Abstract
Protein folding is an essential prerequisite for protein function and hence cell function. Kinetic and thermodynamic studies of small proteins that refold reversibly were essential for developing our current understanding of the fundamentals of protein folding mechanisms. However, we still lack sufficient understanding to accurately predict protein structures from sequences, or the effects of disease-causing mutations. To date, model proteins selected for folding studies represent only a small fraction of the complexity of the proteome and are unlikely to exhibit the breadth of folding mechanisms used in vivo. We are in urgent need of new methods – both theoretical and experimental – that can quantify the folding behavior of a truly broad set of proteins under in vivo conditions. Such a shift in focus will provide a more comprehensive framework from which to understand the connections between protein folding, the molecular basis of disease, and cell function and evolution.
Keywords: protein folding in vivo, aggregation, kinetic stability, molecular chaperones, co-translational folding, multi-domain proteins
Our current understanding of protein folding
Proteins are central to all cellular events: they catalyze chemical reactions, do mechanical work, and perform structural roles in the cell. While proteins are synthesized as long linear polymers of amino acids, each must fold into a specific three-dimensional structure in order to perform its cellular function. The precise sequence of amino acids has a profound influence on the structure of a protein, and hence its function. Indeed, pioneering experiments by Anfinsen in the 1950s showed that the structure of a protein can be determined exclusively by the sequence of amino acids in the polypeptide chain [1]. Therefore in theory when provided with the sequence of a new protein we should be able to predict its structure, which can help predict its function. Moreover, a complete understanding of all physical principles that shape protein folding and structure would enable the de novo design of novel protein structures and functions and accurate predictions of the effects of disease-causing mutations on protein structure and folding.
Anfinsen's observations were the wellspring of the protein folding field, and intense effort has been devoted to determining precisely how a full-length polypeptide chain can quickly refold to form its native structure with high efficiency. Similar to Anfinsen, most efforts focus on studying a purified protein that is first chemically denatured to form an enormous ensemble of rapidly interconverting flexible conformations (the unfolded ensemble), then diluted away from the denaturant in order to enable the protein to refold and regain its biologically active structure (the native state) (Box 1). These refolding experiments can be used to measure protein folding rates and also protein stability, calculated as the free energy of folding (ΔGofolding) [2]. An important prerequisite for these measurements is the identification of experimental conditions under which measurable concentrations of reactants and products (the unfolded ensemble and native state) exist at equilibrium, and hence is restricted to the subset of proteins that unfold and refold reversibly on an experimentally measurable time scale.
Our focus on proteins that refold reversibly after chemical denaturation means that the subset of proteins whose folding properties are well understood share many structural and folding features. Many of these proteins are small (<100 aa), single-domain, monomeric, marginally stable (ΔGofolding ~ -15-30 kJ/mol) and fold quickly (msec-sec time scale) via concerted mechanisms that lack well-populated intermediate conformations [3-9]. This is not surprising, as it has been shown that proteins that are large, multimeric, and/or fold slowly via long-lived, partially folded intermediate structures are more likely to misfold and aggregate [10-13], and are therefore unsuitable for calculating ΔGofolding (Box 1).
Detailed studies of proteins that refold reversibly after chemical denaturation have enabled descriptions of many fundamental features of refolding mechanisms, including measurements of typical rates for global hydrophobic collapse and secondary structure formation [14]. Such small proteins are also more amenable to computer simulations than large proteins, and hence continue to provide an invaluable bridge between experimental results and the development of protein folding simulations and theory [15, 16]. Our extensive investigations of these model proteins have now revealed common features of their refolding mechanisms [14, 17, 18], including general correlations between folding rate and the length [5] or structural complexity [3] of the protein, and some common folding intermediates including the ‘molten globule’, a globally collapsed conformation with significant secondary structure but little or no stable tertiary structure that is often observed shortly after dilution of an unfolded protein from denaturant [19, 20]. These and other common mechanistic features identified to date have led to speculation that protein folding in general is now a well-understood phenomenon [21-24]. However, with few notable exceptions [25-28], our current state of knowledge has produced only modest progress towards accurate ab initio predictions of protein structure from sequence, design of novel proteins, or treatments for the molecular basis of protein folding diseases.
Why is protein folding still a “problem”?
