Abstract
Here, we present a study regarding the phylogenetic positions of two enigmatic earwig lineages whose unique phenotypic traits evolved in connection with ectoparasitic relationships with mammals. Extant earwigs (Dermaptera) have traditionally been divided into three suborders: the Hemimerina, Arixeniina, and Forficulina. While the Forficulina are typical, well-known, free-living earwigs, the Hemimerina and Arixeniina are unusual epizoic groups living on molossid bats (Arixeniina) or murid rodents (Hemimerina). The monophyly of both epizoic lineages is well established, but their relationship to the remainder of the Dermaptera is controversial because of their extremely modified morphology with paedomorphic features. We present phylogenetic analyses that include molecular data (18S and 28S ribosomal DNA and histone-3) for both Arixeniina and Hemimerina for the first time. This data set enabled us to apply a rigorous cladistics approach and to test competing hypotheses that were previously scattered in the literature. Our results demonstrate that Arixeniidae and Hemimeridae belong in the dermapteran suborder Neodermaptera, infraorder Epidermaptera, and superfamily Forficuloidea. The results support the sister group relationships of Arixeniidae+Chelisochidae and Hemimeridae+Forficulidae. This study demonstrates the potential for rapid and substantial macroevolutionary changes at the morphological level as related to adaptive evolution, in this case linked to the utilization of a novel trophic niche based on an epizoic life strategy. Our results also indicate that the evolutionary consequences of the transition to an ectoparazitic mode of living, which is extremely rare in earwigs, have biased previous morphology-based hypotheses regarding the phylogeny of this insect group.
Introduction
The insect order Dermaptera (earwigs) comprises nearly 2000 extant described species [1], [2] in 11 families [3]. The monophyly of this group is well supported by morphological traits including the unsegmented forceps-like cerci of adults that assist in predation, mating, and wing folding [4], and also by the presence of holocentric chromosomes [5]. The Dermaptera had been traditionally divided into three suborders, the Hemimerina, Arixeniina, and Forficulina [see 5]–[9], and all three suborders were treated as monophyletic [3], [5], [10], [11]. But studies published during the last decades indicated that the Hemimeridae and Arixeniidae are highly specialized lineages within the traditional Forficulina [12]–[18].
According to the traditional concept of three suborders, the majority of earwig species were placed in the suborder Forficulina, which contains a wide variety of diverse lineages [19]. They are ‘typical earwigs’, i.e., free-living, feeding on plants or insects, oviparous or ovoviviparous [20], [21], and equipped with strong cerci. Some members of the Forficulina have complex wings and specialized wing-locking mechanisms on their tegmina, whereas others, like those in the Anisolabididae, are secondarily wingless [10]. According to the traditional concept, the Forficulina included nine families:: the Anisolabididae, Apachyidae, Chelisochidae, Diplatyidae, Forficulidae, Karschiellidae, Labiduridae, Pygidicranidae, and Spongiphoridae [3].
Hemimerids comprise 11 species, all commensal inhabitants of the fur of giant murid rats of the genera Beamys Thomas, 1909 and Cricetomys Waterhouse, 1840 in sub-Saharan Africa [22]. They are viviparous, which significantly increases the chances that the nymph finds the proper host. Synapomorphies supporting the monophyly of hemimerids are associated with adaptations to epizoic life. These include specialized grooves on the legs that allow the earwig to attach its legs close to the host body, loss of wings and eyes, and straight, narrow cerci [11], [22].
Arixeniids comprise five species occurring in Indonesia, the Philippines, and the Malay peninsula, and are associated with the molossid genus Cheiromeles Horsfield, 1824. Bats in this genus are characterized by their almost completely naked appearance, marked throat sack, and wing pouches that enable relatively advanced quadrupedal locomotion [22]. Arixeniid earwigs feed on skin and gland secretions of their hosts, but being frequently found also on guano in caves and trees, they are less closely associated with their hosts than are hemimerids. All arixeniids are viviparous. Synapomorphies include the absence of wings, reduced eyes, and the presence of thin, pubescent cerci that presumably serve a sensory function.
The systematic position of both epizoic lineages (the Hemimerina and Arixeniina) has been discussed throughout the history of their study. Walker [23], when he described the first Hemimerus, H. taploides Walker, 1871, placed this species in the Gryllidae. Saussure [24] erected the order Diploglossata to include Hemimerus and concluded that the new order should stand between Orthoptera and Thysanura. Sharp [25] even tried to place hemimerids among the Coleoptera near Platypsilidae, but three years later [26] created the family Hemimeridae, giving it equal rank in the orthopteroid stock to the Forficulidae, which at that time was used for all earwigs. Burr [27] considered the Hemimerina to represent a suborder of Dermaptera. This position was confirmed by many subsequent studies of external morphology [18], [21]–[30]. Popham [31] elevated the suprageneric status of Hemimerina to ordinal status, but Giles [8] challenged that change and reestablished the Hemimerina as a suborder within the Dermaptera.
When the genus Arixenia and the family Arixeniidae were described by Jordan [32], the author pointed out that if Hemimerus justified the creation of a separate suborder, a 3rd suborder of Dermaptera would have to be erected for the reception of the Arixenia. Some authors disagreed with Jordan, and Handlirsch [33] placed the Arixeniidae in the suborder Forficulina. Giles [8] considered the Arixeniina a suborder of Dermaptera and, based on external morphology, pointed out that 6 of 283 characters are common to the Forficulina and the Hemimerina but do not occur in the Arixeniina, that 12 characters are common to the Forficulina and Arixeniina but do not occur in the Hemimerina, and that the Arixeniina differ in 20 characters from the Hemimerina. However, these characters may be plesiomorphic, and the unique hemimerid and arixeniid characters may be independently derived as a result of an epizoic lifestyle. Alternatively, one might suspect that many of the features that make epizoic groups seem more primitive than Forficulina could be due to secondary reduction [12].
