Abstract
The aim of this work was to investigate the association between polymorphisms located at the HSP90AA1 ovine gene promoter and gene expression rate under different environmental conditions, using a mixed model approach. Blood samples from 120 unrelated rams of the Manchega sheep breed were collected at three time points differing in environmental conditions. Rams were selected on the basis of their genotype for the transversion G/C located 660 base pairs upstream the gene transcription initiation site. Animals were also genotyped for another set of 6 SNPs located at the gene promoter. Two SNPs, G/C−660 and A/G−444, were associated with gene overexpression resulting from heat stress. The composed genotype CC−660-AG−444 was the genotype having the highest expression rates with fold changes ranging from 2.2 to 3.0. The genotype AG−522 showed the highest expression levels under control conditions with a fold change of 1.4. Under these conditions, the composed genotype CC−601-TT−524-AG−522-TT−468 is expected to be correlated with higher basal expression of the gene according to genotype frequencies and linkage disequilibrium values. Some putative transcription factors were predicted for binding sites where the SNPs considered are located. Since the expression rate of the gene under alternative environmental conditions seems to depend on the composed genotype of several SNPs located at its promoter, a cooperative regulation of the transcription of the HSP90AA1 gene could be hypothesized. Nevertheless epigenetic regulation mechanisms cannot be discarded.
Introduction
Current concern about global warming and its effects over agricultural and livestock production systems have opened novel scientific opportunities to study the adaptation of organisms to new and harsher environmental conditions. In this context, the study of the genetic basis of traits linked with adaptation and fitness has great importance. Heat is one of the main sources of stress which has an important impact in livestock production. The genetic variability underlying animal’s thermo tolerance could be exploited in livestock breeding programs to achieve animals that could cope with the effects of heat stress over productive and functional traits. Among the livestock animals, sheep (Ovis aries), is one of the oldest domesticated species [1]. It is widely distributed throughout the world due to its high plasticity and adaptability to withstand poor nutrient diets and tolerance to extreme climatic conditions [2], [3]. Sheep is thus an interesting biological material to study the genetic basis of thermo-tolerance. There is some literature about heat stress effects over physiological and productive traits in cattle [4], [5], [6] and sheep [7], [8], [9], [10]. Also, at the molecular level, genes involved in the heat stress response have been described [11], [12], [13], [14]. Among them, those encoding heat shock proteins have been the most studied. However, in sheep there are few works regarding this topic.
Heat shock proteins (HSPs) [15], play a fundamental role in the maintenance of cellular homeostasis, under both physiological and stress conditions [16]. HSPs are organized into several families according to their molecular size (kDa). HSPs are highly conserved across species [17], particularly the 90 kDa heat shock protein (HSP90) [18]. In eukaryotes there are two major isoforms of HSP90 constituted by gene duplication [19]: the inducible form, HSP90α and the constitutive form, HSP90β. The HSP90AA1 ovine gene (DQ983231) which encodes the HSP90α protein has been sequenced, mapped and characterized in sheep by Marcos-Carcavilla et al. [20]. In their work 34 polymorphisms (12 in the coding region, 14 in the promoter -Figure S1- and 8 in the intron 10) were detected. Further on, also a new INDEL (insertion/deletion) was observed at the promoter [21] (Figure S1). The transversion G/C located at position −660 in the gene promoter was associated with resistance/susceptibility to scrapie [22], with sperm DNA fragmentation in rams [23] and with the adaptation pattern of different sheep breeds to the thermal conditions in where they are reared [24]. In this last study, this polymorphism was associated with differences in the transcription rate of the HSP90AA1 gene, being either the causal mutation which putatively modifies a transcription factor binding site or in linkage disequilibrium with the causal mutation. However, the study was based on a limited number of animals and on standard basic statistical methods used to analyze quantitative real-time PCR (qPCR) data and to asses differences in expression levels of alternative genotypes under control and heat stress conditions.
In general, qPCR has provided a powerful tool for quantifying gene expression. Nevertheless, it makes necessary to carefully consider some technical and analytical factors to ensure reproducible and accurate measurements and not lead to misinterpretations [25]. However, commonly, several of these essential procedures have been widely ignored. Those technical factors include the initial sample amount, RNA recovery and RNA purity and integrity [26], [27] among others. Some factors considered as analytical are the selection of the suitable housekeeping gene(s) (HK), the experimental design [28], the statistical method used, etc.
Traditional statistical analyses have been restricted to pair-wise comparisons of treatments in which Cq (quantification cycle) values of GOIs (genes of interest) were previously normalized using standard HKs. This kind of approach does not allow to include technical and biological effects having influence over gene expression data. The joint analysis of GOIs and HKs data can lead to a better partition of such sources of variation [29] and allows checking HK stability and subsequent normalization of GOIs. Mixed model methodology makes possible this kind of approach, giving the possibility of including systematic and random effects, and interactions among them. They constitute a powerful tool in qPCR analyses including more than two treatments and multiple experimental factors [30], [31].
The objectives of this study were to 1) confirm gene expression differences observed for alternative genotypes of the G/C−660 transversion of the HSP90AA1 gene promoter using more animals and a wider range of climatic conditions that those in [12]; 2) study the effect over the expression levels of another six polymorphisms (A/C−601, G/A−528, G/T−524, A/G−522, G/T−468 and A/G−444) selected on the basis of their population genotype frequencies (Table S1) and relative position regarding SNP G/C−660; and 3) to simultaneously select the best HK and analyze expression data using a mixed model statistical approach that includes technical and biological sources of variation.
Materials and Methods
Ethics Statement
The current study was carried out under a Project License from the INIA Scientific Ethic Committee. Animal manipulations were performed according to the Spanish Policy for Animal Protection RD 53/2013, which meets the European Union Directive 86/609 about the protection of animals used in experimentation. We hereby confirm that the INIA Scientific Ethic Committee, which is the named IACUC for the INIA, specifically approved this study.
Animals belong to an artificial insemination centre, were raised in small groups in different barns and fed according to their necessities.
Linkage Disequilibrium Analysis and Haplotype Determination
Animal material, nucleic acid isolation, DNA amplification and SNPs genotyping
Peripheral whole blood samples were collected from 103 animals of the Manchega Spanish sheep breed in order to analyse linkage disequilibrium among the 7 SNPs of interest located at the HSP90AA1 promoter. Animals were grouped in 48 parent-offspring trios. Trios consist of 10 sires, 48 dams and 1 offspring per pair (all females). Three of the dams were also daughters from another trio. Genomic DNA was extracted from lymphocytes according to the salting out procedure [32]. The polymerase chain reaction was performed from 100 ng of genomic DNA using CERTAMP complex amplifications kit chemistry (Biotools, Madrid, Spain) with specific primers (Forward: 5′CGAGGCTCTGGCAGGCACTTGTTG3′ and Reverse: 5′ GCCGCCGTTCCCA GCCCTACCT 3′). A 499 bp fragment of the promoter containing SNP−660 and 6 more SNPs (−601, −528, −524, −522, −468, −444) was obtained. The resulting PCR fragment was purified with ExoSAP-IT (USB Corporation, OH, USA) and sequenced with specific primers (shown above).
Linkage disequilibrium estimation
PLINK software [33] was used to estimate linkage disequilibrium among all pairs of the 7 SNPs measured as r2, the squared correlation based on genotypic allele counts [34]. Hardy-Weinberg equilibrium exact test and observed and expected heterozygosities for each SNP were also calculated using PLINK.
Detection of Putative Transcription Factor (TF) Binding Sites in Ovine HSP90AA1 Promoter
Putative TF binding sites were predicted using TESS [35] (keeping default settings) and ALGGEN-PROMO [36], [37] (limiting to mammal transcription factors) softwares.
Expression Analysis
Animal material
In order to confirm the association of the HSP90AA1 polymorphism (G/C−660) previously associated with the adaptation to different thermal conditions in sheep previously described [24], 428 unrelated rams of Manchega Spanish sheep breed were genotyped (same protocol and primers as described in [24]). All animals belonged to an artificial insemination centre, and therefore they were reared under the same environmental and management conditions. A total of 120 out of 428 rams were selected based on their genotype: 40 CC−660, 40 GC−660 and 40 GG−660. Genomic DNA from these 120 animals was used to genotype the previosly defined 499 bp amplicon of the HSP90AA1 promoter. Genotype frequencies are shown in Table 1.
