATA/T) | 250 (ATA/T) | 280 (ATG/T) | 298 (ATG/T) | 275 (ATA/TA) | 274 (ATG/TAA) | 268 (ATG/T) | 288 (ATG/TAA) | 283 (ATG/T) | 286 (ATG/T) | 279 (TTG/TAG) | 291 (TTG/TAA) | 327 (TTG/TAG) | 286 (TTG/T) | 288 (GTG/TAG) |
| Nadh5 | 1677 (ATA/TAA) | 1686 (ATG/TAG) | 1680 (TTG/TAG) | 846 (ATA/T)* | 1638 (ATT/TAG) | 1680 (ATG/TAA) | 1612 (ATC/TAA) | 1641 (GTG/TAG) | 1536 (ATG/TAG) | 1671 (TTG/TAG) | 1680 (TTG/TAG) | 1671 (TTG/TAG) | 1656 (GTG/TAG) | 1665 (ATA/TAG) | 1671 (TTG/T) | 1701 (ATG/TAG) |
| Nadh6 | 489 (TTG/TAA) | 493 (ATT/TA) | 498 (ATA/TAG) | 141 (ATG/T)* | 468 (ATG/TAA) | 453 (ATG/TAA) | 459 (ATG/TAA) | 465 (TTG/TAA) | 468 (ATT/TAA) | 468 (ATT/TAA) | 456 (TTG/TAG) | 474 (TTG/TAA) | 468 (ATA/TAG) | 483 (ATT/TAA) | 480 (ATT/TAG) | 462 (TTG/TAA) |
| ATP6 | 648 (ATG/TAG) | 559 (ATT/T) | 652 (ATG/T) | 246 (ATG/T)* | 634 (ATG/T) | 657 (ATG/TAA) | 643 (ATG/T) | 645 (TTG/TAG) | 645 (ATG/TAA) | 642 (TTG/TAA) | 643 (ATG/T) | 643 (ATG/T) | 641 (ATA/TA) | 643 (GTG/T) | 645 (GTG/TAG) | 642 (ATG/TAG) |
| ATP8 | 166 (ATA/T) | 162 (ATG/TAG) | 159 (GTG/TAG) | 98 (CTT/TAG)* | 168 (ATG/TAG) | 123 (TTG/TAA) | 153 (ATG/TAA) | 147 (ATG/TAA) | 138 (ATT/TAA) | 153 (ATG/TAA) | 186 (ATG/TAG) | 151 (ATG/T) | 151 (ATG/T) | 157 (GTG/T) | 159 (ATG/TAG) | 153 (ATG/TAA) |
| N° tRNAs | 22 | 22 | 22 | 22 | 22 | 22 | 22 | 25 | 24 | 22 | 22 | 22 | 22 | 22 | 22 | 22 |
The size of each genome, gene and the POR are in bp. Start and stop codons for protein-coding genes are indicated in parentheses.
Partial sequences.
Protein-coding Genes, Transfer RNAs, Ribosomal RNA Genes and Genome Organization
The size of the 13 protein-coding genes is similar to those described for other Eupulmonata species (Table 2), and their nucleotide composition reveals low GC content, where the lowest values were estimated for the NADH and ATP genes (Table S1). The most frequent start codon is ATG in six genes, followed by ATA in five genes and TTG in two genes (Table 2). On the other hand, five protein-coding genes (COX1, COX3, NADH4, NADH5, NADH6) use TAA as a stop codon, four (NADH1, NADH2, NADH4L and ATP6) use TAG, and other four (COX2, Cytb, NADH3 and ATP8) use TXX (Table 2).
From the 22 annotated tRNAs 18 were identified with DOGMA [33], whereas the other four (tRNA-S1, tRNA-I, tRNA-K and tRNA-W) were identified with ARWEN v.1.2 [35] and tRNAscan-SE v.1.21 [36]. All tRNAs are spread over the entire genome, and are located on both strands (Fig. 2), and their length varies from 54 to 64 nucleotides. On the other hand, ribosomal RNAs showed similar characteristics to other Pulmonata rRNAs. Nevertheless, the large subunit (16S-rRNA) is shorter than those reported in other Pulmonata species (Table 2), and is encoded on the major strand, whereas the small subunit (12S-rRNA) is encoded on the minor strand (Fig. 2).