Why is it that, despite extensive study and the emergence of common features, our ability to predict the folding behavior of a protein from its sequence is still so limited? We wondered whether the intense focus on proteins that unfold and refold reversibly in vitro, which provide valuable tools for building the foundation of our understanding of folding mechanisms, might nevertheless hamper the development of a broader, more comprehensive understanding of protein folding. Unfortunately, the majority of proteins in a proteome cannot refold reversibly after dilution from a chemical denaturant. Instead, once taken out of the cellular environment, chemically denatured and diluted into a buffer designed to mimic physiologically relevant conditions (pH, salt, etc.), most proteins misfold and aggregate (Box 1) [29, 30]. Such proteins are rarely used as folding models, despite their widespread appearance in proteomes.
Typical folding models provide incomplete representations of proteomes
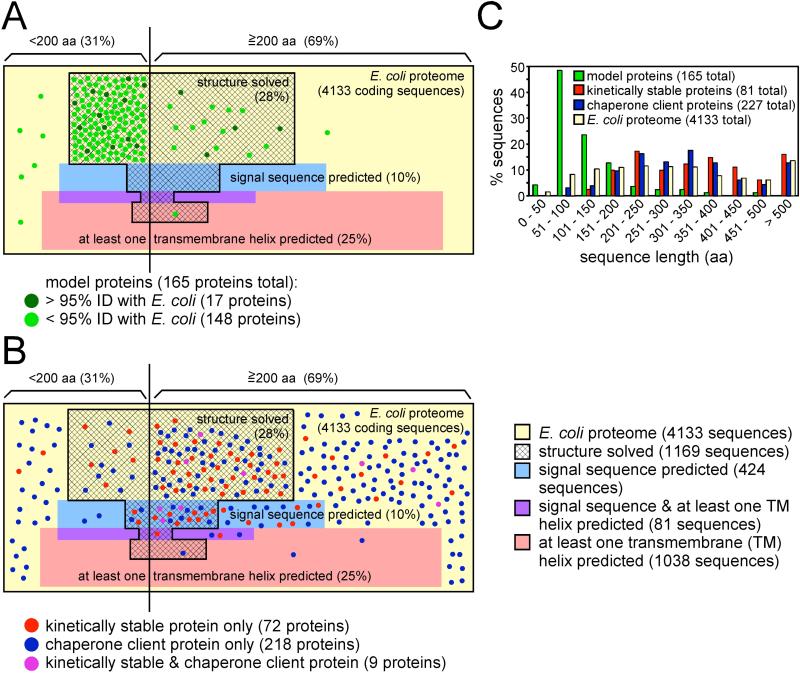
Can we extrapolate from our current understanding of protein folding behavior to describe truly general features of protein folding? An accurate extrapolation requires that our model proteins represent the diversity of proteins from across entire proteomes. To address this question, we compared structural properties of 165 non-redundant proteins commonly used to draw general conclusions regarding protein folding mechanisms [3-9, 23] (Supplementary Table 1) to properties of the well-characterized Escherichia coli proteome (Figures 1A, 2). This comparison revealed that protein folding models capture only a small subset of protein structural diversity, shared by only 8.4% of the E. coli proteome. The diversity of our model proteins therefore does not accurately represent the diversity of the proteome of even a small, simple organism. This discrepancy is most apparent for transmembrane proteins, but proteome diversity is under-represented even amongst water-soluble proteins, particularly those that are large and/or multimeric (Figures 1C, 2). Of course, proteins in a proteome can be classified using other properties beyond those shown here, but we consistently observed that current protein folding models fail to adequately sample the diverse properties of the proteome regardless of the metric used. In addition, there is a growing appreciation that the cellular environment introduces additional challenges – including higher temperatures and protein concentrations – not present during in vitro refolding experiments [31, 32]. Paradoxically, despite these challenges more proteins fold efficiently in vivo than in vitro [29, 30]. This indicates that the properties that enable folding to proceed efficiently in vivo are not necessarily sufficient to enable efficient refolding in vitro.
Figure 1. While typical protein folding models exhibit properties not representative of the E. coli proteome, emerging techniques can capture a broader set of proteins.