Table 1. Genotype frequencies of the SNPs located at the HSP90AA1 promoter in 120 rams of Manchega sheep breed.
| SNPs | ||||||||
| −660 | −601 | −528 | −524 | −522 | −468 | −444 | N | Freq.% |
| CC | AC | AA | GT | GG | GT | GG | 2 | 1.63 |
| CC | AC | AA | TT | GG | TT | GG | 1 | 0.81 |
| CC | AC | AG | GT | GG | GT | GG | 1 | 0.81 |
| CC | CC | AA | GT | GG | GT | GG | 1 | 0.81 |
| CC | CC | AA | TT | AG | TT | GG | 3 | 2.44 |
| CC | CC | AA | TT | GG | GT | GG | 1 | 0.81 |
| CC | CC | AA | TT | GG | TT | AA | 1 | 0.81 |
| CC | CC | AA | TT | GG | TT | AG | 1 | 0.81 |
| CC | CC | AA | TT | GG | TT | GG | 26 | 21.14 |
| CC | CC | GA | TT | AG | TT | GG | 2 | 1.63 |
| CC | CC | GA | TT | GG | TT | GG | 2 | 1.63 |
| CC | CC | GG | TT | GG | TT | GG | 1 | 0.81 |
| GC | AC | AA | GT | GG | GT | GG | 1 | 0.81 |
| GC | AC | GA | GT | GG | GT | GG | 4 | 3.25 |
| GC | CC | AA | TT | GG | TT | AG | 1 | 0.81 |
| GC | CC | AA | TT | GG | TT | GG | 2 | 1.63 |
| GC | CC | GA | GT | GG | GT | GG | 1 | 0.81 |
| GC | CC | GA | TT | AG | TT | GG | 5 | 4.07 |
| GC | CC | GA | TT | GG | TT | GG | 21 | 17.08 |
| GC | CC | GG | TT | GG | TT | GG | 4 | 3.25 |
| GG | CC | AA | TT | GG | TT | AG | 1 | 0.81 |
| GG | CC | AA | TT | GG | TT | GG | 1 | 0.81 |
| GG | CC | GA | TT | GG | TT | AG | 2 | 1.63 |
| GG | CC | GA | TT | GG | TT | GG | 9 | 7.32 |
| GG | CC | GG | TT | GG | TT | GG | 29 | 23.58 |
Peripheral whole blood samples from the 120 rams were collected in 3 time points, corresponding to different climatic conditions in a dry region of central Spain (Ciudad Real). The 3 time points were in March, when environmental temperature conditions are mild, and in July and August when heat stress temperatures occur. Hereafter, we will refer to the March collection as the control. The temperature humidity index (THI) equation proposed by Marai et al. [8] was used as another indicator of thermal stress. This index combines both temperature and relative humidity. The enviromental parameters for the 3 points in time are shown in Table 2.
Table 2. Climate parameters at day of samples collection, and average values of the five days previous to samples collection.
| Time points | Treatment name | AvT | MaT | MiT | Rh/100 | Rhmax/100 | THIavr | THImax | AvT-5daysBC | MaT-5daysBC | Rh-5daysBC/100 | Rhmax-5daysBC/100 | THIavr-5daysBC | THImax-5daysBC |
| 23/03/2010 | control | 11.6 | 19.9 | 3.8 | 0.69 | 0.93 | 11.87 | 19.77 | 12.7 | 19.1 | 0.70 | 0.89 | 12.86 | 18.95 |
| 05/07/2010 | July | 26.8 | 35.0 | 16.8 | 0.39 | 0.64 | 24.47 | 32.69 | 25.2 | 33.5 | 0.49 | 0.83 | 23.49 | 32.49 |
| 03/08/2010 | August | 24.7 | 34.4 | 16.6 | 0.49 | 0.89 | 23.08 | 33.74 | 27.2 | 36.4 | 0.39 | 0.72 | 24.78 | 34.52 |
From: Manzanares (Ciudad Real) Meteorological Station, coordinates 654m-38° 59′47N-03° 22′23W (http://crea.uclm.es/siar).
AvT = average temperature (°C).
MaT = maximum temperature (°C).
MiT = minimum temperature (°C).
Rh/100 = relative humidity (%)/100.
Rhmax/100 = maximum relative humidity (%)/100.
THIavr = THI calculated with the average temperature and relative humidity.
THImax = THI calculated with the maximum temperature and the maximum relative humidity.
AvT-5daysBC = Average mean temperature of the five days previous to collection (°C).
MaT-5daysBC = Average maximum temperature of the five days previous to collection (°C).
Rh-5daysBC/100 = Average relative humidity of the five days previous to collection (%)/100.
Rhmax-5daysBC/100 = Average maximum relative humidity of the five days previous to collection (%)/100.
THIavr-5daysBC = THI calculated with the average temperature and relative humidity of the 5 days before collection.
THImax-5daysBC = THI calculated with the average maximum temperature and maximum relative humidity of the 5 days before collection.
Temperature humidity index (THI) calculated as THI = T°C – ((0.31–0.31RH) (T°C-14.4). T = temperature in °C; RH = relative humidity in %/100 [8].
Total RNA isolation and cDNA synthesis
Total RNA was isolated from 9 ml of whole blood using the LeukoLock kit (Ambion, Inc., TX, USA), following manufacturers instructions. RNA concentration was determined using a NanoDrop ND-1000 UV/Vis spectrophotometer (Nanodrop Technologies, Inc., DE, USA). Degradation of RNA samples was assessed with the Agilent 2100 bionalyzer (Agilent Technologies Hewlett-Packard-Str.8 76337 Waldbronn, Germany) in RNA Nano Chips, following manufacturers instructions. RIN (RNA Integrity Number) values were obtained. cDNA was synthesized using the ImProm-II™ Reverse Transcription System (Promega Corp., WI, USA).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
qRT-PCR was performed on all samples collected. Three HKs were tested, MDH1, SDHA and HSP90AB1. MDH1 and SDHA became the most stable HK pair for the heat stress response in sheep under similar conditions [38]. Also the HSP90AB1 gene was included as HK candidate since its expression is ubiquitous, less inducible and more constitutive than that of the HSP90AA1 gene [39], [40]. Primers were designed with NetPrimer software (Biosoft International, CA, USA), and are listed in Table 3 together with amplicon sizes and CG content. Primers were designed avoiding possible genomic DNA amplifications. In silico specificity of the amplicons was screened by BLAST searches.
Table 3. Primers and efficiencies of the qPCR reactions.
| Gene | Forward primer (5′-3′) Reverse primer (5′-3′) | Ampliconsize (bp) | Efficiencies | Amplicon % bases and GC content |
| HSP90AA1 | CCACTTGGCGGTCAAGCATT AAGGAGCTCGTCTTGGGACAA | 80 | 1.951 | A/22.50 G/25.00 C/31.25 T/21.25GC content 47.50 |
| MDH1 | GGTCAAATTGCATATTCACTACTA ACCATCCAGGACACCCATCAT | 117 | 1.883 | A/23.07 G/20.51 C/32.48 T/23.93GC content 43.58 |
| SDHA | GGCATCCCCACCAACTACA TACACCACCTCAAAGCCCCG | 134 | 2.000 | A/35.55 G/29.62 C/17.03 T/17.77GC content 65.17 |
| HSP90AB1 | TACATCACTGGTAAGAGCAAAGA TACACCACCTCAAAGCCCCG | 81 | 1.950 | A/37.03 G/18.52 C/22.22 T/22.22GC content 55.55 |
qPCR amplification reactions were performed from 100 ng of cDNA using LightCycler® 480 SYBR Green I Master kit (Roche, Switzerland). Reactions were run in triplicate on a LightCycler® 480 (Roche, Switzerland) following manufacturer’s cycling parameters. Dissociation curves were performed for each gene to check primer specificity and to confirm the presence of a unique PCR product. The corresponding mRNA levels were measured and analyzed by their Cq.
To estimate PCR efficiencies, standard curves based on 6 serial dilutions (1/20 from a departure concentration of 50 ng/µl) of a cDNA stock (a cDNA mixture of more than 121 samples accounting for the 3 genotypes and the 3 time points) were performed. Efficiencies (E) were calculated from the slope of curves as in Rasmussen and coworkers’ [41]. Estimated E for each gene are shown in Table 3.
Statistical Procedures
Statistical analysis of RIN values
A mixed model was fitted by using the MIXED procedure of the SAS statistical package [42] for determining factors affecting RIN values. RIN value of all samples were included as a dependent variable. Fixed effects included were genotype G/C−660 (G) - 3 levels: CC, GC and GG -; date of collection (D) - 3 levels: Control, July and August -; group of sample processing (GP) - 4 levels corresponding to the barn where a group of animals were located and sampled - and the interaction date of collection x group of sample processing (DxGP) were included as fixed effects. The barn needs to be included because it is related to the period of time between samples collection and processing. The animal (A) was included as random effect. Goodness of fit statistics AIC (Akaike’s Information Criterion) and BIC (Schwarz’s Bayesian Criterion) were used as criteria for model selection. A type III fixed effects test was used to determine significance of the effects included in the model. P<0.05 was established as threshold for statistical significance.