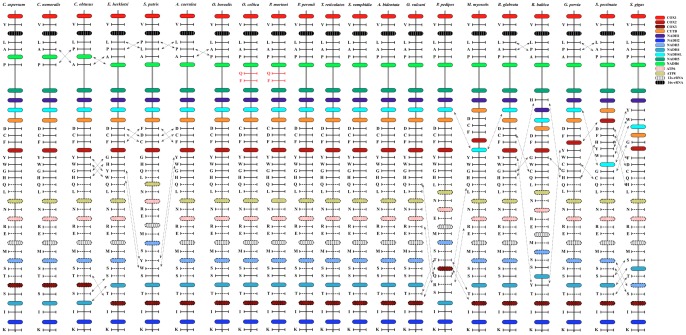
The gene order on the C. aspersum mitochondrial genome is the same as previously described for Cepaea nemoralis (Fig. 3). However, few differences were found when it was compared to C. obtusus, in which two tRNAs (tRNA-A and tRNA-P) are switched (Fig. 3). Similarly, when it was compared to E. herklotsi, S. putris and A. caerulea (i.e., Stylommatophora), some additional switches were detected in nine tRNAs (tRNA-D, tRNA-C, tRNA-F, tRNA-Y, tRNA-W, tRNA-G, tRNA-H, tRNA-T, tRNA-S2) and one protein-coding gene (NADH 4) (Fig. 3). Almost all of these differences are conserved in other Pulmonata species with the exception of tRNA-P that is inverted (Fig. 3).
Figure 3. Linear representation of the gene order and t/rRNA locations in seven Pulmonata mollusc species.
Mitochondrial genomes scaled to 100%. tRNAs are encoded in single letter code according to the amino acid they represent: A, Ala; G, Gly; P, Pro; T, Thr; V, Val; S, Ser; R, Arg; L, Leu; F, Phe; N, Asn; K, Lys; D, Asp; E, Glu; H, His; Q, Gln; I, Ile; M, Met; Y, Tyr; C, Cys; W, Trp). Boxes with shadow represent the reverse direction.
Intra-specific Divergence of C. aspersum mtDNA Genomes
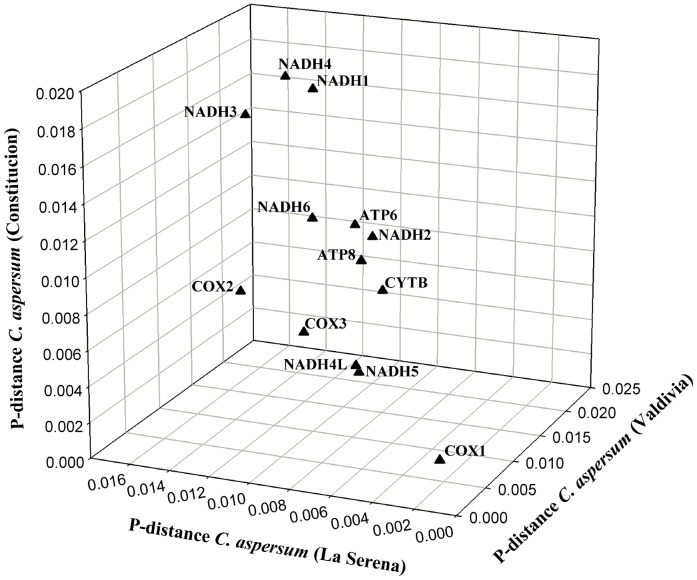
The intra-specific divergence analysis for each of the 13 protein-coding genes revealed that COX1 has the lowest genetic differentiation among the three mitochondrial genomes (p-distance = 0.0019±4×10−4; Mean ± SD). The most divergent genes were NADH1, NADH3 and NADH4 with an average p-distance value of 0.0150. The other nine protein-coding genes showed intermediate values ranging from 0.0055 to 0.0120 (Fig. 4). When the large AT-rich intergenic spacer was included in the analysis, it almost doubled the p-distance values of most divergent protein-coding genes (i.e., p-distance = 0.025±2×10−3; Mean ± SD), showing high intra-specific genetic differentiation among populations.
Figure 4. Intra-specific divergence of mitochondrial protein-coding genes in Cornu aspersum.
P-distance corresponds to the number of base differences per site from averaging over all sequence pairs between genomes.
