Abstract
Objective
A negative relationship between total bilirubin concentration (TBili) and CVD risk has been documented in a series of epidemiological studies. In addition, TBili is thought to be under strong genetic regulation via the UGT1A gene family, suggesting it may be a heritable CVD risk factor. However, few studies directly relate TBili-associated UGT1A variants to CVD severity or outcome. This study replicated the genetic association for TBili in the Diabetes Heart Study (DHS), and examined the relationships of TBili-associated SNPs with measures of subclinical CVD and mortality.
Methods
This investigation included 1220 self-described European American (EA) individuals from the DHS, a family-based study examining risk for macrovascular complications in type 2 diabetes (T2D). Genetic associations with TBili were examined using the Affymetrix Genome-wide Human SNP Array 5.0 and the Illumina Infinium Human Exome beadchip v1.0. Subsequent analyses assessed the relationships of the top TBili-associated SNPs with measures of vascular calcified plaque and mortality.
Results
A genome-wide association study (GWAS) detected 18 SNPs within the UGT1A gene family associated with TBili at p<5×10-8. The top hit was rs887829 (p=8.67×10-20). There was no compelling evidence of association between the top TBili-associated SNPs and vascular calcified plaque (p=0.05-0.88). There was, however, evidence of association with all-cause mortality (p=0.0004-0.06), the top hit being rs2741034.
Conclusion
These findings support a potential role for UGT1A genetic variants in risk for mortality in T2D. Further quantification of the extent of CVD risk conferred by UGT1A gene family variants in a high risk cohort with T2D is still required.
Keywords: bilirubin, genetics, cardiovascular disease, vascular calcified plaque
1. INTRODUCTION
Bilirubin, the end product of heme catabolism in the systemic circulation [1], has been documented as a risk factor for cardiovascular disease (CVD). The association of low total bilirubin concentrations (TBili) with increased CVD risk was initially reported in an investigation involving an asymptomatic, male population (n=877) with <35% having prevalent coronary artery disease (CAD) defined by angiography [2]. This negative association was subsequently confirmed in a smaller familial CAD sample (161 CAD cases and 155 controls) [3] and has been further replicated and confirmed in both larger and prospective investigations (see Schwertner and Vitek [4] for review of key studies), including in populations with clinical hyperbilirubinemia [5]. The increased risk for CVD associated with lower bilirubin concentrations has also been observed in the context of subclinical CVD with a meta-analysis, utilizing data from male subjects only, supporting a clear negative association between serum bilirubin concentrations and severity of atherosclerosis [6]. Significant negative associations have also reported from studies of Asian populations utilizing both coronary artery calcified plaque (n=637) [7, 8] and indices of carotid intima-media thickening (n=172) [9].
The mechanisms by which TBili contributes to reduced CVD risk are likely to include both antioxidant actions [1, 4], including the prevention of LDL oxidation [10, 11], and anti-inflammatory effects [12]. Whether the TBili-CVD risk paradigm is similarly relevant in patients with type 2 diabetes (T2D), a disease characterized by excess CVD-mortality, increased inflammation [13, 14] and where abnormal liver function measures have been previously reported [15], has not been thoroughly investigated. One small study in a diabetic sample of European origin (n=80) supports the negative association between TBili and carotid intima media thickness [16]. More recently, TBili concentrations were found to be negatively associated with lower limb amputation, reflecting peripheral vascular disease, in the Fenofibrate Intervention and Event Lowering in Diabetes Study [17]. Finally, the report of a negative relationship between TBili and glycated hemoglobin levels in a Korean sample with T2D [18] supports the need for further investigation of the TBili-CVD risk paradigm in diabetic populations.
Bilirubin concentrations are thought to be under strong genetic regulation. Polymorphism of a TA repeat sequence in the UGT1A1 gene promoter (UGT1A1*28; rs8175347) has been identified as an important risk factor in Gilbert Syndrome, a benign clinical hyperbilirubinemia [19], with increasing repeat length associated with decreased gene promoter activity [20]. More recently, genome-wide association studies (GWAS) have also shown strong and reproducible associations between single nucleotide polymorphism (SNPs) in the UGT1A gene cluster and TBili concentrations [21-23]. However, despite the multiple epidemiological studies linking TBili to CVD risk, few existing reports directly relate the TBili-associated UGT1A gene family variants to measures of CVD severity or outcome. Association between the UGT1A1*28 microsatellite and coronary heart disease has been investigated in both the Rotterdam [24] and Framingham offspring [11] studies, with conflicting results. We are unaware of any similar studies involving the GWAS-identified bilirubin-associated UGT1A SNPs. As such, we sought to replicate the TBili-UGT1A association in the Diabetes Heart Study (DHS), a family based study examining risk for macrovascular complications in T2D, and examine the relationships of the TBili-UGT1A associated SNPs with measures of subclinical CVD and mortality.