HK selection
HK selection among HSP90AB1, MDH1 and SDHA genes followed the strategy from Serrano et al. [38]. As amplification efficiencies of some genes were <2 (<100%), Cq data were transformed using the equation proposed by Steibel et al. [29] to rescale Cq values.
The equation of the mixed model used was the following:
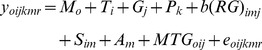 |
(1) |
where yoijkmr is the transformed Cq data of the jth gene, from the rth well, in the kth plate, collected from de mth animal under the ith treatment; Mo is the fixed effect of the oth genotype; Ti is the fixed effect of ith treatment; Gj is the fixed effect of the jth gene; Pk is the effect of the kth plate; b(RG)imnj is the interaction between the RIN value of the mith sample and the jth gene, where b is the regression coefficient of RIN x gene variable on Cq; Sim is the random effect of the biological sample 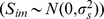 ; Am is the random effect of the animal from where samples were collected
; Am is the random effect of the animal from where samples were collected 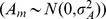 ; MTGoij is the random interaction effect among the oth genotype, the ith treatment and the jth gene
; MTGoij is the random interaction effect among the oth genotype, the ith treatment and the jth gene 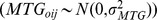 ; eoijkmr is the random residual. Gene specific residual variance (heterogeneous residual) was fitted to the gene by treatment effect
; eoijkmr is the random residual. Gene specific residual variance (heterogeneous residual) was fitted to the gene by treatment effect 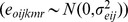 .
.
Expression stability values were obtained by calculating the Mean Square Error (MSE), which was defined as in [38].
Analysis of expression results
Statistical analysis of gene expression was carried out following the method proposed by Steibel et al. [29]. As amplification efficiencies for HSP90AA1, HSP90AB1 and MDH1 genes were <2, Cq data were transformed as aforementioned. The mixed model fitted was:
| (2) |
where effects were as in model 1, except that in this case the MTG factor was included in the model as fixed effect and the residual variance was heterogeneous for the gene effect) .
To test differences, diffGOI, in the expression rate of alternative genotypes and to obtain fold change (FC) values from the estimated MTG differences, the approach suggested in [29] was used. Significance of diffGOI estimates was determined with the t statistic.
Also asymmetric 95% confidence intervals (up and low) were calculated for each FC value by using the standard error (SE) of diffGOI:
| (3) |
| (4) |
Only contrasts between genotypes expression data that remained significant after the Holm-Bonferroni correction and with a FC >1 are going to be discussed. Supplementary Tables (Tables S2, S3 and S4) show estimates, standard errors, FC and confidence intervals (FCup-FClow) of significant contrasts between genotypes in each treatment. Also FCs are graphically represented in Figures 1, 2 and 3 where segments indicate 95% confidence interval.
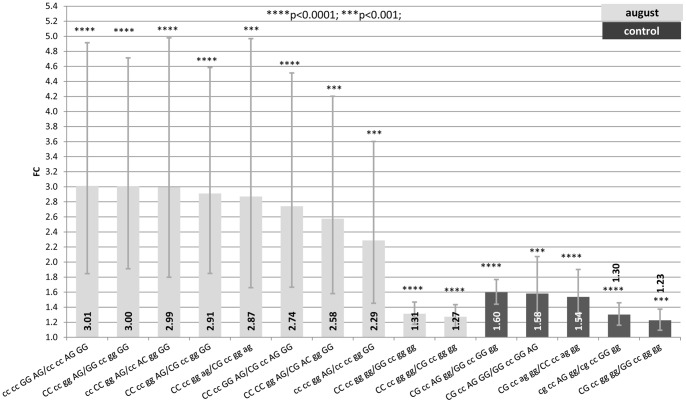
Figure 1. Fold change (FC) for the contrast among alternative genotypes G/C−660-C/A−601-G/A−522-A/G−444 of the HSP90AA1 promoter within each treatment (Control, July and August) normalized by HSP90AB1.
Segments indicate the 95% confidence interval (FCup-FClow). In abscissa the FC, in ordinate genotype contrasts. Asterisk over each bar indicates the significance level of the contrasts.
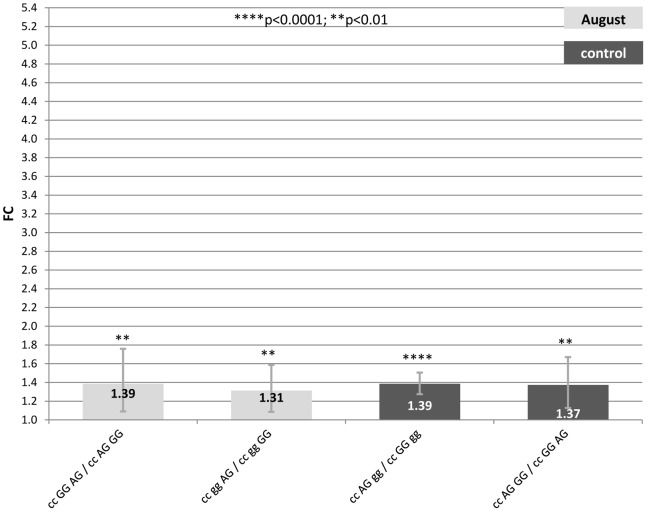
Figure 2. Fold change (FC) for the contrasts among alternative genotypes A/C−601-G/A522-A/G−444 of the HSP90AA1 promoter within each treatment (Control, July and August) normalized by HSP90AB1.
Segments indicate the 95% confidence interval (FCup-FClow).In abscissa the FC, in ordinate genotype contrasts. Asterisk over each bar indicates the significance level of the contrasts.
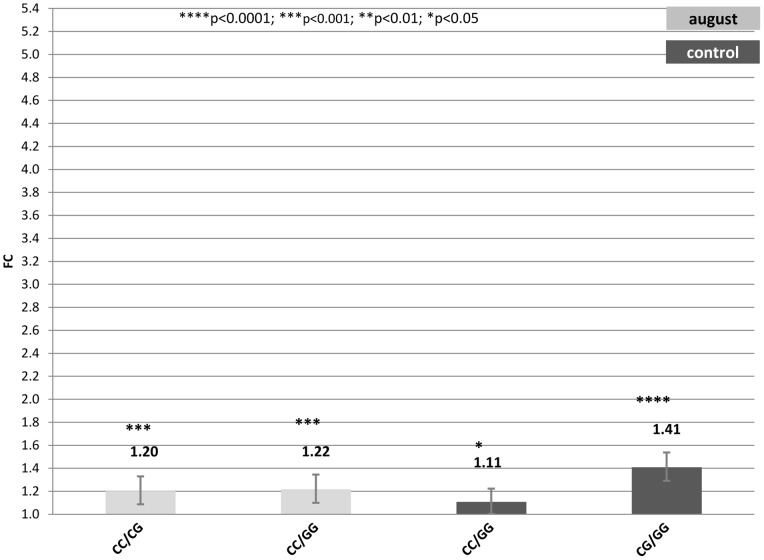
Figure 3. Fold change (FC) for the contrast among alternative genotypes G/C−660 of the HSP90AA1 promoter within each treatment (Control, July and August) normalized by HSP90AB1.
Segments indicate the 95% confidence interval (FCup-FClow).In abscissa the FC, in ordinate genotype contrast. Asterisk over each bar indicates the significance level of the contrasts.
Results
Linkage Disequilibrium Analysis
Results from Hardy-Weinberg equilibrium exact test and expected and observed heterozygosities are shown in Table S5. No deviations from the Hardy-Weinberg equilibrium were observed for any of the SNPs genotyped on population composed by trios. The average expected and observed heterozygosities were 0.256 and 0.306, respectively. The SNPs with the lowest allele frequencies were A/G−522 and A/G−444 (0.018 and 0.053, respectively).
Results from linkage disequilibrium analyses are shown in Table S6. The analysis revealed the existence of two blocks of SNPs segregating jointly. One block was constituted by the SNPs at positions −660 and −528 (r2 = 0.95), and the other was integrated by the SNPs mapped at −601, −524 and −468 (r2 between them ranged from 0.92 to 1.00). Linkage disequilibrium between other SNP pairs showed values lower than 0.06, ranging from 0.001 to 0.059.