Phylogenetic Reconstruction
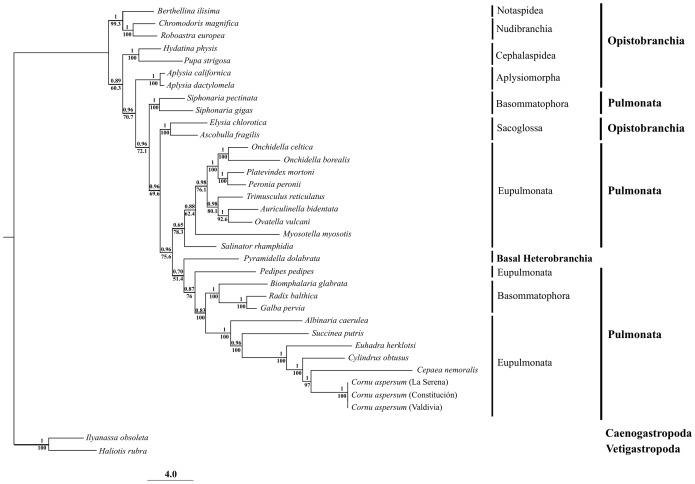
The Best Partition Scheme (BPS) for the concatenated alignment of the 13 protein-coding genes (12573 base positions) had 10 subsets partitions (Table S2). This BPS, and the estimated models of molecular evolution were used for both Bayesian and Maximum Likelihood analyses, which produced identical topologies, where most of the clades were strongly supported (Fig. 5). Phylogenetic reconstruction recovered Basommatophora as polyphyletic group, whereas Eupulmonata and Pulmonata as paraphyletic groups (Fig. 5). Bayesian and Maximum Likelihood analyses both recovered a clade containing the three C. aspersum genomes, with high support (Fig. 5), which in turn shares a most recent common ancestor with Cepaea nemoralis, followed by Cylindrus obtusus and Euhadra herklotsi, supporting the monophy of Helicidae (Fig. 5). Our analysis also supports the sister group relationship between Helicidae and the representative species of the family Succineidae (Succinea putris) and Clausiliidae (Albinaria caerulea), supporting the monophyly of the Stylommatophora clade (Fig. 5).
Figure 5. Maximum likelihood phylogram describing phylogenetic relationships among euthyneuran species (pulmonates and opisthobranchs).
Numbers above the node correspond to Bayesian posterior probabilities, and those below the nodes to maximum likelihood bootstrap support. Sequences from Ilyanassa obsoleta and Haliotis rubra were the outgroup used to root the tree.
Discussion
General Features of the C. aspersum Mitochondrial Genome
In gastropods the length of the mitochondrial genomes usually vary from 13 to 17 kb (e.g., 13670 bp in Biomphalaria glabrata, 17575 bp in Diodora aspera), however, there are some exceptions such as the ribbed limpet Lottia digitalis, with a length of 26835 bp [43]. Here we found that the length of the mitochondrial genome of C. aspersum is similar to the other Eupulmonata species examined (Table 2). Although the mitochondrial genomes of gastropods are very compact [3], they contain intergenic non-coding regions of variable size that are responsible for differences in genome size [16]. In this regard, C. aspersum, C. nemoralis and C. obtusus have an AT-rich intergenic region, that contains the putative origin for mitochondrial DNA replication (POR), which is almost five times longer than other Eupulmonata species. This difference is probably due to insertions of tandem repeats in the POR region [44], which can be caused by gene rearrangements experienced by these species. Accordingly our results show that the POR region of C. aspersum, C. nemoralis and C. obtusus are located between COX3 and tRNA-S1 (i.e., First Serine), whereas in the other Pulmonata species is located between COX3 and tRNA-I (Fig. 3).
The overall AT-content of the mitochondrial genome of C. aspersum is higher in comparison to C. nemoralis and C. obtusus (similar to that one observed in E. herklotsi and A. caerulea) but lower in comparison to S. putris (Table 2). Nevertheless, the average AT-content of the whole Stylommatophora clade is higher in comparison to other Eupulmonata orders (Table 2). The high AT-content is a common feature of most animal mitochondrial genomes [3], and it has been suggested that is the result of directional mutation pressure due to the lower energy requirement for opening the DNA strands with high AT content [45].