2. METHODS
2.1 Study Design and Sample
This investigation included 1,220 self-described European American (EA) individuals from 475 families comprising the Diabetes Heart Study (DHS) cohort. Briefly, the DHS includes siblings concordant for T2D, but without advanced renal insufficiency. When possible, unaffected siblings were also recruited. T2D was clinically defined as diabetes developing after the age of 35 years and treated with insulin and/or oral agents, in the absence of historical evidence of ketoacidosis. Ascertainment and recruitment have been previously described in detail [25, 26].
Study protocols were approved by the Institutional Review Board at Wake Forest School of Medicine, and all participants provided written informed consent prior to participation. Participant examinations were conducted in the General Clinical Research Center of the Wake Forest Baptist Medical Center, and included interviews for medical history (including self-reported history of prior CVD events or intervention) and health behaviors, anthropometric measures, resting blood pressure, electrocardiography, fasting blood sampling for laboratory analyses, and spot urine collection. Standard laboratory analyses included fasting glucose, glycated hemoglobin (HbA1C), blood lipids and measures of liver function, including total bilirubin concentrations, alkaline phosphatase (ALP), serum glutamic oxaloacetic transaminase (SGOT) and serum glutamic pyruvate transaminase (SGPT). Total (conjugated and ungonjugated) bilirubin concentrations were determined using the timed-endpoint diazo method with 2,5-dicholophenyl diazonium. Measures of subclinical CVD included coronary artery calcified plaque (CAC), carotid artery calcified plaque (CarCP) and infra-renal abdominal aortic calcified plaque (AACP), measured using fast-gated helical CT scanners, with calcium scores calculated as previously described [27, 28]. Not all measurements were available for all participants.
2.2 Mortality
Vital status was determined for all participants from the National Social Security Death Index maintained by the United States Social Security Administration. For those participants confirmed as deceased, length of follow-up was determined from the date of the initial study visit to date of death. For deceased participants, copies of death certificates were obtained from relevant county Vital Records Offices to confirm cause of death. For all other participants the length of follow-up was determined from the date of the initial study visit to the end of 2011. Cause of death was categorized based on information contained in death certificates as CVD-mortality (myocardial infarction, congestive heart failure, cardiac arrhythmia, sudden cardiac death, peripheral vascular disease, and stroke) or either cancer, infection, end-stage renal disease, accidental, or other (including obstructive pulmonary disease, pulmonary fibrosis, liver failure and Alzheimer's dementia).
2.3 Genetic Analysis
Genomic DNA was purified from whole blood samples using the PUREGENE DNA isolation kit (Gentra, Inc., Minneapolis, MN). DNA concentration was quantified using standardized fluorometric readings on a Hoefer DyNA Quant 200 fluorometer (Hoefer Pharmacia Biotech Inc., San Francisco, CA). Genotyping was completed in two stages (i) using the Affymetrix genotyping platform and the Affymetrix Genome-wide Human SNP Array 5.0 (Affymetrix, CA, USA) (the GWAS set) and (ii) using the Illumina Infinium Human Exome beadchip v1.0 (Illumina, Ca, USA) (the Exome set).
For the GWAS set, genotype calling was completed using the BRLLM-P algorithm in Genotyping Console v4.0 (Affymetrix). Samples failing to meet an intensity quality control (QC) threshold (n=4) were not included for genotype calling and those failing to meet a minimum acceptable call rate of 95% (n=3) were excluded from further analyses. An additional 39 samples were included as blind duplicates within the genotyping set to serve as quality controls; the concordance rate for these blind duplicates was 99.0 ± 0.72% (mean ± SD).
For the Exome set, genotype calling was completed using Genome Studio Software v1.9.4 (Illumina). Samples failing to meet a minimum acceptable call rate of 98% (n=3) were excluded from further analyses. An additional 58 samples were included as blind duplicates within the genotyping set to serve as quality controls; the concordance rate for blind duplicates was 99.9 ± 0.0001% (mean ± SD).
Following genotype calling, exploratory analyses of genotype data were performed using PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) and identified samples with poor quality genotype calls, gender errors, or unclear/unexpected sibling relationships; all of these were excluded from further analysis. A total of 1,180 samples with available genotyping data in both the GWAS and Exome sets were analyzed. For the GWAS set, exclusion criteria for SNP performance included call rate <95% (n=11,085), Hardy-Weinberg Equilibrium p-value <1×10-6 (n=332), and minor allele frequency <0.01 (n=57,382); 371,951 SNPs were retained for analysis. For the Exome set, exclusion criteria for SNP performance included call rate <99% (n=972), monomorphic SNPs (n=157,754) and Hardy-Weinberg Equilibrium p-value <1×10-6 (n=26); 88,483 SNPs were retained for analysis.