Statistical Analysis of RIN Values
The model including the interaction DxGP as a fixed effect and the animal as a random effect, showed the lowest values for the goodness of fit criteria (AIC and BIC). Estimated animal and residual variance were 0.11 and 2.25, respectively. Type III fixed effects test showed a highly significant (p<0.0001) effect of DxGP on RIN values but no significant effect were observed for G, D and GP on the trait.
Thus, as RIN values only depend on the order in which samples were processed after their collection, it can be included as a systematic effect in the statistical model used to analyse expression data.
Best HKs
Given the linkage disequilibrium results for the 7 SNPs, the “genotype” included as an effect in the mixed model to test the stability of genes was G/C−660-A/C−601-A/G−522-A/G−444. Table 4 shows MSE values obtained for each gene within treatments and across genes. HSP90AB1 was in all cases the most stable gene, followed by HSP90AA1. Therefore, HSP90AB1 was selected as the only HK to normalize the expression results of HSP90AA1. The highest stability values for all genes corresponded to samples collected in August and lowest stability values corresponded to the control samples.
Table 4. Minimum square error (MSE) within and across treatments for HSP90AA1, HSP90AB1, MDHA and SDHA genes.
| treatment | gene | MSE within treatment | MSE within gene |
| August | HSP90AB1 | 2.51 | |
| August | HSP90AA1 | 3.17 | |
| August | SDHA | 3.84 | |
| August | MDH1 | 9.81 | |
| July | HSP90AB1 | 3.27 | |
| July | HSP90AA1 | 6.07 | |
| July | SDHA | 7.46 | |
| July | MDH1 | 9.92 | |
| control | HSP90AB1 | 4.62 | 4.62 |
| control | HSP90AA1 | 6.39 | 6.39 |
| control | MDH1 | 8.20 | 9.92 |
| control | SDHA | 9.94 | 9.94 |
Environmental Conditions and Statistical Analysis of Gene Expression
As it is shown in Table 2 the maximum temperatures for days of samples collection in July, 35.0°C, and August, 34.4°C, exceeded the sheep thermoneutral zone, but this is not the case for the average temperatures, 26.8°C and 24.7°C, respectively. For both time points, average and maximum THI values occurred in the zones of severe and extreme heat stress [43]. The THImax was one unit higher in August than in July. Also in August the THImax, calculated over the five days before collection, was two units higher than those of July. For the control time point, temperatures and THI values indicated no heat stress conditions.
Raw Cq values for all genes in each treatment are shown in Figure 4. Under control conditions, Cq values for the HSP90AA1, HSP90AB1, MDH1 and SDHA genes were 26.2, 26.6, 29.4 and 30.3, respectively. Smaller Cq values were observed for all genes in samples collected under high temperatures (July and August). They were 25.7 for both chaperones and 28.8 and 29.7 for MDH1 and SDHA, respectively. Variability in the expression rate of all genes was higher in samples collected under control conditions.
Figure 4. Distribution of Quantification Cycle (Cq) values for the target gene HSP90AA1 and the reference genes HSP90AB1, MDH1 and SDHA.
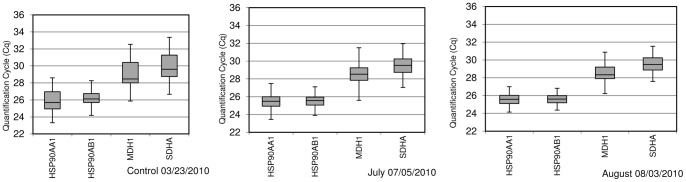
They were obtained by qPCR from samples collected at three different time points (Control, July and August) along the year. Boxes show the range of Cq values within each gene and treatment; the centre line indicates the median; extended vertical bars show standard deviation of the mean.
Based on linkage disequilibrium results, 3 sets of genotypes were selected to carry out expression studies. Genotypes considered in separate analyses were: 1) G/C−660-A/C−601-A/G−522-A/G−444, 2) A/C−601- A/G−522-A/G−444, 3) G/C−660.
Overall outcomes from fitting the mixed model
Test type III of fixed effects shows high significant F values (p<0.0001) for the MTG, P and b(RG) effects in all sets of genotypes. Estimates of the P effect levels (74) averaged 0.73 and ranged between 0.01 to 1.41 Cq. Regression coefficient estimates relating covariation of Cq and RIN values for each gene, were −0.28, −0.47, −0.55 and −0.67 for HSP90AB1, HSP90AA1, MDH1 and SDHA, respectively.
Estimates of animal, sample and residual variances were very similar for the 3 sets of genotypes studied. Animal variance was very small and ranged between 0.03 and 0.05. Sample variance (1.40) was 2.8 times higher than the animal effect one. HSP90AB1 showed a residual variance (0.0086) 34 times lower than the one of HSP90AA1 (0.27) and 53 times lower than the one of SDHA (0.46). MDH1 had the highest residual variance (1.34).
1) Genotype G/C−660-A/C−601- A/G−522-A/G−444
Table S2 and Figure 1, show results for contrasts among G/C−660-A/C−601-A/G−522-A/G−444 genotypes under the different climatic conditions considered.
For samples collected in August, CC−660-CC−601-GG−522-AG−444 showed the highest expression rate (differences in FC from 2 to 3) when comparing with other 8 genotypes. Seven of these 8 genotypes were GG−444, highlighting the importance of this position for the expression rate of the gene under heat stress conditions independently of the other three SNPs. Two significant contrasts pointed out the effect of SNP−660 in terms of expression efficiency. CC−660 showed differences in FC of 1.27 and 1.31 when comparing with GC−660 and GG−660, respectively. It is important to note that contrasts showing differences in FC from 2.3 to 3.0 were those with wider confidence intervals which indicate higher estimated standard errors. This is due to the fact that in many of these contrasts, composed genotypes are at very low frequencies. In some cases only one animal exhibit the genotype compared. For comparisons where FC differences ranged between 1.2 and 1.3, confidence intervals were narrower due to the higher frequencies of the genotypes compared.
For samples collected in July, no comparisons between alternative genotypes were significant after the Holm-Bonferroni correction.
For the 5 significant contrasts of control samples the loose of the effect over the expression rate of the SNP−444 in the gene promoter and the change in the expression rank of the −660 genotypes were the most important effects observed. In 4 contrasts the GC−660-CC−601-AG−522-GG−444 genotype showed differences in FC from 1.3 to 1.6 when compared with alternative genotypes at −660 and −522. The superiority in terms of expression rate seems to depend in this case on the combination of genotypes existing at −660 and −522. Thus, the double heterozygous GC−660-AG−522 showed higher expression levels than GG−660-GG−522 (1.58–1.60), CC−660-AG−522 (1.54) and GC−660-GG−522 (1.30). Finally, the SNP−601 did not seem to be involved in the regulation of the basal expression of the gene. In this case, differences in the confidence intervals were smaller since genotypes compared had higher frequencies.
2) Genotype A/C−601- A/G −522-A/G−444
To determine the impact of SNP−660 over the expression rate of the gene under the different environmental conditions previously described [24], we have tested a genotype of three SNPs excluding SNP−660.
Table S3 and Figure 2, show results for contrasts among A/C−601- A/G −522-A/G−444 genotypes under the different climatic conditions considered.
Regarding the high decrease in the magnitude of the FC differences for the 4 significant comparisons, we can confirm the critical influence of SNP−660 over the gene expression rate. Surprisingly, higher differences in FC were detected for contrasts in control than in August, revealing a more important effect of these SNPs on the basal expression of the HSP90AA1 gene under control conditions.
In samples collected in August, the two significant contrasts with the highest expression rate implied the AG−444 genotype in all cases. Again, AG−444 showed higher expression rate (FC = 1.3–1.4) over GG−444 under heat stress conditions. Genotypes at −601 and −522 did not show any clear effect. No significant contrast was found among genotypes from samples collected in July.
Once more, under mild temperatures the SNP−444 lost its effect over the HSP90AA1 expression which appeared under heat stress conditions. As in the case above mentioned, AG−522 showed a positive effect over the expression of the gene comparing with GG−522 (FC = 1.37–1.39).
3) Genotype G/C−660
Table S4 and Figure 3, show results for contrasts among G/C−660 genotypes under the different climatic conditions considered.
Contrasts of this genotype showed lower differences in FC than that observed for the previous 2 sets of genotypes. Two comparisons were significant in samples collected in August. Differences in FC were 1.22 and 1.20 for the contrasts CC−660 vs. GG−660 and CC−660 vs. GC−660, respectively, showing the superiority of the CC genotype at position −660 over the other genotypes in terms of gene expression rate under heat stress conditions.