Gene Order and Content
Given that the mitochondrial genome of pulmonate snails and slugs contain very little intergenic non-coding sequences and/or gene overlaps [11], [12], gene rearrangements are very rare considering that they would most likely disrupt some of the genes involved, rendering them non-functional. As a result of this, gene order in these species is well conserved [5]. It has been described that tRNA genes are more prone to switch their position than larger protein-coding and rRNA genes [5], [11], [16]. This pattern is observed in our study where C. aspersum has identical gene order and content to C. nemoralis, but both have small differences in comparison to the other stylommatophorans. These differences are explained by the inversions of some tRNA genes, and the NADH4 gene (Fig. 3). Most of these inversions are maintained within Eupulmonata, but comparisons with Basommatophora revealed additional rearrangements that involve two additional tRNAs, and protein-coding genes (COX2 and NADH4L) [4], [16].
Here we found that the size of the protein-coding, tRNA and rRNA genes is conserved within Eupulmonata (Table 2). The start and stop codon of the protein-coding genes in C. aspersum shows similarities to those of the other species analyzed. The presence of incomplete stop codons in C. aspersum seems to occur frequently in protein-coding genes of most of the mollusc mitochondrial genomes sequenced to date [4], [12], [16], [46], and it has been suggested that the transcripts of those genes would be modified to form a complete stop codon via post-transcriptional polyadenylation [47].
Intra-specific Divergence of Mitochondrial Protein-coding Genes in C. aspersum
Intra-specific divergence analysis for each of the 13 protein-coding genes in C. aspersum revealed the existence of genes with high (NADH1, NADH3 and NADH4) intermediate (COX2, COX3, Cytb, NADH2, NADH4L, NADH5, NADH6, ATP6 and ATP8) and low (COX1) degrees of genetic differentiation (Fig. 4). In agreement with our results it has been shown that the COX1 gene is one of the most conserved protein-coding genes in the mitochondrial genome of all metazoans [48], [49], while the most variable are the subunits of the NADH oxidoreductase and ATP synthase complexes [50]–[52]. The relatively low divergence in the mitochondrial genes that belong to the cytochrome c oxidase, the complex that catalyze the transfer of electrons from cytochrome c to oxygen, is most probably explained because this complex is the rate limiting step of the electron transport chain [53].
Phylogenetic Analysis
The Bayesian and Maximum Likelihood phylogenetic tree topologies were identical, and clearly reject the hypothesis that Pulmonata is a monophyletic group (Fig. 5). This result is consistent with previous studies that place Pulmonata as a paraphyletic group within Euthyneura [4], [11], [16]. Additionally, our phylogenetic analysis recovered Basommatophora as a polyphyletic group, whereas Eupulmonata is a paraphyletic group (Fig. 5). The four species of onchidiids (Onchidella celtica, O. borealis, Platevindex mortini and Peronia peronii), were recovered as a highly supported monophyletic group (Fig. 5), sister to a clade containing Trimusculus reticulatus and two species of ellobiids (Auriculinella bidentata and Ovatella vulcani). The other two species of ellobiids (Myostella myosotis and pedipes pedipes) are recovered in two different clades within pulmonata (Fig. 5). Similarly to previous studies, Pyramidella dolabrata was found more closely related to pulmonates than to other Euthyneurans [4], [54]. On the other hand, the clade containing the newly sequenced C. aspersum share a most recent common ancestor with the banded snail C. nemoralis as expected given the current taxonomic classification [55], and also based on previous molecular studies [56], [57]. This clade was recovered as sister to Cylindrus obtusus and Euhadra herklotsi, supporting the monophy of Helicidae. Our analysis also supports the sister group relationship between Helicidae, and representative species from the family Succineidae and Clausiliidae, thus supporting the monophyly of Stylommatophora (land snails and slugs). This is in agreement with previous morphological [14], [55] and molecular studies [4], [11], [16].
Conclusion
Pulmonata mitochondrial genomes display highly conserved structure and composition. However, with the sequencing of C. aspersum some general features of the mitochondrial genomes of Eupulmonata have been elucidated. Particularly, changes in gene order within Helicidae, due to the inversions of some tRNA genes and the NADH4 gene, could be related to the longer size of the POR region in this clade compared to the other Eupulmonata species. The inclusion of the newly published sequence of the mitochondrial genome of the land snail Cylindrus obtusus [58] confirms the aforementioned characteristics for the family Helicidae, and together with C. aspersum, contribute to our understanding of the evolution of mtDNA within Stylommatophora and Pulmonata. Additionally, our phylogenetic results are consistent with the findings of Grande et al. [16] and White et al. [4] about the phylogenetic relationships among gastropods, and the monophyly of the lineage that clusters the land snails and slugs (i.e., Stylommatophora clade). Finally, our intraspecific comparisons revealed that COX1 gene is one of the most conserved protein-coding genes in the mitochondrial genome of C. aspersum, while the most variable are the subunits of the NADH oxidoreductase complex.