2.4 Statistical Analysis
Summary statistics were calculated including means and standard deviations (SD) for continuous variables, and count and percentages for categorical variables. Continuous variables were transformed prior to analysis to approximate conditional normality. Due to the inclusion of related individuals in this study, marginal models with incorporation of generalized estimating equations (GEE) were used to describe associations between the TBili and continuous measures of subclinical CVD (CAC, CarCP and AACP). In order to compare the relative effect, dependent variables (CAC, CarCP, AACP) were standardized for analysis of phenotypic associations. Analysis of the univariate associations between TBili and measures of vascular calcified plaque were undertaken initially. Each of these associations was then adjusted for (i) age, sex and BMI (partially adjusted) and (ii) age sex, BMI, T2D affected status, hypertension, current smoking and dyslipidemia (fully adjusted). Cox proportional hazards models with sandwich-based variance estimation were used to examine the relationships between TBili and all-cause mortality, as well as between TBili and either CVD-mortality or non-CVD mortality. Cox proportional hazards models were partially and fully adjusted as described above. The same analyses were repeated restricting to the T2D affected individuals only. SAS version 9.2 (SAS Institute Inc., Cary, NC) was used for the analysis. Statistical significance was accepted a p<0.05.
Genetic association analyses were performed using variance components methods as implemented in Sequential Oligogenic Linkage Analysis Routines (SOLAR) version 6.3.4 (Texas Biomedical Research Institute, San Antonio, Tx, USA) to account for family relationships. For genome-wide TBili association an additive model of inheritance was assumed with adjustment for age, sex and T2D affected status. The relationship between the top TBili-associated SNPs (p<5×10-8) with measures of vascular calcified plaque and mortality were examined, also under an additive model, with adjustment for age, sex and T2D affected status. These analyses were repeated with additional adjustment for TBili to further account for the potential contribution of TBili to the phenotypic variance. All analyses were repeated restricting to the T2D affected individuals only. Genome-wide significance was accepted at p<5×10-8.
3. RESULTS
The demographic and clinical characteristics of the DHS cohort are presented in Table 1. As anticipated in a T2D-enriched sample, a predominance of traditional CVD risk factors were evident including high body mass index (BMI), hypertension, dyslipidemia and prior CVD events. Scores for CAC, CarCP and AACP reflect a substantial burden of subclinical CVD in these diabetes-affected individuals and their siblings. Average total bilirubin concentrations were 13.9 ± 5.1 μmol/L and were within the laboratory reference range (3.4-20.5 μmol/L). Average values for other measured liver enzymes (ALP, SGOT, SGPT) were also within established laboratory reference ranges. This cohort was followed for 8.2 ± 2.6 yrs (mean ± SD) over which time 247 (20.9%) participants were deceased, 107 (9.1%) from CVD causes.
Table 1.
Demographic characteristics of the 1,180 DHS participants with genotype data.
| Mean ± SD or % | Median (inter-quartile range) | |
|---|---|---|
| Demographic Information | ||
| Age (years) | 62.1 ± 9.3 | 62.6 (55.7-69.1) |
| Gender (% female) | 53.5% | |
| Type 2 diabetes affected (%) | 83.7% | |
| Diabetes Duration (years) | 10.5 ± 7.2 | 8 (5-14) |
| % smoking (current or past) | 58.7% | |
| Self-reported history of prior CVD | 38.9% | |
| Deceased | 20.9% | |
| Deceased (CVD causes) | 9.1% | |
|
Body Composition | ||
| Height (cm) | 168.6 ± 9.7 | 168.2 (161.0-175.7) |
| Weight (kg) | 90.5 ± 20.3 | 88.1 (76.0-103.9) |
| BMI (kg/m2) | 31.8 ± 6.5 | 30.7 (27.2-35.6) |
|
Medications | ||
| Lipid Lowering (%) | 44.7% | |
| Anti-hypertensive (%) | 62.6% | |
|
Blood Pressure | ||
| Systolic BP (mmHg) | 139 ± 19 | 138 (126-151) |
| Diastolic BP (mmHg) | 73 ± 10 | 72 (66-80) |
| Hypertension (%) | 85.6% | |
|
Blood Biochemistry | ||
| Glucose (mg/dL) | 139.4 ± 55.4 | 127 (102-164) |
| Hemoglobin A1C (%) | 7.3 ± 1.8 | 6.9 (6.1-8.1) |
| Total Cholesterol (mg/ dL) | 186.6 ± 42.6 | 183.0 (158.8-209.3) |
| HDL-cholesterol (mg/dL) | 43.1 ± 12.6 | 41 (35-50) |
| LDL-cholesterol (mg/dL) | 104.9 ± 32.7 | 102 (81-125) |
| Triglycerides (mg/dL) | 201.0 ± 132.7 | 171 (116-242) |
| Total Bilirubin (umol/L) | 13.9 ± 5.1 | 13.7 (10.3-15.4) |
| ALP (U/L) | 78 ± 25 | 76 (61-91) |
| SGOT/AST (U/L) | 27 ± 12 | 27 (20-30) |
| SGPT/ALT (U/L) | 24 ± 13 | 24 (17-27) |
|
Vascular Imaging – Calcified Plaque (CP) | ||
| Coronary Artery CP | 1672 ± 3179 | 348 (33-2043) |
| Carotid Artery CP | 313 ± 671 | 44 (0-297) |
| Abdominal Aortic CP | 11158 ± 15939 | 4020 (550-16040) |
In unadjusted analyses a positive association was observed between TBili and both CAC and CarCP. This was negated following adjustment for traditional CVD risk factors including age, sex, BMI, T2D affected status, current smoking, hypertension and dyslipidemia (Table 2). There were no significant associations between TBili and either all-cause, CVD-mortality, or non-CVD mortality. These relationships were essentially unchanged when analyses were restricted to T2D affected individuals only (Supplementary Material, Table 1).