In July samples, no comparison had statistic significance. However, under control temperatures, 2 contrasts showed significant differences in FC, CC−660 vs. GG−660 with a FC equal to 1.11 and GC−660 vs. GG−660 with a FC of 1.41.
Thus, when considering only SNP−660, results indicated no differences in HSP90AA1 expression rate across treatments, but the existence of such differences across genotypes.
In order to understand better the results obtained, contrasts involving SNP−601 and SNP−522, were carried out (Figure S2 and Figure S3). In the first analysis, alternative genotypes of A/C−601-A/G−522 were compared. Only one significant contrast was found under control conditions. A FC value of 1.4 was observed for the contrast CC−601-AG−522 vs. CC−601-GG−522, confirming the effect of A/G−522 over the basal expression of the gene under mild temperatures. SNP−601, did not seem to play any clear role in the HSP90AA1 expression rate.
In the second additional analysis, genotypes of G/C−660-A/G−522 were compared to elucidate the relevance of SNP−522 under heat stress conditions. In this case, significant contrasts were found for control samples and those collected in August. The importance of SNP−660 in August was again revealed. CC−660 had higher expression rate than GC−660 and GG−660 (FC = 1.24 and 1.27, respectively). Under control conditions the effect of SNP−522 was clear (FC = 1.28 at least). Changes in the behavior of G/C−660 under control conditions were observed as well. In this case, GC−660 was superior to CC−660 and GG−660 as it was showed in previous analyses.
Discussion
In this study, we aimed to confirm the results obtained previously [24], that showed an association of the SNP−660 of the ovine HSP90AA1 gene promoter with the expression levels of this gene under different environmental temperatures, using a higher amount of samples. We also aimed to increase the information available of the biological process underlying this type of stress conditions. With these purposes we have studied new polymorphisms found at the promoter region, which would affect the expression rate of the gene not only in heat stress events as it was firstly thought, but also modulating its basal expression.
RIN Effect
Best conservation and minimum degradation processes are critical points when sampling commercial livestock animals for expression studies. The degree of RNA degradation in the samples affects gene expression measurements. We have established that RIN values depended neither on the source of biological sample (the animal) nor on the environmental conditions surrounding samples collection. The only factor having a significant effect on RIN values was the period of time occurring between blood extraction and blood processing in LeukoLock platforms for each time point (DxGP). The higher this period of time was, the higher the RNA was degraded (lower values of RIN). Therefore, we proposed to include RIN values as a fixed effect or as a covariate in the statistical model used to analyze expression differences. In fact, the same results were obtained using both approaches (data not shown).
RIN values affect Cq of samples depending on amplicon size [44]. The higher the amplified DNA fragment is, the higher the probability is to be broken down. The length of the amplified product was more correlated with RIN values than expected. Amplicon sizes were in the range of 70 to 250 bp (Table 3) for which Fleige and Pfaffl [26] indicates a more or less independence of qPCR products and RNA quality. Furthermore, DNA CG content did not seem to affect RNA stability as it has been previously described, where lower CG degree content was correlated with higher RIN values [45]. SDHA amplicon was the one with the highest CG content (65%) but was the most affected by RNA degradation. Only for MDH1, which has the lowest CG content and a high RIN effect, this relationship seems to be true. These results reveal that the effect of RNA integrity over both the GOI and the HK should be taken into account in expression analyses.
HK Selection
A crucial aspect revealed in this work, is the need to test the stability of the candidate HKs and the GOI simultaneously. MDH1 and SDHA were previously selected [38] among 16 candidates tested, as the most stable pair in similar conditions to those evaluated here. HSP90AA1 was not included in that experiment. In the present work, we have verified that the GOI, HSP90AA1, is much more stable than the two previously selected HKs, MDH1 and SDHA. Therefore, none of them can be used to normalize the GOI expression data. The constitutive counterpart of the HSP90AA1 gene, HSP90AB1, showed the best stability values within and between treatments (Table 4), and it was chosen as the HK to normalize the expression data of the HSP90AA1. Differences in the stability of both chaperone genes might be due to the effect of the SNPs existing at the promoter of the HSP90AA1 [13] [40] and to the more inducible behavior of this gene.
qPCR Experimental Design and Statistical Methods for Expression Data Analyses
When a great number of samples and treatments are included in a qPCR study the experimental design is important since qPCR plates have a limited capacity (96 or 384 wells). In our design, plates contained a randomized set of animals, treatments, genotypes, RIN values and genes to avoid estimation biases. The repetition of one or more samples in all plates connects the plate’s system allowing to remove technical nuisance from this source of variability and to compare results from all plates. We have confirmed that the plate effect is an important source of variability since differences in Cq among plates can reach values up to 1.4.
Traditional statistical methods to analyze qPCR data was restricted to pair-wise comparisons of treatments in which expression data from GOIs are previously normalized with one or more HKs. This kind of approach does not include systematic nor random effects and their interactions that could affect expression results. In the linear mixed model used in this study, GOI and HKs data are simultaneously analyzed.This model includes different sources of biological and technical variation (i.e. plate, RIN, genotypes, genes, and interactions among them) as fixed or random effects. Fitting this model let us checking HK stability, normalization of GOI data with the most stable HK(s) and test the linear hypothesis of the existence of different expression levels of the HSP90AA1 gene depending on the genotype of the mutations located at its promoter and on diverse environmental conditions.
Environmental Conditions and Gene Expression
Sheep are believed to be one of the most resistant species to climatic extremes, especially to high environmental temperatures. Environmental conditions in Ciudad Real often exceed sheep thermo neutral zone which is comprised between 5°C and 25°C [46]. As expected, expression results differed between heat stress and control conditions. However, unexpectedly, differences in expression rate among genotypes were observed in samples collected in August but not in July. The scarce differences in climatic parameters existing between August and July collects did not explain the observed differences between these time points in terms of FC. The higher THImax values at collection time and during 5 days before collection in August than in July could be the clue to such differences. Other environmental factors here unknown such as wind speed, number of hours over the comfort temperature, insulation, etc. included in Fanger’s comfort equation [47] would have also contributed in such differences.
Significant differences among A/C−601-A/G−522-A/G−444 and G/C−660 among genotypes, found for August and control samples but not for July ones would be explained by the existence of a transition in the expression state of the gene between the basal transcription and the heat stress response. Changes in the expression ranking of G/C−660 observed between control and August samples, and also for other SNPs in a less evident way, would support this hypothesis. Also, since the heat stress response is not a permanent state, in terms of gene expression, even when heat shock conditions are still present, acclimatization processes cannot be discarded as possible source of differences found in samples collected in July and August [48].
Expression Analysis and Genotype Comparisons
Our initial hypothesis was that differences in the expression rate of the gene with different G/C−660 genotypes would be observed only under heat stress conditions [24]. Surprisingly, after including a higher amount of samples and a set of 6 additional polymorphisms located also at the HSP90AA1 promoter, the existence of expression differences under non heat stress situations was confirmed as well. Thus, polymorphisms located in the promoter of HSP90AA1 affect not only its expression rate as response to heat shock but also its basal transcription levels.
Differences in the expression rate found for the contrasts among alternative genotypes for the SNPs studied here suggest that the transcription of this gene may be multiply regulated by cross-talk of various transcription factors, as it was pointed out for this gene in human [39]. Although much of the heat-induced gene expression can be explained by HSF1, a perfect correlation between its binding and induction has not been found [12]. Signal transduction cascades activated by p53, Jak and Ras pathways via HSF1 binding to the heat-shock response element (HSE) and integrating to modulate HSP transcription have been reported [49]. Additional positive or negative factors may modulate the transcriptional induction of HSF1-bound genes. Moreover, eukaryotic gene expression is tightly regulated at many levels, and can vary its regulation complexity [50]. The core promoter (TATA box, initiator –INR- and downstream promoter element –DPE-), is the essential part. Next to the core, proximal enhancers as cis-control elements (i.e. CCAAT box, GC box, B recognition element (BRE) and STRE elements) might be acting. Upstream, distal enhancers (hormone responsive elements -HRE- and nuclear factor element –NFE-) and a huge diversity of regulators that recruit a cascade of more transcription factors contribute to gene transcription regulation [51], [52]. The SNPs studied in this work are located enough upstream to the beginning of the transcription initiation to consider them as binding sites of these co-regulators, or distal enhancers that do not directly activate the transcription of the gene but modulate its expression.