Supporting Information
Compositional bias of the % GC content in the 13 protein-coding genes of the Pulmonata mitochondrial genomes used for the phylogenetic reconstruction.
(DOCX)
Best Partition Scheme (BPS) and best-fit models of molecular evolution for the subsets partitions of the mitochondrial protein-coding genes alignment. The likelihood score (lnL) and the Bayesian Information Criterion (BIC) value were -123862 and 248843 respectively.
(DOCX)
Acknowledgments
The authors thank Dr. Alice Nguyen for her critical review of this paper.
Funding Statement
This work was funded by a grant to RFN from the Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 1090423). JDGE was supported by a doctoral fellowship from CONICYT Chile. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Blier PU, Breton S, Desrosiers V, Lemieux H (2006) Functional conservatism in mitochondrial evolution: insight from hybridization of arctic and brook charrs. J Exp Zool Part B 306: 425–432 doi:10.1002/jez.b [DOI] [PubMed] [Google Scholar]
- 2.Scheffler IE (2007) Mitochondria. Wiley-Liss, editor Canada.
- 3. Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. White T, Conrad MM, Tseng R, Balayan S, Golding R, et al. (2011) Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetic relationships. BMC Evol Biol 11: 295 doi:10.1186/1471-2148-11-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kilpert F, Podsiadlowski L (2006) The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genomics 7: 241 doi:10.1186/1471-2164-7-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knudsen B, Kohn AB, Nahir B, Mcfadden CS, Moroz LL (2006) Complete DNA sequence of the mitochondrial genome of the sea-slug, Aplysia californica?: Conservation of the gene order in Euthyneura. Digestion 38: 459–469 doi:10.1016/j.ympev.2005.08.017 [DOI] [PubMed] [Google Scholar]
- 7. Hatzoglou E, Rodakis GC, Lecanidou R (1995) Complete Sequence and Gene Organization of the Mitochondrial Genome of the Land Snail Albinaria coerulea. Genetics 140: 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamazaki N, Ueshima R, Terrett JA, Yokobori SI, Kaifu M, et al. (1997) Evolution of Pulmonate Gastropod Mitochondrial Genomes: Comparisons of Gene Organizations of Euhadra, Cepaea and Albinaria and Implications of Unusual tRNA Secondary Structures. Genetics 145: 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kurabayashi A, Ueshima R (2000) Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: systematic implication of the genome organization. Mol Biol Evol 17: 266–277. [DOI] [PubMed] [Google Scholar]
- 10. Grande C, Templado J, Cervera JL, Zardoya R (2002) The Complete Mitochondrial Genome of the Nudibranch Roboastra europaea (Mollusca?: Gastropoda ) Supports the Monophyly of Opisthobranchs. Mol Biol Evol 19: 1672–1685. [DOI] [PubMed] [Google Scholar]
- 11. Grande C, Templado J, Cervera JL, Zardoya R (2004) Molecular phylogeny of Euthyneura (Mollusca: Gastropoda). Mol Biol Evol 21: 303–313 doi:10.1093/molbev/msh016 [DOI] [PubMed] [Google Scholar]
- 12.Simison WB, Boore JL (2008) Molluscan Evolutionary Genomics. In: Ponder W, Lindberg D, editors. Phylogeny and Evolution of the Mollusca. Berkeley: University of California Press. 447–461.
- 13. Dayrat B, Conrad M, Balayan S, White T, Albrecht C, et al. (2011) Phylogenetic relationships and evolution of pulmonate gastropods (Mollusca): new insights from increased taxon sampling. Mol Phylogenet Evol 59: 425–437 doi:10.1016/j.ympev.2011.02.014 [DOI] [PubMed] [Google Scholar]
- 14.Dayrat B, Tillier S (2002) Evolutionary relationships of euthyneuran gastropods (Mollusca): a cladistic re-evaluation of morphological characters. Zool J Linn Soc-Lond: 403–470.
- 15.McArthur A, Harasewych M (2003) Molecular systematics of the major lineages of the Gastropoda. In: Lydeard C, Lindberg D, editors. Molecular Systematics and Phylogeography of Mollusks. Washington: Smithsonian Books. 140–160.