(A) The 4133 proteins from E. coli str. K-12 substr. MG1655 (NC_000913.2) were used to construct a proportional Venn diagram, with each unit area in the yellow rectangle corresponding to one E. coli protein coding sequence. These sequences were divided by length (< or ≥200 aa) and analyzed for the presence of an N-terminal signal sequence (http://www.cbs.dtu.dk/services/SignalP) (blue shading), one or more transmembrane α-helices (http://www.cbs.dtu.dk/services/TMHMM) (pink shading), both a signal sequence and a transmembrane α-helix (purple shading) and/or a PDB entry with >95% sequence identity to at least some portion of the protein sequence (hatched area). Note that this map underestimates the complexity of the proteome, as each protein coding sequence from E. coli genome is treated as a separate monomeric protein. A set of 165 non-redundant model proteins used to study protein folding (<95% sequence identity) [3-9] was also analyzed. Each protein is indicated by a green point proportional to the size of one E. coli coding sequence. Seventeen of the model proteins have >95% sequence identity to an E. coli protein (dark green points); the remaining 148 model proteins are from other organisms (light green points). In some cases these models represent individual domains or fragments taken from larger proteins, but as it is known that removal from a larger protein context can change folding behavior [33, 44] (see text), the size of the studied domain is used here. (B) Subsets of proteins identified by proteome-wide screens designed to select other, non-traditional folding behavior were categorized as described for the 165 folding models and compared to the properties of the E. coli proteome as in panel (A). Kinetically stable proteins (red points) were identified by protease resistance [43] or resistance to moderate concentrations of sodium dodecyl sulfate (SDS) [42], yielding 81 non-redundant E. coli proteins. E. coli chaperone client proteins (blue points) represent both DnaK substrates (category “enriched” in [61]) and GroEL substrates (“class IV” in [60]), resulting in a set of 227 proteins. Proteins present in both sets (kinetically stable and chaperone client) are indicated as purple points. Note that there is only one protein in common between the folding models (panel (A)) and kinetically stable and/or chaperone client proteins: maltose binding protein, a kinetically stable protein [43]. (C) Size distribution for each protein group shown in panels (A) and (B), sorted by sequence length.
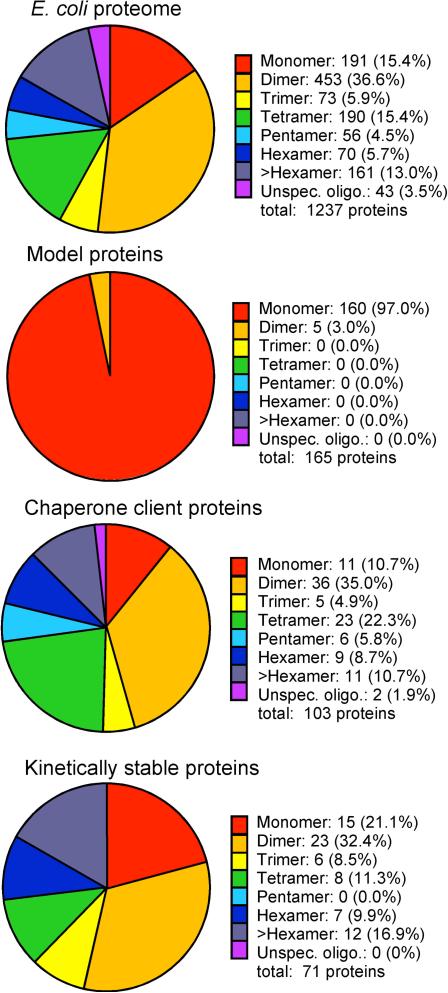
Figure 2. Protein folding models are biased towards monomeric proteins.
The multimerization state of each group of proteins shown in Figure 1 (E. coli proteome, protein folding models, kinetically stable proteins, chaperone client proteins) was determined. For the E. coli proteome, subunit assignments in the Uniprot database were used (30% of proteins in the E. coli proteome have assignments; 1236 proteins). Multimerization state for the 165 non-redundant protein folding models was assigned based on reported multimerization state in the protein folding literature. The multimerization state is indicated for 71 of the 81 kinetically stable proteins identified in ref. [42, 43]. The multimerization state of the chaperone client proteins was assigned using the Uniprot database. 103 of the 227 non-redundant chaperone client proteins have a subunit assignment in the Uniprot database.