Table 2.
Associations between total bilirubin and measures of vascular calcified plaque and mortality (unadjusted and adjusted models) presented as beta (β) estimate and standard error (SE) for continuous traits and hazard ratio (HR) and 95 % confidence interval (CI) for discrete traits.
| Unadjusted | Partially Adjusted* | Fully Adjusted† | ||||
|---|---|---|---|---|---|---|
| β (SE) | p-value | β (SE) | p-value | β (SE) | p-value | |
| CAC | 0.784 (0.201) | <0.0001 | 0.121 (0.178) | 0.50 | 0.163 (0.180) | 0.37 |
| CarCP | 0.553 (0.200) | 0.0056 | 0.158 (0.181) | 0.38 | 0.188 (0.181) | 0.30 |
| AACP | 0.199 (0.234) | 0.39 | -0.205 (0.209) | 0.33 | -0.111 (0.212) | 0.60 |
| HR (CI) | p-value | HR (CI) | p-value | HR (CI) | p-value | |
|---|---|---|---|---|---|---|
| All-cause mortality | 2.06 (0.85-5.00) | 0.11 | 1.29 (0.51-3.25) | 0.60 | 1.59 (0.63-4.01) | 0.32 |
| CVD mortality | 1.53 (0.40-5.89) | 0.54 | 0.89 (0.23-3.57) | 0.89 | 0.87 (0.23-3.34) | 0.84 |
| Non-CVD mortality | 1.60 (0.86-2.99) | 0.14 | 1.35 (0.72-2.55) | 0.35 | 1.55 (0.82-2.92) | 0.18 |
CAC = coronary artery calcified plaque, CarCP= carotid artery calcified plaque. AACP = abdominal aortic calcified plaque
adjusted for age, sex, BMI
adjusted for age, sex, BMI, T2D affected status, current smoking, hypertension, dyslipidemia
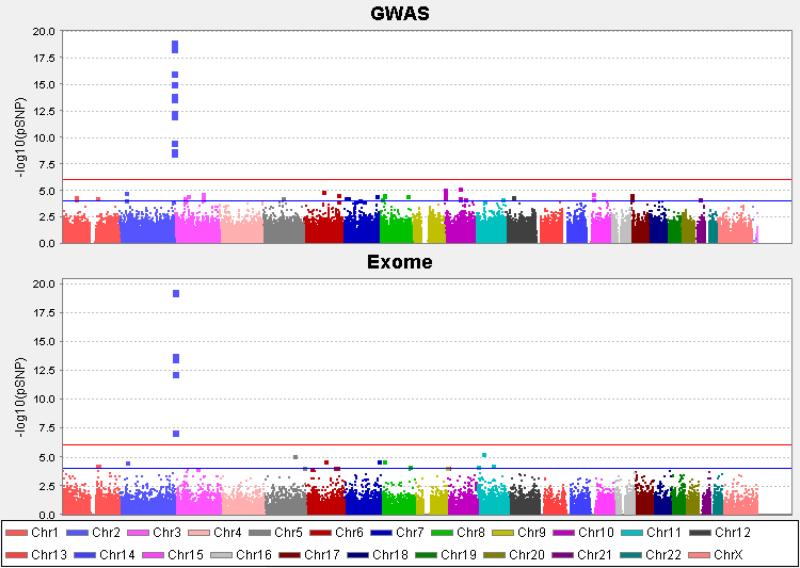
Genetic association analysis revealed 14 SNPs within the UGT1A gene cluster to be associated with TBili at p<5×10-8 for the GWAS set (Figure 1a); the most strongly associated SNP was rs887829 (p=8.67×10-20). For the Exome set, six SNPs within the UGT1A gene cluster were also associated with TBili at p<5×10-8 (Figure 1b). Two of these SNPs (rs887829 and rs6759892) were contained within the GWAS set. All associated SNPs were in at least moderate linkage disequilibrium (LD; r2=0.2-0.8) with the lead GWAS SNP rs887829 (Supplementary Material, Figure 1).