- 16. Grande C, Templado J, Zardoya R (2008) Evolution of gastropod mitochondrial genome arrangements. BMC Evol Biol 8: 61 doi:10.1186/1471-2148-8-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wade CM, Mordan PB (2000) Evolution within the gastropod molluscs?; using the ribosomal RNA gene-cluster as an indicator of phylogenetic relationships. J Mollus Stud 66: 565–570. [Google Scholar]
- 18. Klussmann-Kolb A, Dinapoli A, Kuhn K, Streit B, Albrecht C (2008) From sea to land and beyond–new insights into the evolution of euthyneuran Gastropoda (Mollusca). BMC Evol Biol 8: 57 doi:10.1186/1471-2148-8-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arnaud JF, Madec L, Guiller A, Deunff J (2003) Population genetic structure in a human-disturbed environment?: a case study in the land snail Helix aspersa (Gastropoda?: Pulmonata ). Heredity 90: 451–458 doi:10.1038/sj.hdy.6800256 [DOI] [PubMed] [Google Scholar]
- 20. Peltanová A, Petrusek A, Kment P, Juřičková L (2012) A fast snail’s pace: colonization of Central Europe by Mediterranean gastropods. Biol Invasions 14: 759–764 doi:10.1007/s10530-011-0121-9 [Google Scholar]
- 21. Guiller A, Coutellec-Vreto MA, Madec L, Deunff J (2001) Evolutionary history of the land snail Helix aspersa in the Western Mediterranean: preliminary results inferred from mitochondrial DNA sequences. Mol Ecol 10: 81–87. [DOI] [PubMed] [Google Scholar]
- 22.Barker GM (2001) The Biology of Terrestrial Molluscs. Hamilton, New Zealand: Landcare Research Hamilton.
- 23.Roth B, Sadeghian P (2006) Checklist of the land snails and slugs of California. vol. 3. Santa Barbara: Santa Barbara Museum of Natural History Contributions in Science.
- 24. Guiller A, Madec L (2010) Historical biogeography of the land snail Cornu aspersum: a new scenario inferred from haplotype distribution in the Western Mediterranean basin. BMC Evol Biol 10: 18–37 doi:10.1186/1471-2148-10-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naya DE, Catalán T, Artacho P, Gaitán-Espitia JD, Nespolo RF (2011) Exploring the functional association between physiological plasticity, climatic variability, and geographical latitude: lessons from land snails. Evol Ecol Res 13: 1–13. [Google Scholar]
- 26. Artacho P, Silva A, Poulin E, Nespolo RF (2006) Phenotypic and genetic variability in a latitudinal gradient in Populations of the garden land snail, Helix aspersa, in Chile. Integr Comp Biol 46: E167–167 doi:10.1093/icb/icl057 [Google Scholar]
- 27. Gaitán-Espitia JD, Scheihing R, Poulin E, Artacho P, Nespolo RF (2013) Mitochondrial phylogeography of the land snail Cornu aspersum?: tracing population history and the impact of human-mediated invasion in austral South America. Evol Ecol Res 15: 1–18. [Google Scholar]
- 28. Gaitán-Espitia JD, Bruning A, Mondaca F, Nespolo R (2013) Intraspecific variation in the metabolic scaling exponent in ectotherms: Testing the effect of latitudinal cline, ontogeny and transgenerational change in the land snail Cornu aspersum. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 165: 169–177 doi:10.1016/j.cbpa.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 29. Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, et al. (2000) A Whole-Genome Assembly of Drosophila. Science 287: 2196–2204 doi:10.1126/science.287.5461.2196 [DOI] [PubMed] [Google Scholar]
- 30.Madison W, Madison D (2005) MacClade 4: Analysis of phylogeny and character evolution. [DOI] [PubMed]
- 31. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, et al.. (2010) Geneious.