The fundamental physical and chemical principles known to govern protein refolding in vitro will of course also apply to larger, more complex proteins in the cellular environment. However, if the protein folding problem were truly solved – if we understood the whole of the physics and chemistry that underlies protein folding – we would now be capable of designing de novo a protein structure of any size or complexity, and predicting the effects of disease-causing mutations on protein structure and folding. The fact that we cannot (see e.g. [33]) indicates that our current knowledge is insufficient to solve the problem. In our opinion, closer attention should be paid to proteins currently regarded as unusual outliers as their mechanistic features, which might be considered unusual amongst our current folding models, could represent footholds for the development of a more comprehensive picture of the diversity of folding mechanisms used across the proteome as a whole. The purpose of this article is to highlight five common perceptions regarding protein folding mechanisms developed from studies of small model proteins that in our opinion are unlikely to scale to the folding behavior of all proteins, with the intent to inspire new technology development and research approaches – both in vitro and in vivo – to close these gaps and increase the predictive power of protein folding research.
(1) Do most proteins globally unfold/refold during their lifetime?
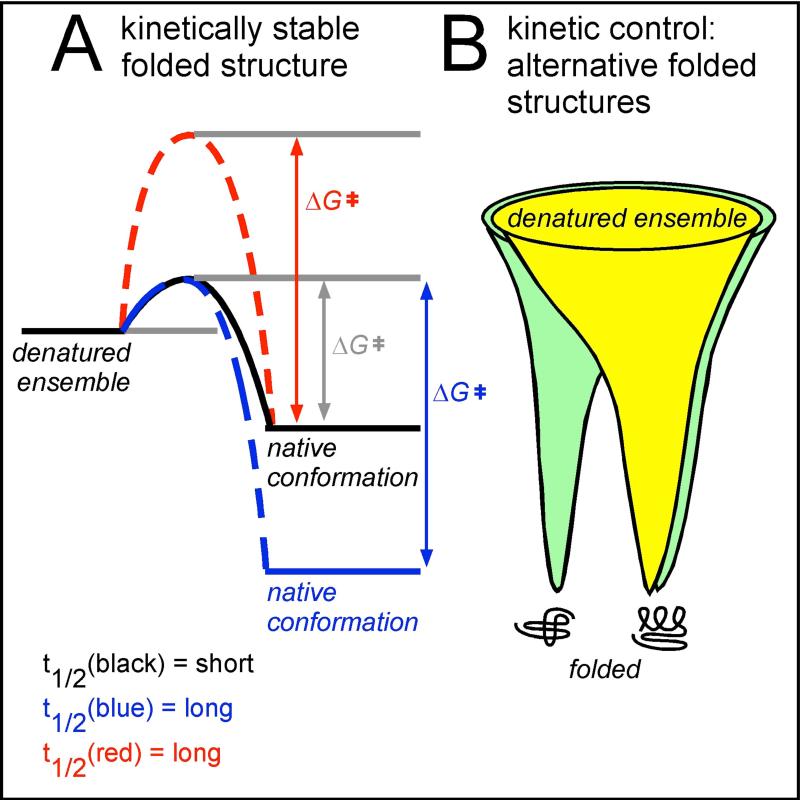
For reasons described above, there has been a strong emphasis on the study of model proteins that unfold and refold reversibly. These proteins tend to be only marginally stable and have fast unfolding and refolding kinetics. These features could mean that these proteins will populate the unfolded state several times over their lifetime in the cell (Figure 3A). The marginal stability and fast kinetics of most model proteins has focused considerable attention on the structural properties of proteins in their chemically denatured ensemble as a model for their unfolded state [34] and the transition between unfolded and native states. However, most proteins do not refold reversibly. Instead, global (or even partial) unfolding can lead to misfolding and aggregation (and/or degradation in vivo) [35]. This suggests that proteins in general will employ strategies to avoid unfolding over their lifetime in the cell. Protein unfolding can be avoided by increasing protein stability, but this tends to rigidify the native structure, which can reduce flexibility needed for binding and catalysis. An alternative option is to increase the energy barrier between the native and denatured states, creating proteins that are kinetically stable but thermodynamically marginally stable, or even less stable than the unfolded state (Figure 3A). Transthyretin [36], P22 tailspike [37] and GFP [38] are three well-studied examples of proteins with high kinetic stability. In vitro, each of these proteins exhibits pronounced hysteresis and hence does not refold reversibly from a chemically denatured state. However, all three fold efficiently in vivo, indicating the cellular environment can alter the energy landscape for folding to arrive at a kinetically trapped native structure. Detailed investigations into the folding mechanisms of these proteins have provided valuable insights into the kinetic competition between folding versus aggregation, including uncovering the effects of conformational changes within a single polypeptide chain on aggregation propensity [39], introducing the concept of folding mutations that can alter partitioning between folding and aggregation without altering the stability of the native state [40] and highlighting the role of folding during translation as a mechanism to increase folding yield in vivo [41].