The role of the SNP G/C−660 in the transcription of the gene under heat stress has been confirmed through analysis in which only this mutation is tested. However, results from the analyses of composed genotypes G/C−660-A/C−601-A/G−522-A/G−444 and A/C−601-A/G−522-A/G−444 revealed a cooperative relationship among several SNPs in terms of transcription efficiency. Thus, alternative genotypes of SNP−660-SNP−444 seem to affect the expression of the gene in response to heat stress and those of SNP−660-SNP−522 the basal transcription of HSP90AA1, which may occur under climatic conditions comprising comfort temperatures. Under heat stress conditions, the superiority of CC−660 over GC−660 and GG−660 and of GC−660 over GG−660 indicated an additive effect for this mutation. However, for the control samples, the GC−660 genotype was superior to CC−660 and GG−660. The effect of the SNP−444 was less clear due to the low frequencies of the AG−444 and AA−444 genotypes; however, for the SNP located at −522, more clear conclusions can be extracted.
Several putative TFs have been predicted (Table S7 and Table S8) for the presence of C and A at SNPs −660 and −444, respectively. Some TFs that could co-activate gene expression as distal enhancers only with CC−660 were NFI/CTF (Nuclear factor I or CCAAT box-binding transcription factor) and VDR (Vitamin D receptor) together with RxR-alpha (Retinoid X receptor Alpha). The last two TFs form a heterodimer which attracts a complex of co-activators proteins. This complex links the heterodimer to the initiation complex formed at the TATA box, promoting the transcription machinery [53]. Both TFs bind putatively at the sequence around the SNP−660, and VDR only when this position is C. For AG−444 a heat shock element that could bind a heat shock factor was predicted for the presence of the A nucleotide [39].
The presence of an INDEL of two adenines (AA) located at position -704 in the promoter [21] completely linked, at least in this breed with SNP−660, must also be considered in the expression regulation of the gene under heat stress conditions. Thus, CC−660 animals are also homozygous for the AA insertion (AA/AA), animals with GC−660 are heterozygous ins/del (AA/−) and animals with GG−660 are homozygous for the AA deletion (−/−). This INDEL (AA) was located within a putative glucocorticoid receptor (GR) transacting factor binding site. The AA deletion (−/−) created a GR transcription site. It has been pointed out that glucocorticoids can suppress the heat shock response in stressed cells by inhibiting the action of the heat shock factor 1 (HSF1) [54]. Therefore, this mutation would be the responsible of the expression differences observed for SNP−660. Because of the high linkage disequilibrium between the SNPs at −660 and −528 (R2 = 0.95) the possible effect of the SNP−528 over the transcription rate of the gene under heat stress conditions is masked by the first, and therefore no conclusions could be extracted from this position.
Under control conditions, A/G−522 seems to have a predominant effect over the transcription rate of the gene, being AG−522 (AA−522 was not found in these samples) superior than GG−522. Due to the proximity of this mutation to the SNPs located at −528 (5pb) and −524 (1pb) TF binding sites were predicted for a sequence containing the three SNPs (Table S7 and Table S8). Several putative TF binding sites linked to the presence of adenine at −522 and thymine at −524 were found. Among them, the stress response element (STRE) [39] and the JunD (functional component of the AP1 -activator protein 1- transcription factor complex) related with transcription coactivator activity, oxidative stress response [55] and spermatogenesis [56] seemed to be closer to the HSP90AA1 functions. The TF c-Fos stimulates transcription of genes containing AP-1 regulatory elements and was predicted for the sequence AtagTcA for the SNPs at −528, −524 and –522. In our samples animals with AG−522 were always CC-601-TT−524-TT−468 and in most cases (70%) CC-601-AG−528-TT−524-TT−468. Two putative TF binding sites implying the presence of cytosine were found for A/C−601, HES-1 (hairy and enhancer of split-1) [57], which can act as a repressor or activator, and USF1 (Upstream stimulatory factor 1) [58], that has been found to be involved in the stress-activated signaling cascade [59] and in the cessation of Sertoli cell proliferation and differentiation to spermatozoids [60]. For the SNP−468 one interesting homolog of the human ZNF395 binding Sp1 was found for maize [61] linked to the response to oxidative stress (Table S7).
Most stress-response genes are regulated in a concordant manner with respect to transcript levels and translational efficiency. A strong overall correlation has been observed between transcriptional/translational induction of genes and induction of the corresponding proteins [62]. During environmental stress in fission yeast, most mRNAs are regulated both at transcription and translation level but only up-regulated mRNAs showed a strong correlation with protein expression, while down-regulated mRNAs showed no such correlation [62]. Therefore changes in the expression rate of the HSP90AA1 here observed as a function of environmental temperatures and genotypes at the SNPs located in its promoter will be accompanied by changes in the amount of protein produced. Thus, those genotypes which showed higher expression levels under heat shock also will display higher protein amounts. Despite the low magnitude of the changes in HSP90AA1 expression rate observed, even a small proportion may be significant as HSP90 is one of the most abundant proteins in most cells [39]. Higher amounts of HSP90α protein would increase its capacity to exert its protective role over the effects caused by heat stress at the cellular level. Effects of changes in the protein amount as consequence of several stress sources must be studied in tissues in which HSP90α predominates, such as brain and testis [39]. In this context, our results for two of the known functions of the HSP90α, refolding proteins with aberrant conformation [39] and spermatogenesis and meiotic progression in testis [63], confirm this hypothesis. GG−660 was related with higher sperm DNA fragmentation values in rams under heat stress conditions [23] and also to lower scrapie incubation period in sheep (missfolding stress) [22].
Future studies will focus on testing TF interaction at binding sites where polymorphisms of the HSP90AA1 promoter are located, by employing in vitro techniques of EMSA (Electrophoretic Mobility Shift Assay). In addition, an epigenetic regulation of the HSP90AA1 expression cannot be discarded. Methylation of CpG islands located at gene promoters is a well known mechanism of expression regulation and gene silencing [64]. A CpG island has been predicted for the promoter of this gene and some of the polymorphisms here analyzed are susceptible to be methylated (SNPs located at −660, −601, −528 and −522). Therefore, future research will be also focused in the study of the methylation pattern of alternative genotypes of the SNPs containing cytosine by bisulfite sequencing techniques.
Supporting Information
Sequence and polymorphisms of the ovine HSP90AA1 gene promoter (DQ983231). Intron sequence in lower case and exons in capital letters. Primers sequences used to amplify the 499pb fragment are highlighted in dark. SNPs are in square brackets. The 7 SNPs of interest included in the 499pb amplicon sequenced (−660, −601, −528, −524, −522 and −444) are in grey. INDELs are in brackets. Putative methylated SNPs are also circled. Initiation of transcription (TATA box) and translation (ATG) in bold. A HSE already detected is underlined. Modified from Marcos-Carcavilla and coworkers [20].
(TIF)
Fold change (FC) for the contrast among alternative genotypes G/C−660-G/A−522 of the HSP90AA1 promoter within each treatment (Control, July and August) normalized by HSP90AB1. Segments indicate the 95% confidence interval (FCup-FClow). In abscissa the FC, in ordinate genotype contrasts. Asterisk over each bar indicates the significance level of the contrasts.
(TIF)
Fold change (FC) for the contrast among alternative genotypes A/C−601-G/A−522 of the HSP90AA1 promoter within each treatment (Control, July and August) normalized by HSP90AB1. Segments indicate the 95% confidence interval (FCup-FClow). In abscissa the FC, in ordinate genotype contrasts. Asterisk over each bar indicates the significance level of the contrasts.
(TIF)
SNP frequencies of the polymorphisms found at the HSP90AA1 promoter as described in Marcos-Carcavilla’s work [24] .
(XLSX)
Genotype contrasts of G/C−660-A/C−601-A/G−522-A/G−444. Estimates, standard error (SE), t values and p values of significant contrasts among genotypes G/C−660-A/C−601-A/G−522-A/G−444 of the HSP90AA1 gene promoter normalized with HSP90AB1 in each treatment (Control, July and August). Also Fold change (FC) and the 95% FC confidence interval (FCup-FClow) are included.
(XLSX)
Genotype contrasts of A/C−601-A/G522-A/G−444. Estimates, standard error (SE), t values and p values of significant contrasts among genotypes A/C−601-A/G522-A/G−444 of the HSP90AA1 gene promoter normalized with HSP90AB1 in each treatment (Control, July and August). Also Fold change (FC) and the 95% FC confidence interval (FCup-FClow) are included.
(XLSX)
Genotype contrasts of SNP G/C−660. Estimates, standard error (SE), t values and p values of significant contrasts among genotypes G/C−660 of the HSP90AA1 gene promoter normalized with HSP90AB1 in each treatment (Control, July and August). Also Fold change (FC) and the 95% FC confidence interval (FCup-FClow) are included.