Popham [7], [34], [35] proposed a classification of the Dermaptera based on characters of the reproductive anatomy and primarily on the characters of the male external genitalia. Although he produced a large number of detailed drawings of genitalia, abdomens, and other structures, his proposed phylogeny was not based on a formal character analysis. Popham [7], [34], [35] considered arixeniids to be sister to the Spongiphoridae. Popham’s [7] hypothesis has not been generally accepted because it was based on an informal analysis rather than on a cladistic treatment of coded characters. Haas [10] performed the first quantitative phylogenetic analysis of the Dermaptera using morphological characters derived from 10 forficuline earwig taxa representing all nine recognized forficuline families and three blattid outgroups. Neither hemimerids nor arixeniids were included in his analysis. Haas & Kukalova-Peck [9] expanded on the Haas [10] matrix by including additional wing structure and venational characters and additional taxa, but hemimerids and arixeniids were again omitted. Klass [12] studied in detail the exoskeleton, musculature, and nervous system of the female abdomen of Hemimerus vosseleri Rehn & Rehn, 1935, and his results suggested that hemimerids are related to Apachyidae. The author [12] pointed out that hemimerids probably have many paedomorphic features in the thorax, in the female genital region, and in the cerci, which are thread-like.
Grimaldi & Engel [14] summarized information concerning the evolution of the Dermaptera mainly based on paleontological material. According to their concept, the Dermaptera consists of two primitive and extinct suborders, the Archidermaptera and Eodermaptera, and one recent suborder, the Neodermaptera. The latter is divided into the infraorder Protodermaptera, which contains the families Karscheiellidae, Diplatyidae, and Pygidicranidae, and the infraorder Epidermaptera, which contains the remaining eight families, including the Hemimeridae, nesting near the family Apachyidae, and the Arixeniidae, nesting near the Spongiphoridae (but the positions of the epizoic lineages are based on the poorly supported analyses of Popham [7], Haas & Kukalova-Peck [10], and Klass [12]). This classification was also adopted by Engel & Haas [15].
Karyotypic characters have not clarified the phylogeny of the suborders because the chromosome count (2n) ranges from 10 to 37 within the Forficulina [12], [36] and is 7 in the Arixeniina [37] and 60 in the Hemimerina [38].
Unfortunately, most of the recent systematic research on the Dermaptera has neglected the epizoic lineages altogether because fresh specimens for DNA analysis were unavailable [39]–[42]. Four molecular studies on Dermaptera have provided some additional insight into earwig phylogeny, but all lacked samples of hemimerids and arixeniids. Wirth et al. [39] sequenced 684 bp of cytochrome oxidase II across six dermapteran species. Guillet & Vancassel [40] reconstructed a phylogeny of 15 earwig species in four families based on l6S mitochondrial ribosomal sequence. Colgan et al. [41] included the widest selection of taxa and phylogenetic markers to date. Their analysis included 12 dermapteran taxa representing seven families and four outgroup taxa. They generated partial sequences from cytochrome oxidase I, 16S, 18S, and 28S. Kamimura [42] reconstructed an earwig phylogeny with representatives of seven families by using partial sequences of the mitochondrial 16S and nuclear 28S rRNA genes. The only study to consider the affinity of hemimerids in the phylogeny of the Dermaptera was that by Jarvis et al. [11], which was based on the sequences of the large ribosomal subunit 28S rRNA, small ribosomal subunit 18S rRNA, histone-3 (H3) nuclear DNA, and 43 morphological characters. The results indicated that the epizoic hemimerids are sister to Forficulidae+Chelisochidae but not to the remaining dermapterans.
Recently, Tworzydlo et al. [14], [16], [17] studied the ovary structure and initial stages of oogenesis in 15 representatives of several dermapteran taxa, including the epizoic Arixenia esau Jordan, 1909. The interpretation of the results in an evolutionary context supports the monophyly of the Dermaptera and the inclusion of the arixeniids in the clade Forficuloidea containing Spongiphoridae, Chelisochidae, and Forficulidae.
Here, we present a phylogenetic analysis of relationships between Arixenia, Hemimerus, and the remaining Dermaptera using 18S and 28S rRNA and histone-3 sequences. Our aim was to clarify the phylogeny of these morphologically modified epizoic earwigs and to establish their position within the Dermaptera based on molecular data.
Materials and Methods
Collecting of Samples
We used Arixenia esau Jordan, 1909 as representative of the Arixeniidae and Hemimerus hanseni Sharp, 1895 as representative of the Hemimeridae. Arixenia esau was collected in a cave in the Mulu area, Sarawak, Malaysia (3°57′N, 114°49′E) on 19 January 2009. The specimens were collected individually on rocks under the colony of the naked bulldog bat Cheiromeles torquatus Horsfield, 1824 and were preserved in pure ethanol. Hemimerus hanseni was collected in Big Babanki, Northwest Cameroon (6°7′N, 10°15′E) on 10 February 2007. The specimens were collected individually from the hairs of the Gambian pouched rat Cricetomys gambianus Waterhouse, 1840 that had been killed by automobiles and randomly found on the road. The material was preserved in 70% ethanol. A. esau and H. hanseni are not protected in the countries where they were collected, are not protected by any international law, and were not collected in protected areas. The taxa were unequivocally identified using the criteria in Nakata & Maa [15] and Rehn & Rehn [35]. The studied specimens are kept in the collection of Department of Biology and Ecology, University of Ostrava.
Sequencing
Total genomic DNA was isolated from the femur of the leg with a Qiagen DNeasy Tissue kit (Qiagen) and following the manufacturer’s protocol. Three nuclear markers (18S and 28S ribosomal DNA and histone-3) were analyzed using primers published by Whiting [43] and Jarvis et al. [11] and listed in Table 1. PCR reactions were performed in 20-µl volumes containing 1×Taq buffer, 2.5 mM MgCl2, 200 µM dNTPs, 0.6 µM primers, 1 U Taq polymerase (Promega), and 50 ng of template DNA. The thermal protocol included predenaturation (95°C, 3 min); 10 cycles of denaturation (94°C, 60 s), annealing (54°C with a decrease of 0.5°C in each cycle, 90 s), and extension (72°C, 75 s); followed by 20 cycles identical to the previous 10 but with an annealing temperature of 48°C; and a final extension (72°C, 4 min). PCR products were examined on a 1% agarose gel, and amplified DNA was isolated from the gel using the QIAquick Gel Extraction Kit (Qiagen) or from a PCR mixture using the QIAquick PCR Purification kit (Qiagen). Purified PCR products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Invitrogen) and were analyzed on a 3130 Genetic Analyzer (Applied Biosystems). Results were viewed and edited using the program Chromas Lite v. 2.01 (http://www.technelysium.com.au/chromas_lite.html), and sequences were submitted to GenBank (Table 2).