Figure 3. Examples of diversity amongst protein folding mechanisms.
(A): Most proteins currently used as folding models are marginally stable (black), meaning that their folded lifetime (t1/2) is short. Lifetime can be increased in two ways. The native structure can be stabilized thermodynamically, increasing the energetic difference between the denatured ensemble and the native structure (increasing ΔGofolding, blue). Alternatively, the energetic barrier separating the denatured ensemble and the native conformation (ΔG‡) can be increased (red); this will preserve the (low) thermodynamic stability but increase the folded state lifetime. Increasing the energy barrier yields kinetically stable proteins, which can be identified by proteome-wide folding screens [42, 43] (see also Figure 1B). (B): Proteins fold from an ensemble of unfolded states, represented by the wide top of a protein folding funnel. In simple model systems (yellow), the funnel has one energy minimum, the native conformation. However, some proteins have a more complex energy landscape and can adopt alternative folded structures (green). These two folded structures may interconvert, or features of the cellular environment may stabilize a subset of early folding intermediates, resulting in a biased accumulation of one structure versus the other(s).
Many kinetically stable proteins are resistant to unfolding in the presence of moderate concentrations of sodium dodecyl sulfate (SDS) [42] and/or to protease digestion [43]. Proteome-wide screens developed to identify proteins with these characteristics have revealed that kinetically stable proteins sample some properties of the proteome more effectively than typical folding models (Figure 1B, 2 and Supplementary Table 2) [42, 43]. Indeed, these screens underrepresent small (<200 aa) proteins, which might reflect a lower propensity for kinetic stability among small proteins or a bias within the screen against smaller proteins, similar to technical limitations that impose a bias against transmembrane proteins. Traditionally, kinetically stable proteins are rarely selected as folding models because refolding from a chemically denatured state in vitro can require these proteins to populate long-lived, partially folded conformations that may be prone to aggregation. We suggest that focusing our efforts to understanding the structural features that confer kinetic stability, and the mechanisms used to populate such native structures in vivo, provides an opportunity to explore a folding strategy used by a diverse set of proteins that can reduce aggregation in vivo.
(2) Do larger, multi-domain proteins fold like their component domains?
The vast majority of proteins used for folding studies are both monomeric and consist of a single structural domain (Figures 2, 4). By contrast, larger proteins often consist of multiple domains. This modular architecture makes it tempting to hypothesize that larger proteins will fold via a hierarchical mechanism whereby local contacts first determine the folding properties of individual domains, followed by the formation of inter-domain interactions. This model assumes that the complexities of multi-domain protein folding can be reduced to the independent folding of the constituent domains. While such behavior has been observed for some proteins, for others the placement of a domain within the context of a larger multi-domain protein can significantly alter the energy landscape for folding [44], making it impossible to predict the effects of disease-causing mutations or other sequence alterations from studies of isolated domains alone [33]. For example, while yeast phosphoglycerate kinase (PGK) can be studied as two separate, independently-folding domains – both of which appear in the list of 165 model proteins (Supplementary Table 1) – the E. coli PGK homolog has a nearly identical structure but unfolds five orders of magnitude more slowly, and its C-terminal domain cannot adopt its native structure in isolation [45]. Even for proteins comprised of seemingly “independent” domains connected by flexible linkers, contextual effects can alter folding efficiency. For example, Ig domains within titin form a beads-on-a-string architecture that was shaped by gene duplication events. Concomitant folding of two adjacent titin domains increases the likelihood of non-native inter-domain interactions akin to domain swapping, which occur faster than the folding rate and therefore retard accumulation of the native structure [46]. Similar non-native inter-domain interactions reduce the folding rate for calmodulin [47]. Interestingly, evolution has reduced the sequence identity between adjacent titin domains, which reduces the likelihood of misfolding [44].