Figure 1.
Manhattan plots for genotypic associations with total Bilirubin concentrations for the GWAS and Exome data sets
There was no compelling evidence of association between the 18 top TBili-associated SNPs and CAC at the genome-wide level (Table 3), however, following adjustment for TBili, five SNPs trended towards nominal association (p<0.05) (Table 3). There was no evidence supporting association between the 18 top TBili-associated SNPs and either CarCP (p=0.08-0.89) or AACP (p=0.14-0.95). Finally, modest associations were observed between the 18 top TBili-associated SNPs and all-cause mortality (Table 3), but not with CVD-mortality (p=0.04-0.55). The associations with mortality were also enhanced following adjustment for TBili (Table 3). Bilirubin concentrations and all-cause mortality rates by SNP are included in the Supplementary Material (Supplementary Material, Table 2). Patterns were generally similar when analyses were restricted to T2D affected individuals only (Supplementary Material, Table 3).
Table 3.
UGT1A gene cluster SNP associations with total bilirubin concentrations (TBili), coronary artery calcified plaque (CAC) and mortality.
| Association p-values |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | Location | Alleles (Maj/Min) | MAF | Source | TBili* | CAC* | CAC† | All-cause mortality* | All-cause mortality† |
| rs2741023 | 234516714 | upstream | G/A | 0.368 | GWAS | 2.55×10-10 | 0.32 | 0.17 | 0.0006 | 7.1 ×10-05 |
| rs2741029 | 234530160 | intronic | A/C | 0.289 | GWAS | 2.18×10-9 | 0.88 | 0.47 | 0.002 | 0.0004 |
| rs2741034 | 234548814 | intronic | A/G | 0.278 | GWAS | 1.49×10-9 | 0.31 | 0.16 | 0.0004 | 4.5 ×10-05 |
| rs2741045 | 234580140 | intronic | C/T | 0.294 | GWAS | 2.02×10-14 | 0.29 | 0.14 | 0.002 | 0.0002 |
| rs10197460 | 234589190 | intronic | G/T | 0.388 | GWAS | 3.70×10-13 | 0.05 | 0.02 | 0.01 | 0.002 |
| rs6736508 | 234595747 | intronic | G/A | 0.405 | GWAS | 4.71×10-13 | 0.06 | 0.03 | 0.03 | 0.007 |
| rs7563561 | 234598991 | intronic | T/G | 0.406 | GWAS | 7.18×10-13 | 0.06 | 0.03 | 0.03 | 0.006 |
| rs7608175 | 234599089 | intronic | C/G | 0.405 | GWAS | 7.45×10-13 | 0.06 | 0.03 | 0.04 | 0.007 |
| rs6759892 | 234601669 | Misense¶ | T/G | 0.406 | GWAS§ | 7.93×10-13 | 0.05 | 0.02 | 0.03 | 0.007 |
| rs2070959 | 234602191 | Misense¶ | A/G | 0.313 | Exome | 2.48×10-14 | 0.29 | 0.11 | 0.003 | 0.0006 |
| rs1105879 | 234602202 | Misense¶ | A/C | 0.334 | Exome | 1.21×10-14 | 0.27 | 0.11 | 0.01 | 0.003 |
| rs6744284 | 234625297 | intronic | C/T | 0.295 | GWAS | 3.85×10-19 | 0.54 | 0.33 | 0.02 | 0.005 |
| rs11891311 | 234639310 | intronic | G/A | 0.367 | GWAS | 6.71×10-17 | 0.33 | 0.18 | 0.06 | 0.01 |
| rs4663965 | 234650604 | intronic | T/C | 0.453 | GWAS | 1.05×10-14 | 0.29 | 0.19 | 0.03 | 0.01 |
| rs3755319 | 234667582 | intronic | A/C | 0.456 | GWAS | 8.09×10-16 | 0.59 | 0.66 | 0.02 | 0.01 |
| rs887829 | 234668570 | intronic | C/T | 0.312 | GWAS§ | 8.67×10-20 | 0.27 | 0.13 | 0.006 | 0.001 |
| rs6742078 | 234672639 | intronic | C/A | 0.318 | Exome | 4.44×10-20 | 0.41 | 0.19 | 0.007 | 0.001 |
| rs4148325 | 234673309 | intronic | G/A | 0.318 | Exome | 4.44×10-20 | 0.41 | 0.19 | 0.007 | 0.001 |
assuming an additive model of inheritance with adjustment for age, sex and T2D affected status
assuming an additive model of inheritance with adjustment for age, sex and T2D affected status and bilirubin concentrations
SNPs contained in both the GWAS and Exome sets; results reported for GWAS-derived data
SNPs contained in the coding region of the UGT1A6 splice variant
4. DISCUSSION
The current study successfully replicated the association of genetic variants in the UGT1A gene cluster with total bilirubin concentrations in a sample enriched for T2D. Although these variants were not associated with measures of vascular calcified plaque at the genome-wide level, modest association with all-cause mortality was observed. Despite these relationships, direct phenotypic correlations were not evident between TBili and either subclinical CVD, or outcome, after accounting for traditional CVD risk factors. However, evidence from this investigation highlights the need for further quantification of the risk for adverse outcomes conferred by genetic variation in the UGT1A family among individuals with T2D.