Table 1. Primers used for amplification of 18S and 28S ribosomal DNA and histone-3 nuclear DNA (according to[11] and [42]).
| Primer name | Sequence (5' → 3') |
| 18S 1.2 F | TGCTTGTCTCAAAGATTAAGC |
| 18S b5.0 | TAACCGCAACAACTTTAAT |
| 18S a0.7 | ATTAAAGTTGTTGCGGTT |
| 18S b0.5 | GTTTCAGCTTTGCAACCAT |
| 18S a2.0 | ATGGTTGCAAAGCTGAAAC |
| 18S 7R | GCATCACAGACCTGTTATTGC |
| 18S 7F | GCAATAACAGGTCTGTGATGCCC |
| 18S 9R | GATCCTTCCGCAGGTTCACCTAC |
| 28S rD1.2a | CCCSSGTAATTTAAGCATATTA |
| 28S Rd4.2b | CCTTGGTCCGTGTTTCAAGACGG |
| 28S SA | GACCCGTCTTGAAGCACG |
| 28S rD5b | CCACAGCGCCAGTTCTGCTTAC |
| 28S Rd4.8a | ACCTATTCTCAAACTTTAAATGG |
| 28S Rd6.2b | AATAKKAACCRGATTCCCTTTCGC |
| 28S Rd6.2a | GAAAGGGAATCYGGTTMMTATTCC |
| 28S rD7.b1 | GACTTCCCTTACCTACAT |
| Hex AF | ATGGCTCGTACCAAGCAGACGGC |
| Hex AR | ATATCCTTGGGCATGATGGTGAC |
Table 2. Information about specimens and GenBank accession numbers.
| Family | Subfamily | Species | 18s | 28s | H3 |
| Anisolabididae | Carcinophorinae | Euborellia femoralis (Dohrn, 1863) | AY707326, AY707349 | AY707373, AY707393 | AY707429 |
| Anisolabididae | Carcinophorinae | Thekalabis sp. | AY707325, AY707348 | AY707372, AY707392 | AY707428 |
| Anisolabididae | Parisolabiinae | Parisopsalis spryi Burr, 1914 | - | AY144654 | - |
| Apachyidae | Apachyinae | Dendroiketes novaeguineae Boeseman, 1954 | AY521839 | - | - |
| Arixeniidae | - | Arixenia esau Jordan, 1909 | JX399774 | JX399775 | - |
| Chelisochidae | Chelisochinae | Chelisoches morio (Fabricius, 1775) | AY121133 | AY125273 | AY125220 |
| Chelisochidae | Chelisochinae | Chelisoches annulatus Burr, 1906 | AY707323, AY707346 | AY707370, AY707390 | AY707426 |
| Chelisochidae | Chelisochinae | Proreus duruoides Hebard, 1933 | AY707363 | AY707384, AY707404 | AY707439 |
| Chelisochidae | - | - | AY521841 | - | - |
| Forficulidae | Cosmiellinae | Acanthocordax papuanus Günther, 1929 | AY707331, AY707354 | AY707378, AY707398 | - |
| Forficulidae | Ancistrogastrinae | Ancistrogaster sp. | AY144633 | AY144660 | - |
| Forficulidae | Forficulinae | Doru spiculiferum (Kirby, 1891) | AY121131 | AY125272 | AY125218 |
| Forficulidae | Forficulinae | Elaunon bipartitus (Kirby, 1891) | AY707338, AY707361 | AY707382, AY707402 | AY707438 |
| Forficulidae | Forficulinae | Forficula sp. | AY521836 | AY521752 | AY521703 |
| Forficulidae | Forficulinae | Forficula auricularia Linnaeus, 1758 | - | EU426876 | - |
| Forficulidae | Opisthocosminae | Eparchus biroi (Burr, 1902) 1 | AY707324, AY707347 | AY707371, AY707391 | AY707427 |
| Forficulidae | Opisthocosminae | Eparchus biroi (Burr, 1902) 2 | AY521837 | AY521753, AY521754 | - |
| Forficulidae | Opisthocosminae | Opisthocosmia tenuis Rehn, 1936 | AY707321, AY707344 | - | AY707425 |
| Forficulidae | Opisthocosminae | Paratimomenus sp. | AY707322, AY707345 | - | - |
| Hemimeridae | Hemimerinae | Hemimerus sp. | AY707334, AY707357 | - | - |
| Hemimeridae | Hemimerinae | Hemimerus hanseni Sharp, 1895 | JX399776 | - | - |
| Labiduridae | Labidurinae | Forcipula clavata Liu, 1946 | AY707320, AY707343 | - | - |
| Labiduridae | Labidurinae | Forcipula decolyi de Bormans, 1900 | AY707327, AY707350 | AY707374, AY707394 | AY707430 |
| Labiduridae | Labidurinae | Labidura riparia (Pallas, 1773) | AY707333, AY707356 | AY707380, AY707400 | AY707435 |
| Labiduridae | Nalinae | Nala tenuicornis (de Bormans, 1900) | AY707336, AY707359 | - | AY707436 |
| Labiduridae | Nalinae | Nala lividipes (Dufour, 1820) | AY707339, AY707362 | - | - |
| Pygidicranidae | Echinosomatinae | Echinosoma sp. | AY121132 | - | AY125219 |
| Pygidicranidae | Echinosomatinae | Echinosoma micropteryx Günther, 1929 | AY707330, AY707353 | AY707377, AY707397 | AY707433 |
| Pygidicranidae | Echinosomatinae | Echinosoma yorkense Dohrn, 1869 | AY144626 | AY144652 | - |
| Pygidicranidae | Pygidicraninae | Cranopygia ophthalmica (Dohrn, 1862) | AY707340, AY707364 | AY707385, AY707405 | AY707440 |
| Pygidicranidae | Pygidicraninae | Tagalina sp. | AY521838 | AY521756 | AY521704 |
| Spongiphoridae | Labiinae | Labia sp. | AY521840 | AY521761, AY521762 | - |
| Spongiphoridae | Nesogastrinae | Nesogaster aculeatus (de Bormans, 1900) | AY707335, AY707358 | - | - |
| Spongiphoridae | Sparattinae | Auchenomus forcipatus Ramamurthi, 1967 | AY707329, AY707352 | AY707376, AY707396 | AY707432 |
| Spongiphoridae | Sparattinae | Auchenomus sp. | AY707337, AY707360 | - | AY707437 |
| Spongiphoridae | Spongiphorinae | Irdex sp. 1 | AY707365 | AY707386, AY707406 | AY707441 |
| Spongiphoridae | Spongiphorinae | Irdex sp. 2 | AY707366 | AY707387, AY707407 | AY707442 |
| Family | Species | 18s | 28s | H3 | |
| Embioptera | Oligotomidae | Oligotoma nigra (Hagen, 1866) | AY121134 | AY125274 | AY125221 |
| Embioptera | Teratembiidae | Teratembia sp. | AY121135 | AY125275 | AY125222 |
| Grylloblattodea | Grylloblattidae | Galloisana sp. | AY707341, AY707367 | AY707388, AY707408 | AY707443 |