- 33. Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics (Oxford, England) 20: 3252–3255 doi:10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 34. Altschul S, Madden T, Schäffer A, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laslett D, Canbäck B (2008) ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24: 172–175. [DOI] [PubMed] [Google Scholar]
- 36. Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–689 doi:10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Abascal F, Zardoya R, Telford M (2010) TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38: W7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701 doi:10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 40.Silvestro D, Michalak I (2011) RaxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution: doi:10.1007/s13127-011-0056-0
- 41. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22: 2688–2690 doi:10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 42. Ronquist F, Huelsenbeck J (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 43. Simison WB, Lindberg DR, Boore JL (2006) Rolling circle amplification of metazoan mitochondrial genomes. Mol Phylogenet Evol 39: 562–567 doi:10.1016/j.ympev.2005.11.006 [DOI] [PubMed] [Google Scholar]
- 44. Ray D, Densmore L (2002) The crocodilian mitochondrial control region: general structure, conserved sequences, and evolutionary implications. J Exp Zool 294: 334–345 doi:10.1002/jez.10198 [DOI] [PubMed] [Google Scholar]
- 45. Wirth T, Le Guellec R, Veuille M (1999) Directional substitution and evolution of nucleotide content in the Cytochrome Oxidase II gene in earwigs (Dermapteran insects). Mol Biol Evol 16: 1645–1653. [DOI] [PubMed] [Google Scholar]
- 46. Dreyer H, Steiner G (2004) The complete sequence and gene organization of the mitochondrial genome of the gadilid scaphopod Siphonondentalium lobatum (Mollusca). Mol Phylogenet Evol 31: 605–617 doi:10.1016/j.ympev.2003.08.007 [DOI] [PubMed] [Google Scholar]
- 47. Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474 doi:10.1038/290470a0 [DOI] [PubMed] [Google Scholar]
- 48.Brown WM (1985) The mitochondrial genome of animals. In: MacIntyre R, editor. Molecular Evolutionary Genetics. New York, USA: Plenum. 95–130.
- 49. Carlini D, Graves J (1999) Phylogenetic analysis of cytochrome c oxidase I sequences to determine higher-level relationships within the coleoid cephalopods. Bulletin of Marine Science 64: 57–76. [Google Scholar]
- 50. Pesole G, Gissi C, De Chirico A, Saccone C (1999) Nucleotide substitution rate of mammalian mitochondrial genomes. J Mol Evol 48: 427–434. [DOI] [PubMed] [Google Scholar]
- 51. Oliveira D, Raychoudhury R, Lavrov D, Werren J (2008) Rapidly Evolving Mitochondrial Genome and Directional Selection in Mitochondrial Genes in the Parasitic Wasp Nasonia (Hymenoptera?: Pteromalidae). Mol Biol Evol 25: 2167–2180 doi:10.1093/molbev/msn159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Burzyn A, Wenne R (2010) Comparative genomics of marine mussels (Mytilus spp.) gender associated mtDNA: rapidly evolving atp8. J Mol Evol 71: 385–400 doi:10.1007/s00239-010-9393-4 [DOI] [PubMed] [Google Scholar]
- 53. Villani G, Attardi G (2000) In vivo control of respiration by cytochrome c oxidase in human cells. Free Radical Biol Med 29: 202–210. [DOI] [PubMed] [Google Scholar]
- 54. Jörger KM, Stöger I, Kano Y, Fukuda H, Knebelsberger T, et al. (2010) On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evol Biol 10: 323 doi:10.1186/1471-2148-10-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barker GM (2001) Gastropods on land: phylogeny, diversity and adaptive morphology. In: Barker G, editor. The Biology of Terrestrial Molluscs. Wallingford: CABI Publishing. 1–146.
- 56. Wade CM, Mordan PB, Clarke B (2001) A phylogeny of the land snails (Gastropoda?: Pulmonata). Proceedings of The Royal Society B 268: 413–422 doi:10.1098/rspb.2000.1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wade CM, Mordan PB, Naggs F (2006) Evolutionary relationships among the Pulmonate land snails and slugs (Pulmonata, Stylommatophora ). Biol J Linn Soc 87: 593–610. [Google Scholar]
- 58. Groenenberg D, Pirovano W, Gittenberger E, Schilthuizen M (2012) The complete mitogenome of Cylindrus obtusus (Helicidae, Ariantinae) using Illumina Next Generation Sequencing. BMC Genomics 13: 114 doi:10.1186/1471-2164-13-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Compositional bias of the % GC content in the 13 protein-coding genes of the Pulmonata mitochondrial genomes used for the phylogenetic reconstruction.
(DOCX)
Best Partition Scheme (BPS) and best-fit models of molecular evolution for the subsets partitions of the mitochondrial protein-coding genes alignment. The likelihood score (lnL) and the Bayesian Information Criterion (BIC) value were -123862 and 248843 respectively.
(DOCX)