The UGT1A gene cluster association with TBili is biologically plausible. The UGT1A gene cluster encodes the set of uridine diphosphate-glucuronosyltransferase enzymes which are important components of the glucuronidation pathway necessary for the conversion of lipophilic to water soluble molecules for excretion [11, 23] (i.e. conversion of unconjugated to conjugated bilirubin). In this way, the UGT1A gene products contribute to the regulation of bilirubin clearance/excretion [4, 24] and, as such, to total TBili concentrations. Although a limited number of studies have directly examined the interplay between UGT1A genetic variants and CVD risk and/or outcomes [11, 24], we are unaware of any attempts to do this using GWAS identified TBili-associated SNPs. Although able to successfully replicate the association of UGT1A gene cluster with TBili, we did not detect a genome-wide significant relationship between the TBili-associated SNPs and measures of subclinical CVD used in this study. Low penetrance of these genetic variants cannot be discounted here.
Studies that have previously examined the association between UGT1A*28 microsatellite and CVD have produced conflicting results. In the Rotterdam Study, the UGT1A1*28 microsatellite was not significantly associated with coronary heart disease, nor were TBili concentrations associated with disease [24]. In contrast, a subsequent investigation in the Framingham Offspring cohort found an association between the UGT1A1*28 microsatellite (rs8175347) and risk for CVD events. The UGT1A1*28 variant associated with increased bilirubin concentrations (i.e. increased microsatellite length) was also associated with a >50% reduction in risk for CVD events over 24 years of follow-up [11], in keeping with the earlier purported negative relationship. Given the proximity of our lead SNP (rs887829) to UGT1A*28 microsatellite (rs8175347) (311bp apart) and the LD structure across the region (based on available HapMap data), we consider it likely that the identified TBili-associated SNPs from the current study are, at minimum, moderately correlated with the previously identified UGT1A*28 microsatellite (Supplementary Material, Figure 2). In addition, given their correlated nature, this set of 18 SNPs most likely reflect a single association signal from this region with the causal variant yet to be resolved.
Whether the lack of observed association between the UGT1A variants and measures of CVD in the DHS is the result of the T2D-enriched nature of the sample is not clear as the majority of existing studies were not enriched for T2D-affected individuals. When restricting analyses to the DHS T2D-affected individuals only, patterns of association were essentially unchanged although magnitudes were, as expected, generally weaker given the reduction in sample size. There are other differences between this DHS analysis and prior studies. First, the current study utilized measures of vascular calcified plaque, reflecting subclinical CVD, and not verified CVD events as utilized in both the initial epidemiological studies and the Rotterdam [24] and Framingham Offspring [11] cohorts. Use of an intermediate phenotype may be one explanation for the lack of association between both TBili and the UGT1A SNPs, and the chosen measures of CVD. It should be noted however, that the availability of quantitative CAC scores in the DHS should provide enhanced power relative to discrete measures in comparably sized analyses.
The seemingly paradoxical association between the UGT1A SNPs and mortality in the absence of a direct phenotypic association between TBili and mortality warrants further consideration. A potential confounding effect of traditional CVD risk factors on the TBili-CVD negative relationship has been acknowledged previously in the context of causal inference [29]. Given the excess of CVD risk factors in this T2D-enriched sample, this cannot be discounted. In these analyses, adjustment for TBili increased the strength of the observed genetic associations suggesting that TBili itself may account for some of the otherwise unexplained phenotypic variance. Indeed, our failure to observe similar relationships at the phenotypic level may be the result of lack of control for other elements important in the bilirubin pathway, including albumin transport and the ratio of unconjugated to conjugated bilirubin or insufficient adjustment for residual confounding. For example, a recent report from the National Health and Nutrition Examination Survey found that another measure of liver function, SGPT, was associated with diabetes related mortality and diabetes related heart disease mortality [30]. However, we do not observe associations between SGPT and either all-cause or CVD-mortality in the current study (Supplementary Material, Table 5) and do not consider it to be a confounding factor here. Lastly, failure to observe direct phenotypic correlations for TBili may be the result of a threshold effect whereby CVD risk is only conferred at very low TBili concentrations (<10 umol/L), as has been reported previously [2, 4, 6]; less than 15% of the DHS sample have TBili concentrations below this value.