| Grylloblattodea | Grylloblattidae | Grylloblattina djakonovi Bey-Bienko, 1951 | AY707342, AY707368 | AY707389, AY707409 | AY707444 |
| Orthoptera | Rhapidiophoridae | Ceuthophilus utahensis Thomas, 1876 | AY521870 | AY521800 | AY521720 |
| Orthoptera | Tridactylidae | Ellipes minutus (Scudder, 1892) | AY338723 | AY338679 | AY338641 |
| Phasmida | Heteronemiidae | Sceptrophasma langkawicensis Brock & Seow-Choen, 2000 | AY121166 | AY125306 | AY125249 |
| Phasmida | Pseudophasmatidae | Paraphasma rufipes (Redtenbacher, 1906) | AY121160 | AY125300 | AY125244 |
Assembling the Taxon-character Matrix
We used sequences from 35 other earwig species to determine the phylogenetic position of Arixenia esau and Hemimerus hanseni (Table 2). Among these comparative sequences, those from 34 species were obtained from Jarvis et al. [11], and those from one species were obtained from von Reumont et al. [44]. Following Jarvis et al. [11], we chose species from the orders Orthoptera, Phasmida, Embidiina, and Grylloblattodea as outgroups.
Because of large expansion regions in 28S and 18S ribosomal DNA and/or mutations in target sequences of universal primers for histone-3, the PCR amplification of some of these regions in the Dermaptera is problematic [cf. 11]. Sequences were aligned using Multalin [45] under default conditions and were edited in MEGA version 5 [46]. The use of different alignment software, e.g., Mafft [47], generated nearly identical results (data not shown). Regions with low consensus values (resulting from a lack of amplification, a lack of available complementary sequences, and/or a high degree of sequence polymorphisms including indels typical for non-protein coding regions) were excluded to avoid incorrect identification of homologies and/or overweighting of indels [48], [49]. Among the 18S DNA sequences, three regions with low consensus values were excluded: 48 bp at the 5′ end and 33 bp at the 3′ end were excluded because of a large number of missing sequences; a 156-bp region that included parts of the V4 hypervariable region described in De Rijk et al. [50] was excluded because of a combination of missing sequences and alignment failure. Among the 28S DNA sequences and following Jarvis et al. [11], we included a 509-bp region (including the D2 variable region described by De Rijk et al. [51]) at the proximal part (from the 5′ end) and a 378-bp region (including D4) in the middle of the gene, but we used more stringent conditions and therefore did not include the distal part of the gene because only a small number (<35%) of comparative sequences were available. A second alignment was derived from the variant described above by including only those parts with sequence information of Arixenia and Hemimerus. Both alignments are accessible at TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S14160).
Phylogenetic Analyses
A basic description of the genetic variability within the data set was obtained using DnaSP v5 [52]. The data set was divided into three parts corresponding to 18S DNA, 28S DNA, and histone-3 sequences. For each part, the best model of sequence evolution was calculated with the aid of MrModelTest 2.3 [53] using Akaike Information Criterion (AIC).
Bayesian analyses were conducted in MrBayes 3.2.1 [54] using the resultant GTR+I+Γ model. Each analysis consisted of two independent runs for 40 million generations; trees were sampled every 5000 generations, and the first 25% were discarded as burn-in. The AWTY (Are We There Yet?) approach was used to explore the convergence of MCMC in Bayesian inference [55]. The stationarity of the runs was inspected by cumulative plots displaying the posterior probabilities of splits at selected increments over an MCMC run, and the convergence was visualized by comparative plots displaying posterior probabilities of all splits for paired MCMC runs.
Results
The resultant alignment contained 2876 characters, including 1665 bp from 18S, 835 bp from 28S, and 376 bp from H3; 1853 positions in total were variable. The shortened alignment contained 2365 characters, including 1585 bp from 18S and 780 bp from 28S; 1531 positions in total were variable. Based on AIC in MrModelTest and ModelTest analyses, the GTR+I+Γ model was chosen as the best in all three independent analyses for individual partitions in both alignments. Within Bayesian phylogenetic inference, two chains converged at similar topologies. The standard deviation of the split frequencies reached values lower than 0.01 during the analysis. The stationarity was reached after approximately 2×107 generations (Fig. 1).
Figure 1. Results of the exploration of MCMC convergence using the AWTY (Are We There Yet?) approach.
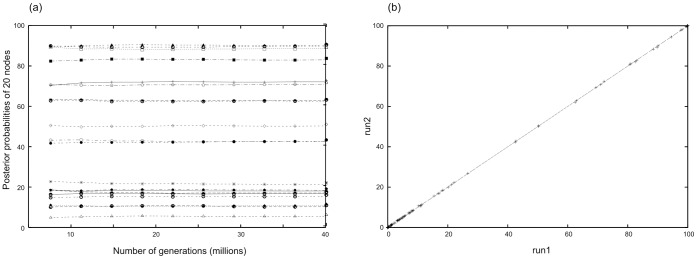
(a) Cumulative plot of the posterior probabilities of 20 splits at selected increments over one of two MCMC runs. (b) Comparative plot of posterior probabilities of all splits for paired one and two MCMC runs.