(XLSX)
Hardy-Weinberg equilibrium analysis of the 7SNPs of interest at HSP90AA1 gene promoter on trios population.
(XLSX)
LD analyses from trios population with PLINK software. Linkage disequilibrium values between each couple of SNPs are shown. The couples of SNPs in bold show a high degree of linkage disequilibrium between them.
(XLSX)
Predicted transcription factor binding sites from TESS software.
(XLSX)
Predicted transcription factor binding sites from ALGENN-PROMO software.
(XLSX)
Acknowledgments
We thank the AGRAMA breeders association, CERSYRA Valdepeñas and INIA for providing the biological samples and Professor Juan Pedro Steibel (Michigan State University) for his statistic advice. Also to Dr Villanueva for her suggestions and revision.
Funding Statement
RTA2009-00098-00-00 INIA project (Subprograma de Investigación Fundamental orientada a los Recursos y Tecnologías Agrarias) and FEDER (Fondo Europeo de Desarrollo Regional) funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pedrosa S, Uzun M, Arranz JJ, Gutierrez-Gill B, Primitivo FS, et al. (2005) Evidence of three maternal lineages in near eastern sheep supporting multiple domestication events. Proceedings of the Royal Society B-Biological Sciences 272: 2211–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maijala K (1997) Genetic aspects of domestication, common breeds and their origin. In: Piper LR, A., editor. The genetics of sheep. 13–49.
- 3.Kijas JW, Townley D, Dalrymple BP, Heaton MP, Maddox JF, et al.. (2009) A Genome Wide Survey of SNP Variation Reveals the Genetic Structure of Sheep Breeds. Plos One 4. [DOI] [PMC free article] [PubMed]
- 4. Morton JM, Tranter WP, Mayer DG, Jonsson NN (2007) Effects of environmental heat on conception rates in lactating dairy cows: Critical periods of exposure. Journal of Dairy Science 90: 2271–2278. [DOI] [PubMed] [Google Scholar]
- 5. Sanchez JP, Misztal I, Aguilar I, Zumbach B, Rekaya R (2009) Genetic determination of the onset of heat stress on daily milk production in the US Holstein cattle. Journal of Dairy Science 92: 4035–4045. [DOI] [PubMed] [Google Scholar]
- 6. Aguilar I, Misztal I, Tsuruta S (2009) Genetic components of heat stress for dairy cattle with multiple lactations. Journal of Dairy Science 92: 5702–5711. [DOI] [PubMed] [Google Scholar]
- 7. Finocchiaro R, van Kaam J, Portolano B, Misztal I (2005) Effect of heat stress on production of Mediterranean dairy sheep. Journal of Dairy Science 88: 1855–1864. [DOI] [PubMed] [Google Scholar]
- 8. Marai IFM, El-Darawany AA, Fadiel A, Abdel-Hafez MAM (2007) Physiological traits as affected by heat stress in sheep - A review. Small Ruminant Research 71: 1–12. [Google Scholar]
- 9. Marai IFM, El-Darawany AA, Fadiel A, Abdel-Hafez MAM (2008) Reproductive performance traits as affected by heat stress and its alleviation in sheep. Tropical and Subtropical Agroecosystems 8: 209–234. [Google Scholar]
- 10. Sevi A, Caroprese M (2012) Impact of heat stress on milk production, immunity and udder health in sheep: A critical review. Small Ruminant Research 107: 1–7. [Google Scholar]
- 11. Favatier F, Bornman L, Hightower LE, Gunther E, Polla BS (1997) Variation in hsp gene expression and Hsp polymorphism: Do they contribute to differential disease susceptibility and stress tolerance? Cell Stress & Chaperones 2: 141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM (2004) The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Molecular Biology of the Cell 15: 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collier RJ, Collier JL, Rhoads RP, Baumgard LH (2008) Invited review: Genes involved in the bovine heat stress response. Journal of Dairy Science 91: 445–454. [DOI] [PubMed] [Google Scholar]
- 14. Charoensook R, Gatphayak K, Sharifi AR, Chaisongkram C, Brenig B, et al. (2012) Polymorphisms in the bovine HSP90AB1 gene are associated with heat tolerance in Thai indigenous cattle. Tropical Animal Health and Production 44: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Becker J, Craig EA (1994) Heat-shock proteins as molecular chaperones. European Journal of Biochemistry 219: 11–23. [DOI] [PubMed] [Google Scholar]
- 16. Sorensen JG, Kristensen TN, Loeschcke V (2003) The evolutionary and ecological role of heat shock proteins. Ecology Letters 6: 1025–1037. [Google Scholar]
- 17. Huang LH, Wang HS, Kang L (2008) Different evolutionary lineages of large and small heat shock proteins in eukaryotes. Cell Research 18: 1074–1076. [DOI] [PubMed] [Google Scholar]
- 18.Chen B, Zhong DB, Monteiro A (2006) Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. Bmc Genomics 7. [DOI] [PMC free article] [PubMed]
- 19. Krone PH, Sass JB (1994) HSP 90 alpha and HSP 90 beta genes are present in the zebrafish and are differentially regulated in developing embryos. Biochemical and Biophysical Research Communications 204: 746–752. [DOI] [PubMed] [Google Scholar]
- 20. Marcos-Carcavilla A, Calvo JH, Gonzalez C, Moazami-Goudarzi K, Laurent P, et al. (2008) Structural and functional analysis of the HSP90AA1 gene: distribution of polymorphisms among sheep with different responses to scrapie. Cell Stress & Chaperones 13: 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oner Y, Calvo JH, Serrano M, Elmaci C (2012) Polymorphisms at the 5′ flanking region of the HSP90AA1 gene in native Turkish sheep breeds. Livestock Science 150: 381–385. [Google Scholar]
- 22. Marcos-Carcavilla A, Moreno C, Serrano M, Laurent P, Cribiu EP, et al. (2010) Polymorphisms in the HSP90AA1 5′ flanking region are associated with scrapie incubation period in sheep. Cell Stress & Chaperones 15: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramón M, Salces-OrtizJ, Garde J, González C, Sánchez-Moreno N et al.. (2012) Effect of environmental temperature and Hsp90aa1 genotype over sperm DNA fragmentation in rams.; 10–21 September, 2012; Les Diablerets, Switzerland.
- 24. Marcos-Carcavilla A, Mutikainen M, Gonzalez C, Calvo JH, Kantanen J, et al. (2010) A SNP in the HSP90AA1 gene 5′ flanking region is associated with the adaptation to differential thermal conditions in the ovine species. Cell Stress & Chaperones 15: 67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dheda K, Huggett JF, Chang JS, Kim LU, Bustin SA, et al. (2005) The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Analytical Biochemistry 344: 141–143. [DOI] [PubMed] [Google Scholar]
- 26.Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the real-time qRT-PCR performance. Molecular aspects of medicine 27. [DOI] [PubMed]
- 27. Bustin SA, Vandesompele J, Pfaffl MW (2009) Standardization of qPCR and RT-qPCR. Genetic Engineering & Biotechnology News 29: 40–43. [Google Scholar]
- 28. Auer PL, Doerge RW (2010) Statistical Design and Analysis of RNA Sequencing Data. Genetics 185: 405–U432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steibel JP, Poletto R, Coussens PM, Rosa GJM (2009) A powerful and flexible linear mixed model framework for the analysis of relative quantification RT-PCR data. Genomics 94: 146–152. [DOI] [PubMed] [Google Scholar]
- 30.Lim A, Steibel JP, Coussens PM, Grooms DL, Bolin SR (2012) Differential gene expression segregates cattle confirmed positive for bovine tuberculosis from antemortem tuberculosis test-false positive cattle originating from herds free of bovine tuberculosis. Veterinary medicine international 2012. [DOI] [PMC free article] [PubMed]
- 31.Arceo ME, Ernst CW, Lunney JK, Choi I, Raney NE, et al.. (2012) Characterizing differential individual response to porcine reproductive and respiratory syndrome virus infection through statistical and functional analysis of gene expression. Frontiers in genetics 3. [DOI] [PMC free article] [PubMed]
- 32. Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting dna from human nucleated cells. Nucleic Acids Research 16: 1215–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. Theoretical and Applied Genetics 38: 226–231. [DOI] [PubMed] [Google Scholar]
- 35.Schug J (2008) Using TESS to predict transcription factor binding sites in DNA sequence. Current protocols in bioinformatics/editoral board, Andreas D Baxevanis [et al] Chapter 2. [DOI] [PubMed]
- 36. Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, et al. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18: 333–334. [DOI] [PubMed] [Google Scholar]
- 37. Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, et al. (2003) Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Research 31: 3651–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serrano M, Moreno-Sanchez N, Gonzalez C, Marcos-Carcavilla A, Van Poucke M, et al.. (2011) Use of Maximum Likelihood-Mixed Models to select stable reference genes: a case of heat stress response in sheep. Bmc Molecular Biology 12. [DOI] [PMC free article] [PubMed]
- 39. Csermely P, Schnaider T, Soti C, Prohaszka Z, Nardai G (1998) The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacology & Therapeutics 79: 129–168. [DOI] [PubMed] [Google Scholar]
- 40. Deuerling E, Patzelt H, Vorderwulbecke S, Rauch T, Kramer G, et al. (2003) Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Molecular Microbiology 47: 1317–1328. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer, C, Nakagawara, K, eds, editor. Rapid Cycle Real-time PCR, Methods and Applications Springer Press, Heidelberg;. 21–34.