In addition to phenotypic differences, differences in sample size and age of the cohorts could also contribute to the variable associations observed. Both sample size and age were offered as explanations as to why the UGT1A*28 microsatellite was associated with CVD in the larger Framingham (n=1780), but not the smaller Rotterdam study (n=276) [11]. The same factors could also account for the present findings. The mean age at recruitment for the DHS (62 years) is closer to the Rotterdam Study (70 years) than the Framingham Offspring (36 years) and the possibility that the accumulated impact of environmental factors across the life-course masks genetic associations in older cohorts can also not be discounted [11]. The length of follow-up in the DHS was only one-third as long as the Framingham Offspring (~8 years v 24 years), although the overall event rate in Framingham (~19%) was similar to the all-cause mortality rate in the DHS (~21%).
Although neither TBili nor the UGT1A SNPs showed compelling association with subclinical CVD in the current study, we did observe a trend for genotypic associations with all-cause mortality. This finding was strengthened upon adjustment for TBili, supporting a contribution of TBili (either directly or as a surrogate for other unknown factors) to the otherwise unexplained phenotypic variance. There was little evidence supporting association between the UGT1A SNPs and CVD-mortality, perhaps simply the result of the low number of CVD-mortality events and resulting reduction in statistical power. In this context, the TBili association with all-cause mortality perhaps alludes to the more global functions of bilirubin as an anti-oxidant [1, 4, 12].
In conclusion, the current study confirms the previously identified role of the UGT1A gene family in the regulation of bilirubin concentration. Neither TBili nor the TBili-associated UGT1A SNPs were related to measures of subclinical CVD, however, the trend for association with all-cause mortality alludes to a potential contribution of genetic variation in the UGT1A gene family to risk for mortality in patients with T2D. Further quantification of the extent of CVD risk conferred by genetic variation in the UGT1A family in a high risk sample with T2D is still required.
Supplementary Material
Highlights.
UGT1A gene cluster associated with total bilirubin concentrations in the DHS
Bilirubin-associated SNPs were not associated vascular calcified plaque
Bilirubin-associated SNPs were also associated with risk for all-cause mortality
ACKNOWLEDGEMENTS
This study was supported in part by R01 HL67348, R01 HL092301, R01 NS058700 (to DWB) and the General Clinical Research Centre of the Wake Forest School of Medicine (M01 RR07122, F32 HL085989). The authors thank the other investigators, the staff, and the participants of the DHS study for their valuable contributions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Vitek L. The role of bilirubin in diabetes, metabolic syndrome, and cardiovascular diseases. Front Pharmacol. 2012;3:55. doi: 10.3389/fphar.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwertner HA, Jackson WG, Tolan G. Association of low serum concentration of bilirubin with increased risk of coronary artery disease. Clin Chem. 1994;40:18–23. [PubMed] [Google Scholar]
- 3.Hopkins PN, Wu LL, Hunt SC, et al. Higher serum bilirubin is associated with decreased risk for early familial coronary artery disease. Arterioscler Thromb Vasc Biol. 1996;16:250–5. doi: 10.1161/01.atv.16.2.250. [DOI] [PubMed] [Google Scholar]
- 4.Schwertner HA, Vitek L. Gilbert syndrome, UGT1A1*28 allele, and cardiovascular disease risk: Possible protective effects and therapeutic applications of bilirubin. Atherosclerosis. 2008;198:1–11. doi: 10.1016/j.atherosclerosis.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Vitek L, Jirsa M, Brodanova M, et al. Gilbert syndrome and ischemic heart disease: A protective effect of elevated bilirubin levels. Atherosclerosis. 2002;160:449–56. doi: 10.1016/s0021-9150(01)00601-3. [DOI] [PubMed] [Google Scholar]
- 6.Novotny L, Vitek L. Inverse relationship between serum bilirubin and atherosclerosis in men: A meta-analysis of published studies. Exp Biol Med. 2003;228:568–71. doi: 10.1177/15353702-0322805-29. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Fukui M, Tomiyasu K, et al. Low serum bilirubin concentration is associated with coronary artery calcification (CAC). Atherosclerosis. 2009;206:287–91. doi: 10.1016/j.atherosclerosis.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Moon JS, Chang WJ, Lee CH, et al. Relationship between serum bilirubin levels and coronary atherosclerosis in patients with type 2 diabetes. Korean Diabetes Journal. 2008;32:338–345. [Google Scholar]
- 9.Ishizaka N, Ishizaka Y, Takahashi E, Yamakado M, Hashimoto H. High serum bilirubin level is inversely associated with the presence of carotid plaque. Stroke. 2001;32:580–3. doi: 10.1161/01.str.32.2.580-b. [DOI] [PubMed] [Google Scholar]