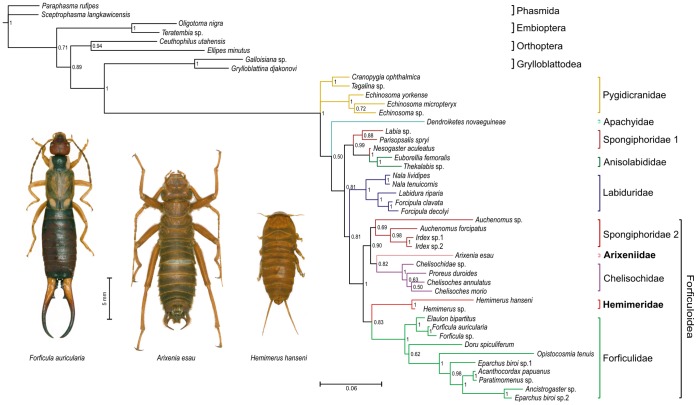
Bayesian analysis of the combined molecular data set produced a moderately resolved tree with several clear patterns (Fig. 2). Pygidicranidae, represented by Cranopygia, Tagalina, and Echinosoma, is unresloved in the tree with clades Cranopygia+Tagalina (PP(posterior probability) = 0.99) and Echinosoma (PP = 1) are monophyletic. Apachyidae is sister to the rest of the Dermaptera taxa except Pygidicranidae (PP = 0.50). Spongiphoridae is a polyphyletic/paraphyletic group with clades provisionally designated Sponiphoridae 1 and Spongiphoridae 2. The paraphyletic clade Spongiphoridae 1/Anisolabididae (PP = 0.99) contains the genera Labia, Parisolabis, Nesogaster, Euborellia, and Thekalabis. The monophyletic clade Spongiphoridae 2 (PP = 0.69) comprising Auchenomus+Irdex is sister to the Arixeniidae+Chelisochidae clade (PP = 0.82). Spongiphoridae 2+Arixeniidae+Chelisochidae+Hemimeridae+Forficulidae form a well-supported monophyletic clade Forficulidae (PP = 0.99), with two branches. Arixeniidae is sister to the Chelisochidae and forms a monophyletic clade with Spongiphoridae 2 (PP = 0.90), and Hemimeridae is sister to the monophyletic Forficulidae (PP = 0.83).
Figure 2. Bayesian phylogram of earwig families based on nuclear sequence data (18S and 28S ribosomal DNA and histone-3).
Numbers above branches indicate posterior probabilities.
Bayesian analysis of the shortened alignment produced a moderately resolved tree similar to that produced by the combined molecular data set (Figure S1). Spongiphoridae 2+Arixeniidae+Chelisochidae+Hemimeridae+Forficulidae consistently form a well-supported monophyletic clade Forficulidae (PP = 0.99), with three polytomic branches: Spongiphoridae 2 (PP = 0.95), Arixeniidae+Chelisochidae (PP = 0.69), and Hemimeridae+Forficulidae (PP = 0.64). Arixeniidae is sister to the Chelisochidae and forms a monophyletic clade, and Hemimeridae is sister to the monophyletic Forficulidae.
Discussion
Molecular Phylogeny of Arixeniids and Hemimerids
The current study is the first to include molecular data for both arixeniids and hemimerids in one phylogenetic analysis. This data set enabled us to apply a rigorous cladistics approach and to test competing hypotheses that were previously scattered in the literature. Based on the results, we conclude that Arixeniidae and Hemimeridae are highly specialized and modified families belonging to the suborder Neodermaptera and infraorder Epidermaptera [11]–[18]. The results placed both groups in the superfamily Forficuloidea and support the sister group relationships of Arixeniidae+Chelisochidae and Hemimeridae+Forficulidae.
The genetic markers used in our study (18S and 28S ribosomal DNA and histone-3) are highly conserved among animals and have been traditionally used in phylogenetic studies [56], [57]. However, complete resolution of the deep branching in the order Dermaptera, which originated in the middle Mesozoic [14], will require analysis of long portions of nuclear DNA. Given the moderate values of posterior probabilities accompanying the nodes for Arixeniidae+Chelisochidae and Hemimeridae+Forficulidae, analysis of long portions of nuclear DNA will be also important for fine-scale resolution of the branching order within the superfamily Forficuloidea. The latter kind of phylogenomic analysis was required, for example, for the fine-scale resolution of mammalian and avian evolutionary histories [58], [59]. Nevertheless, the methods and tests used in our study indicate that our hypotheses regarding the phylogenetic position of groups of our interest, the Arixeniidae and Hemimeridae, are well supported. Our report is the first to use the Bayesian tree-building approach to resolve dermapteran phylogeny. This method is characterized by high power and consistency and is especially useful in crossing deep valleys in a virtual landscape of phylogenetic trees, thereby ensuring an increased probability of reaching global and not only local optima [60]. Exploration of MCMC using AWTY indicates that stationarity and convergence of the chains were reached during particular runs.
The only previous molecular study that included hemimerids [11] indicated that the group nested within the Neodermaptera, but the exact placement was poorly supported. That analysis, which treated parameter values as unity, placed Hemimerus as sister to Forficulidae+Chelisochidae but with relatively low bootstrap and Bremer support. Our study, in contrast, well supports the position of both epizoic Hemimeridae and Arixeniidae in the monophyletic group Forficuloidea representing part of Spongiphoridae +Arixeniidae+Chelisochidae+Hemimeridae+Forficulidae; the Hemimeridae is clearly sister to the monophyletic Forficulidae.
Our results suggest (as do the results of Jarvis et al. [11]) that the Spongiphoridae is not a monophyletic group but rather consists of two distinct lineages. One lineage is paraphyletic and contains the entire Anisolabididae clade, and other lineage is nested in the “Eudermaptera” and is sister to the Chelisochidae. Discussions of relationships within the Dermaptera based on morphological characters have generally focused on the male genitalia and have subdivided the Dermaptera into two infraorders, the Catadermaptera (with two penis lobes) and the Eudermaptera (with one lobe) [61]–[64]. Colgan et al. [41] demonstrated that division of Dermaptera into the “Catadermaptera” and “Eudermaptera” is not supported because the former seems to be paraphyletic. “Eudermaptera” was considered to be monophyletic by these authors. However, polyphyly of Spongiphoridae, which was indicated by Jarvis et al. [11] and was confirmed in this study, is inconsistent with Steinman’s concept of infraorders based on penis characters because it indicates the possibility of repeated evolution of penis lobe reduction during the phylogeny of the Dermaptera.