- 42.Littell RC, Milliken GA, Stroup WW, Wolfinger RD, and Schabenberger O (2006) SAS for Mixed Models. Second Edition, Cary, NC: SAS Institute Inc.
- 43. Marai IFM, Ayyat MS, Abd El-Monem UM (2001) Growth performance and reproductive traits at first parity of New Zealand White female rabbits as affected by heat stress and its alleviation under Egyptian conditions. Tropical Animal Health and Production 33: 451–462. [DOI] [PubMed] [Google Scholar]
- 44. Fleige S, Walf V, Huch S, Prgomet C, Sehm J, et al. (2006) Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnology Letters 28: 1601–1613. [DOI] [PubMed] [Google Scholar]
- 45.Opitz L, Salinas-Riester G, Grade M, Jung K, Jo P, et al.. (2010) Impact of RNA degradation on gene expression profiling. Bmc Medical Genomics 3. [DOI] [PMC free article] [PubMed]
- 46.Curtis SE (1983) Environmental management in animal agriculture. Ames, Iowa 50010: Iowa State University Press.
- 47.Fanger O (1970/1982) Thermal Comfort.
- 48. Basu N, Todgham AE, Ackerman PA, Bibeau MR, Nakano K, et al. (2002) Heat shock protein genes and their functional significance in fish. Gene 295: 173–183. [DOI] [PubMed] [Google Scholar]
- 49.Stephanou A, Latchman DS (2011) Transcriptional modulation of heat-shock protein gene expression. Biochemistry research international 2011. [DOI] [PMC free article] [PubMed]
- 50. Lemon B, Tjian R (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes & Development 14: 2551–2569. [DOI] [PubMed] [Google Scholar]
- 51. Thanos D, Maniatis T (1995) Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83: 1091–1100. [DOI] [PubMed] [Google Scholar]
- 52. Blau J, Xiao H, McCracken S, Ohare P, Greenblatt J, et al. (1996) Three functional classes of transcriptional activation domains. Molecular and Cellular Biology 16: 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bikle DD (2010) Vitamin D: newly discovered actions require reconsideration of physiologic requirements. Trends in Endocrinology and Metabolism 21: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wadekar SA, Li DP, Periyasamy S, Sanchez ER (2001) Inhibition of heat shock transcription factor by GR. Molecular Endocrinology 15: 1396–1410. [DOI] [PubMed] [Google Scholar]
- 55. Mendelson KG, Contois LR, Tevosian SG, Davis RJ, Paulson KE (1996) Independent regulation of JNK/p38 mitogen-activated protein kinases by metabolic oxidative stress in the liver. Proceedings of the National Academy of Sciences of the United States of America 93: 12908–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thepot D, Weitzman JB, Barra J, Segretain D, Stinnakre MG, et al. (2000) Targeted disruption of the murine junD gene results in multiple defects in male reproductive function. Development 127: 143–153. [DOI] [PubMed] [Google Scholar]
- 57. Yan B, Raben N, Plotz PH (2002) Hes-1, a known transcriptional repressor, acts as a transcriptional activator for the human acid alpha-glucosidase gene in human fibroblast cells. Biochemical and Biophysical Research Communications 291: 582–587. [DOI] [PubMed] [Google Scholar]
- 58. Kumari D, Usdin K (2001) Interaction of the transcription factors USF1, USF2, and alpha-Pal/Nrf-1 with the FMR1 promoter - Implications for Fragile X mental retardation syndrome. Journal of Biological Chemistry 276: 4357–4364. [DOI] [PubMed] [Google Scholar]
- 59. Galibert MD, Carreira S, Goding CR (2001) The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced Tyrosinase expression. Embo Journal 20: 5022–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wood MA, Walker WH (2009) USF1/2 Transcription Factor DNA-Binding Activity Is Induced During Rat Sertoli Cell Differentiation. Biology of Reproduction 80: 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lal A, Peters H, St Croix B, Haroon ZA, Dewhirst MW, et al. (2001) Transcriptional response to hypoxia in human tumors. J Natl Cancer Inst 93: 1337–1343. [DOI] [PubMed] [Google Scholar]
- 62.Lackner DH, Schmidt MW, Wu SD, Wolf DA, Bahler J (2012) Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biology 13. [DOI] [PMC free article] [PubMed]
- 63.Grad I, Cederroth CR, Walicki J, Grey C, Barluenga S, et al.. (2010) The Molecular Chaperone Hsp90 alpha Is Required for Meiotic Progression of Spermatocytes beyond Pachytene in the Mouse. Plos One 5. [DOI] [PMC free article] [PubMed]
- 64. Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM (2003) Predicting aberrant CpG island methylation. Proceedings of the National Academy of Sciences of the United States of America 100: 12253–12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence and polymorphisms of the ovine HSP90AA1 gene promoter (DQ983231). Intron sequence in lower case and exons in capital letters. Primers sequences used to amplify the 499pb fragment are highlighted in dark. SNPs are in square brackets. The 7 SNPs of interest included in the 499pb amplicon sequenced (−660, −601, −528, −524, −522 and −444) are in grey. INDELs are in brackets. Putative methylated SNPs are also circled. Initiation of transcription (TATA box) and translation (ATG) in bold. A HSE already detected is underlined. Modified from Marcos-Carcavilla and coworkers [20].
(TIF)
Fold change (FC) for the contrast among alternative genotypes G/C−660-G/A−522 of the HSP90AA1 promoter within each treatment (Control, July and August) normalized by HSP90AB1. Segments indicate the 95% confidence interval (FCup-FClow). In abscissa the FC, in ordinate genotype contrasts. Asterisk over each bar indicates the significance level of the contrasts.
(TIF)
Fold change (FC) for the contrast among alternative genotypes A/C−601-G/A−522 of the HSP90AA1 promoter within each treatment (Control, July and August) normalized by HSP90AB1. Segments indicate the 95% confidence interval (FCup-FClow). In abscissa the FC, in ordinate genotype contrasts. Asterisk over each bar indicates the significance level of the contrasts.
(TIF)
SNP frequencies of the polymorphisms found at the HSP90AA1 promoter as described in Marcos-Carcavilla’s work [24] .
(XLSX)
Genotype contrasts of G/C−660-A/C−601-A/G−522-A/G−444. Estimates, standard error (SE), t values and p values of significant contrasts among genotypes G/C−660-A/C−601-A/G−522-A/G−444 of the HSP90AA1 gene promoter normalized with HSP90AB1 in each treatment (Control, July and August). Also Fold change (FC) and the 95% FC confidence interval (FCup-FClow) are included.
(XLSX)
Genotype contrasts of A/C−601-A/G522-A/G−444. Estimates, standard error (SE), t values and p values of significant contrasts among genotypes A/C−601-A/G522-A/G−444 of the HSP90AA1 gene promoter normalized with HSP90AB1 in each treatment (Control, July and August). Also Fold change (FC) and the 95% FC confidence interval (FCup-FClow) are included.
(XLSX)
Genotype contrasts of SNP G/C−660. Estimates, standard error (SE), t values and p values of significant contrasts among genotypes G/C−660 of the HSP90AA1 gene promoter normalized with HSP90AB1 in each treatment (Control, July and August). Also Fold change (FC) and the 95% FC confidence interval (FCup-FClow) are included.
(XLSX)
Hardy-Weinberg equilibrium analysis of the 7SNPs of interest at HSP90AA1 gene promoter on trios population.
(XLSX)
LD analyses from trios population with PLINK software. Linkage disequilibrium values between each couple of SNPs are shown. The couples of SNPs in bold show a high degree of linkage disequilibrium between them.
(XLSX)
Predicted transcription factor binding sites from TESS software.
(XLSX)
Predicted transcription factor binding sites from ALGENN-PROMO software.
(XLSX)