- 10.Neuzil J, Stocker R. Free and albumin-bound bilirubin are efficient co-antioxidants for alpha-tocopherol, inhibiting plasma and low density lipoprotein lipid peroxidation. J Biol Chem. 1994;269:16712–9. [PubMed] [Google Scholar]
- 11.Lin JP, O'Donnell CJ, Schwaiger JP, et al. Association between the UGT1A1*28 allele, bilirubin levels, and coronary heart disease in the Framingham Heart Study. Circulation. 2006;114:1476–81. doi: 10.1161/CIRCULATIONAHA.106.633206. [DOI] [PubMed] [Google Scholar]
- 12.Vitek L, Schwertner HA. The heme catabolic pathway and its protective effects on oxidative stress-mediated diseases. Adv Clin Chem. 2007;43:1–57. doi: 10.1016/s0065-2423(06)43001-8. [DOI] [PubMed] [Google Scholar]
- 13.Bowden DW, Lange LA, Langefeld CD, et al. The relationship between c-reactive protein and subclinical cardiovascular disease in the Diabetes Heart Study (DHS). Am Heart J. 2005;150:1032–8. doi: 10.1016/j.ahj.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Cox AJ, Agarwal S, Herrington DM, et al. C-reactive protein concentration predicts mortality in type 2 diabetes: The Diabetes Heart Study. Diabet Med. 2012;29:767–70. doi: 10.1111/j.1464-5491.2011.03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris EH. Elevated liver function test in type 2 diabetes. Clinical Diabetes. 2005;23:115–119. [Google Scholar]
- 16.Dullaart RP, Kappelle PJ, de Vries R. Lower carotid intima media thickness is predicted by higher serum bilirubin in both non-diabetic and type 2 diabetic subjects. Clin Chim Acta. 2012;414:161–5. doi: 10.1016/j.cca.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 17.Chan KH, O'Connell RL, Sullivan DR, et al. Plasma total bilirubin levels predict amputation events in type 2 diabetes mellitus: The Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study. Diabetologia. 2013;56:724–36. doi: 10.1007/s00125-012-2818-4. [DOI] [PubMed] [Google Scholar]
- 18.Choi SW, Lee YH, Kweon SS, et al. Association between total bilirubin and hemoglobin A1c in korean type 2 diabetic patients. J Korean Med Sci. 2012;27:1196–201. doi: 10.3346/jkms.2012.27.10.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosma PJ, Chowdhury JR, Bakker C, et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med. 1995;333:1171–5. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- 20.Raijmakers MT, Jansen PL, Steegers EA, Peters WH. Association of human liver bilirubin UDP-glucuronyltransferase activity with a polymorphism in the promoter region of the UGT1A1 gene. J Hepatol. 2000;33:348–51. doi: 10.1016/s0168-8278(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 21.Bielinski SJ, Chai HS, Pathak J, et al. Mayo Genome Consortia: A genotype-phenotype resource for genome-wide association studies with an application to the analysis of circulating bilirubin levels. Mayo Clin Proc. 2011;86:606–14. doi: 10.4065/mcp.2011.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson AD, Kavousi M, Smith AV, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet. 2009;18:2700–10. doi: 10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanna S, Busonero F, Maschio A, et al. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum Mol Genet. 2009;18:2711–8. doi: 10.1093/hmg/ddp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosma PJ, van der Meer IM, Bakker CT, et al. UGT1A1*28 allele and coronary heart disease: The Rotterdam Study. Clin Chem. 2003;49:1180–1. doi: 10.1373/49.7.1180. [DOI] [PubMed] [Google Scholar]
- 25.Bowden DW, Cox AJ, Freedman BI, et al. Review of the Diabetes Heart Study (DHS) family of studies: A comprehensively examined sample for genetic and epidemiological studies of type 2 diabetes and its complications. Rev Diabet Stud. 2010;7:188–201. doi: 10.1900/RDS.2010.7.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowden DW, Lehtinen AB, Ziegler JT, et al. Genetic epidemiology of subclinical cardiovascular disease in the Diabetes Heart Study. Ann Hum Genet. 2008;72:598–610. doi: 10.1111/j.1469-1809.2008.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr JJ, Nelson JC, Wong ND, et al. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: Standardized protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development In young Adults (CARDIA) study. Radiology. 2005;234:35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 28.Carr JJ, Crouse JR, Goff DC, et al. Evaluation of subsecond gated helical ct for quantification of coronary artery calcium and comparison with electron beam ct. AJR Am J Roentgenol. 2000;174:915–21. doi: 10.2214/ajr.174.4.1740915. [DOI] [PubMed] [Google Scholar]
- 29.McArdle PF, Whitcomb BW, Tanner K, et al. Association between bilirubin and cardiovascular disease risk factors: Using mendelian randomization to assess causal inference. BMC Cardiovasc Disord. 2012;12:16. doi: 10.1186/1471-2261-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schooling CM, Kelvin EA, Jones HE. Alanine transaminase has opposite associations with death from diabetes and ischemic heart disease in NAHNES III. Ann Epidemiol. 2012;22:789–98. doi: 10.1016/j.annepidem.2012.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.