Morphological Evolution vs. Molecular Evidence
The morphologies and life histories of the Arixeniidae and Hemimeridae are very different from those of the other Dermaptera. Adaptations to an epizoic life history resulted in the evolution of similar characters and thus the rise of homoplasies in data sets based on morphology.
The supraordinal classification of Grimaldi & Engel [14], which is based on paleontological material, comprises three suborders: the extinct Archidermaptera and Eodermaptera, and the recent Neodermaptera (corresponding to Hemimerina, Arixeniina, and Forficulina together in the traditional view). The internal phylogeny and classification of recent Dermaptera ( = Neodermaptera) has been in constant flux, with dramatically different arrangements of families and superfamilies by contemporaneous authors. Grimaldi & Engel [14] distinguish two infraorders within the Neodermaptera: the Protodermaptera and Epidermaptera. The Protodermaptera is basal including the families Karschielidae, Pygidicranidae, and Diplatyidae (sensu [19]), but may be paraphyletic, e.g., [9], [10]. Although the protodermapterans are well characterized by having ventral sclerites of equal size, carinae on the femora, and a segmented pygidium [10], [14], the validity of the infraordinal status of the Protodermaptera and the phylogeny within the infraorder must be verified by further analyses because the recent phylogenetic studies did not use representative material [11], [39], [42]. In particular, the phylogenetic position of the Karschielidae has not been studied by molecular methods. The Epidermaptera comprises the remainder of the recent families of earwigs. Based on his study of some morphological structures of the abdomen, Klass [12] reported that none of the abdominal characters contradicts a subordinate placement of Hemimerus within the Neodermaptera, and he noted weak support for a close relationship of Hemimerus to Apachyus (Apachyidae).
Haas [10] and Haas and Kukalova-Peck [9] performed extensive quantitative phylogenetic analysis of the Dermaptera based mainly on wing characters (especially the articulation and folding pattern of hind wings). Some of the characters seem to be apomorphic, but these characters cannot be used for the Hemimeridae and Arixeniidae because both of these epizoic groups have secondarily reduced wings. Recently, Tworzydlo et al. [14], [16]–[18] studied the ovary structure and initial stages of oogenesis of several dermapteran taxa, including the epizoic Arixenia esau Jordan, 1909. Their interpretation of the results supports the monophyly of the Dermaptera and the inclusion of the arixeniids in the Neodermaptera. Although the shared characters of the ovaries (elongated oviducts and a few, short ovarioles with a low number of follicles) support the position of the Arixeniina within the clade Forficuloidea comprising Chelisochidae+Forficulidae+Spongiphoridae, a placement of this taxon as a sister group of any of them is not strongly supported.
The families Chelisochidae and Forficulidae exhibit morphological autapomorphic characters on tarsomeres in that the third tarsal segment is inserted dorsally on the second tarsal segment and the second tarsal segment is enlarged [10]. The second tarsomere in the Forficulidae is lobed and very wide, while the second tarsomere in the Chelisochidae is not lobed but is enlarged into a peg-shaped process extending under the ventral surface of the third tarsomere [10], [11]. The third tarsal segment of both the Arixeniidae and Hemimeridae is inserted dorsally, which supports their relationship to the Chelisochidae and Forficulidae. The second tarsomere of the Hemimeridae is of the Forficulidae type (it is lobed and wide) and therefore could support the sister position of Hemimeridae+Forficulidae. The second tarsomere of the Arixeniidae is neither lobed nor enlarged but is a conspicuous process extending under the ventral surface of the third tarsomere. We cannot, however, exclude the possibility that the tarsomere characters in the Arixeniidae and Hemimeridae are adaptations to an epizoic life history rather than examples of synapomorphy.
Application of molecular phylogenetics has often generated hypotheses that are incongruent with results derived from a traditional approach based on morphological analyses [58], [59], [65]. Consequently, the “total evidence approach” may be complicated by conflicting signals between molecular and morphological data sets [66]. The phylogenetic reconstructions within a total evidence framework may be biased by the inclusion of phylogenetically misleading data, which are a priori more frequent in morphological data sets because of the greater potential for rapid adaptive change in particular phenotypic traits than in commonly used genetic traits. For example, the reduction of penis lobes, which was considered a synapomorphy in previous reconstructions of Dermaptera phylogeny [10], [12], [61]–[64], may result from parallel evolution, which is typical for reductive character states. From this point of view, molecular markers and corresponding tree-building methods are especially useful in phylogenetic resolution of groups with morphologically highly derived lineages because these methods effectively reveal relationships based on homologies [67], [68], as demonstrated in present study of epizoic lineages of earwigs.
Supporting Information
(a) Bayesian phylogram of earwig families based on nuclear sequence data (18S and 28S ribosomal) and including only those parts that have sequence information for both Arixenia and Hemimerus. Numbers above branches indicate posterior probabilities. Figure S1 (b, c) Results of the exploration of MCMC convergence (of shortened alignment) using the AWTY (Are We There Yet?) approach. (b) Cumulative plot of the posterior probabilities of 20 splits at selected increments over one of two MCMC runs. (c) Comparative plot of posterior probabilities of all splits for paired one and two MCMC runs.
(TIF)
Acknowledgments
We thank Robert Vlk and Boris Rychnovsky (Masaryk University, Brno, Czech Republic) for providing the material of Hemimerus hanseni. The authors thank Dr. Bruce Jaffee for linguistic and editorial improvements. We also thank two anonymous reviewers for helpful comments that improved the manuscript.
Funding Statement
The funding was provided by an Institutional Research Support grant from the Charles University in Prague (reg. no. SVV-2012-265 206) and an Institutional Research Support grant from the University of Ostrava (reg. no. SGS21/PrF/2013). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Popham EJ (2000) The geographical distribution of Dermaptera with reference to continental drift. J Nat Hist 34: 2007–2027. [Google Scholar]
- 2.Deem LS (2012) Dermaptera Species File. Version 1.0/4.0. [retrieval date]. Available: http://Dermaptera.SpeciesFile.org. Accessed 2013 May 31.
- 3. Sakai S (1982) A new proposed classification of the Dermaptera with special reference to the check list of the Dermaptera of the world. Bull Daito Bunka Univ 20: 1–108. [Google Scholar]
- 4. Haas F, Gorb SN, Wootton RJ (2000) Elastic joints in dermapteran hind wings : materials and wing folding. Arthropod Struct Dev 29: 137–146. [DOI] [PubMed] [Google Scholar]
- 5.Rentz DCF, Kevan DKM (1991) Dermaptera. In: Naumann ID, editor. The insects of Australia: a textbook for students and research workers. Ithaca, NY: Cornell University Press. 360–368.
- 6. Steinmann H (1973) A zoogeographical check-list of world Dermaptera. Folia Entomol Hung 26: 145–154. [Google Scholar]
- 7. Popham EJ (1985) The mutual affinities of the major earwig taxa (Insecta, Dermaptera). Z Zool Syst Evol 23: 199–214. [Google Scholar]
- 8. Giles ET (1963) The comparative external morphology and affinities of the Dermaptera. Trans R Entomol Soc Lond 115: 95–164. [Google Scholar]
- 9. Haas F, Kukalova-Peck J (2001) Dermaptera hindwing structure and folding: new evidence for superordinal relationship within Neoptera (Insecta). Eur J Entomol 98: 445–504. [Google Scholar]
- 10. Haas F (1995) The phylogeny of Forficulina, a suborder of the Dermaptera. Syst Zool 20: 85–98. [Google Scholar]
- 11. Jarvis KJ, Haas F, Whiting MF (2005) Phylogeny of earwigs (Insecta: Dermaptera) based on molecular and morphological evidence: reconsidering the classification of Dermaptera. Syst Entomol 30: 442–453. [Google Scholar]
- 12. Klass KD (2001) The female abdomen of the viviparous earwig Hemimerus vosseleri (Insecta: Dermaptera: Hemimeridae), with a discussion of the postgenital abdomen of Insecta. Zool J Linn Soc 131: 251–307. [Google Scholar]
- 13. Engel MS (2003) The Earwigs of Kansas, with a Key to Genera North of Mexico (Insecta: Dermaptera). Trans Kans Acad Sci 106: 115–123. [Google Scholar]
- 14.Grimaldi D, Engle M (2005) Evolution of Insects. Cambridge: Cambridge University Press. 755 p.
- 15. Engel MS, Haas F (2007) Family-Group names for earwigs (Dermaptera). Am Mus Novit American 3567: 1–20. [Google Scholar]
- 16. Tworzydlo W, Bilinski SM, Kocarek P, Haas F (2010) Ovaries and germline cysts and their evolution in Dermaptera (Insecta). Arthropod Struct Dev 39: 360–368. [DOI] [PubMed] [Google Scholar]
- 17. Tworzydło W, Kocarek P, Bilinski SM (2010) Structure of the ovaries and early stages of oogenesis in the parasitic earwig, Arixenia esau (Dermaptera: Arixenina). In: Abstracts of the XXIX Conference on Embryology Plants - Animals – Humans, May 19–21, 2010, Toruń-Ciechocinek, Poland. Acta Biol Cracov Bot 52 (suppl. 1)95. [Google Scholar]
- 18.Tworzydlo W, Lechowska-Liszka A, Kocarek P, Bilinski SM (2012): Morphology of the ovarioles and the mode of oogenesis of Arixenia esau support the inclusion of Arixeniina to the Eudermaptera. Zool Anz. doi:10.1016/j.jcz.2012.11.002
- 19.Sakai S (1987) Phylogenetic and evolutionary information on Dermaptera from the point of view of insect integrated taxonomy. In: Baccetti B, editor. Evolutionary Biology of Orthopteroid Insects, 496–513. New York: Halsted Press.
- 20. Kocarek P (2009) A case of viviparity in a tropical non-parasitizing earwig (Dermaptera Spongiphoridae). Trop Zool 22: 237–241. [Google Scholar]
- 21. Nakata S, Maa TC (1974) A review of the parasitic earwigs (Dermaptera, Arixeniina; Hemimerina). Pac Insects 16: 307–374. [Google Scholar]
- 22.Walker F (1871) Catalogue of the specimens of Dermaptera Saltatoria in the collection of the British Museum. Part. 5. London. 116 p.
- 23. Saussure H (1879) Spicilegia entomologica genevensia. 1. Sur le genre Hemimerus Walk. Mem Soc Hist Nat Geneve 26: 399–420. [Google Scholar]
- 24. Sharp D (1892) Concerning Hemimerus talpoides Walk. Ent Mon Mag 28: 212–213. [Google Scholar]
- 25. Sharp D (1895) Family II. Hemimeridae. In: Harmer SF, Shipley AE (Eds.): The Cambridge Natural History 5: 217–219. [Google Scholar]
- 26.Burr M (1911) Dermaptera. In: Wytsman PAG, editor. Genera Insectorum, Fascicle 122. Bruxelles: Wytsman. 112 p.
- 27. Heymons R (1911) Ueber die Lebensweise von Hemimerus (Orth.). Deut Ent Z 1911: 163–174. [Google Scholar]
- 28. Hansen HJ (1933) External organs in Dermaptera especially Forficulidae. Ent Meddel 18: 349–358. [Google Scholar]
- 29. Rehn JAG, Rehn JWH (1935) A Study of the Genus Hemimerus (Dermaptera, Hemimerina, Hemimeridae). Proc Acad Nat Sci Phila 87: 457–508. [Google Scholar]
- 30. Rehn JAG, Rehn JWH (1937) Further Notes on the Genus Hemimerus (Dermaptera, Hemimerina, Hemimeridae). Proc Acad Nat Sci Phila 89: 331–335. [Google Scholar]
- 31. Popham EJ (1961) On the systematic position of Hemimerus Walker – a case for ordinal status. Proc R Entomol Soc Lond Ser B 30: 19–25. [Google Scholar]
- 32. Jordan K (1909) Description of a new kind of apterous earwig, apparently parasitic on a bat. Novit Zool 16: 313–326. [Google Scholar]
- 33.Hendlirsch A (1925) Ordnung: Dermaptera (Deg.) Kirby (Ohrwurmer). In: Schroder C (Ed.) Handbuch der Entomologie 3, 472–477.
- 34. Popham EJ (1962) The anatomy related to the feeding habits of Arixenia and Hemimerus (Dermaptera). Proc Zool Soc Lond 139: 429–450. [Google Scholar]
- 35. Popham EJ (1965) The functional morphology of the reproductive organs of the common earwig (Forficula auricularia) and other Dermaptera with reference to the natural classification of the order. J Zool 146: 1–43. [Google Scholar]
- 36.Sakai S, Hoshiba H, Imanishi M (1988) Cytotaxonomic studies on the chromosomes of the Dermaptera with special reference to two chelisochid species. In: International Congress of Entomology XVIII, p.62.
- 37. White MJD (1971) The chromosomes of Hemimerus bouvieri Chopard (Dermaptera). Chromosoma 34: 183–189. [DOI] [PubMed] [Google Scholar]
- 38. White MJD (1972) The chromosomes of Arixenia esau Jordan (Dermaptera). Chromosoma 36: 338–342. [DOI] [PubMed] [Google Scholar]
- 39. Wirth T, Le Guellec R, Veuille M (1999) Directional substitution and evolution of nucleotide content in the cytochrome oxidase II gene in earwigs (dermapteran insects). Mol Biol Evol 16: 1645–1653. [DOI] [PubMed] [Google Scholar]
- 40. Guillet S, Vancassel M (2001) Dermapteran life-history evolution and phylogeny with special reference to the Forficulidae family. Evol Ecol Res 3: 441–447. [Google Scholar]
- 41. Colgan DJ, Cassis G, Beacham E (2003) Setting the molecular phylogenetic framework for the Dermaptera. Insect Syst Evol 34: 65–80. [Google Scholar]
- 42. Kamimura Y (2004) In search of the origin of twin penises: molecular phylogeny of earwigs (Dermaptera: Forficulina) based on mitochondrial and nuclear ribosomal RNA Genes. Ann Entomol Soc Am 97: 903–912. [Google Scholar]
- 43. Whiting MF (2002) Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoologica Scripta 31: 93–104. [Google Scholar]
- 44. Von Reumont BM, Meusemann K, Szucsich NU, Dell Ampio E, Gowri-Shankar V, et al. (2009) Can comprehensive background knowledge be incorporated into substitution models to improve phylogenetic analyses? A case study on major arthropod relationships. BMC Evol Biol 9: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucl Acids Res 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Katoh K, Kuma K, Toh H, Miyata T (2004) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nuc Acid Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [DOI] [PubMed] [Google Scholar]
- 49.Warnow T (2012) Standard maximum likelihood analyses of alignments with gaps can be statistically inconsistent. PLOS Curr. Tree of Life: RRN1308. [DOI] [PMC free article] [PubMed]
- 50. De Rijk P, Neefs JM, Van de Peer Y, De Wachter R (1992) Compilation of small ribosomal subunit RNA sequences. Nucl Acids Res 20: 2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. De Rijk P, Van de Peer Y, Chapelle S, De Wachter R (1994) Database on the structure of large ribosomal subunit RNA. Nucl Acids Res 22: 3495–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Librado P, Rozas J (2009) DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 53. Posada D, Crandall KA (2001) Selecting the best-fit model of nucleotide substitution. Syst Biol 50: 580–601. [PubMed] [Google Scholar]
- 54. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilgenbusch JC, Warren DL, Swofford DL (2004) AWTY: A system for graphical exploration of MCMC convergence in Bayesian phylogenetic inference. Available: http://ceb.csit.fsu.edu/awty. Accessed 2013 May 31. [DOI] [PubMed]
- 56. Field KG, Olsen GJ, Lane DJ, Giovannoni SJ, Ghiselin MT, et al. (1988) Molecular phylogeny of the animal kingdom. Science 239: 748–753. [DOI] [PubMed] [Google Scholar]
- 57. Hedges SB, Moberg KD, Maxson LR (1990) Tetrapod phylogeny inferred from 18s and 28s ribosomal RNA sequences and review of the evidence for amniote relationships. Mol Biol Evol 7: 607–633. [DOI] [PubMed] [Google Scholar]
- 58. Murphy WJ, Eizirik E, O’Brien SJ, Madsen O, Scally M, et al. (2001) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348–2351. [DOI] [PubMed] [Google Scholar]
- 59. Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, et al. (2008) A phylogenomic study of birds reveals their evolutionary history. Science 320: 1763–1768. [DOI] [PubMed] [Google Scholar]
- 60. Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294: 2310–2314. [DOI] [PubMed] [Google Scholar]
- 61.Steinmann H (1986) Dermaptera: Catadermaptera I. Tierreich 102. Berlin–New York: Walter de Gruyter. 345 p.
- 62.Steinmann H (1989) Dermaptera Catadermaptera 2. Tierreich 108. Berlin–New York: Walter de Gruyter. 504 p.
- 63.Steinmann H (1990) Dermaptera: Eudermaptera I. Tierreich 106. Berlin–New York: Walter de Gruyter. 558 p.
- 64.Steinmann H (1993) Dermaptera: Eudermaptera II. Tierreich 108. Berlin–New York: Walter de Gruyter. 709 p.
- 65.Patterson C (1987) Molecules and morphology in evolution: conflict or compromise? Cambridge: Cambridge University Press. 229 p.
- 66. Lecointre G, Deleporte P (2005) Total evidence requires exclusion of phylogenetically misleading data. Zoologica Scripta 34: 101–117. [Google Scholar]
- 67.Page RDM, Holmes EC (2000) Molecular evolution: a phylogenetic approach. Oxford: Blackwell science.
- 68.Avise JC (2004) Molecular markers, natural history and evolution. 2nd ed. Sunderland, MA: Sinauer Associates, Inc. 684 p.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Bayesian phylogram of earwig families based on nuclear sequence data (18S and 28S ribosomal) and including only those parts that have sequence information for both Arixenia and Hemimerus. Numbers above branches indicate posterior probabilities. Figure S1 (b, c) Results of the exploration of MCMC convergence (of shortened alignment) using the AWTY (Are We There Yet?) approach. (b) Cumulative plot of the posterior probabilities of 20 splits at selected increments over one of two MCMC runs. (c) Comparative plot of posterior probabilities of all splits for paired one and two MCMC runs.
(TIF)