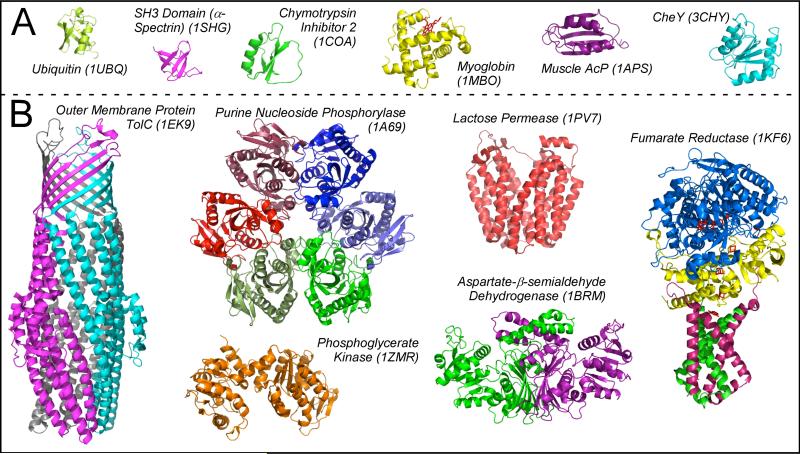
Figure 4. Proteins identified by in vivo assays and folding “outliers” are structurally more complex than typical folding models.
PDB ID codes are indicated in parentheses. Subunits of multimeric proteins are shown in different colors, and cofactors (in myoglobin and fumarate reductase) are shown in red. Most models used to study protein folding (A) are smaller and less complex than proteins representing diverse properties from the E. coli proteome (B). Of the E. coli proteins shown here, purine nucleoside phosphorylase (a hexamer) and phosphoglycerate kinase (a monomer) were identified in the screen for kinetically stable proteins [43], aspartate-β-semialdehyde dehydrogenase (a dimer) was identified in the screen for chaperone client proteins [60], and the outer membrane protein TolC (a trimer) was identified in screens for both kinetic stability and chaperone clients [42, 61]. Lactose permease (a monomer) is an α-helical transmembrane protein. Two subunits of the tetrameric fumarate reductase contain transmembrane α-helices (shown in green and pink). The soluble subunits of fumarate reductase (shown in blue and yellow) were identified in the screens for chaperone clients [60, 61].
A related question is the extent to which protein folding thermodynamics and kinetics are affected by the assembly of subunits into a multimeric protein. Studies of intrinsically disordered proteins have shown that folding and binding can be inter-dependent processes [48, 49]. It will be valuable to adapt these approaches to study more subtle effects of subunit interactions on the thermodynamics that govern the folding of multimeric proteins with ordered subunits. More broadly, the complex and crowded environment of the cell provides numerous opportunities for non-specific binding events not captured during refolding of a purified protein in vitro [50], but our understanding of how protein sequences might have evolved to minimize or modulate these non-native interactions is still in its infancy. We predict that negative design strategies such as these – strategies that suppress aggregation and unproductive non-specific interactions, rather than stabilize the native structure – will play an increasingly important role as we move towards the de novo design of larger, multi-domain and multimeric proteins.
(3) Do most proteins populate only a single native structure?
Anfinsen showed that RNase can refold to its native, active protein structure after chemical denaturation, presumably because this structure represents the global energy minimum on a funnel-like energy landscape (Figure 3B, yellow). Most folding models share this feature of a thermodynamically-controlled folding pathway. But there are now many examples of proteins with multiple, distinct “native” conformations separated by large energy barriers [51, 52] (Figure 3B, green), including α-lytic protease, whose folding properties are well characterized [53] but which is rarely included in general analyses of protein folding behavior. For proteins like α-lytic protease that fold under kinetic control, the conversion between these alternative folded structures is often an essential, regulated component of the functional cycle of these proteins [54-56]. While it is not yet known what fraction of the proteome can populate two or more alternative folded states, our tendency to avoid selecting such proteins as folding models hampers their discovery and hence our understanding of the mechanisms that underlie these folding pathways.
(4) To what extent do molecular chaperones affect protein folding in vivo?
In the cell, proteins fold in an environment that includes molecular chaperones, a class of proteins that has evolved to facilitate the folding and/or suppress the aggregation of other proteins [57-59]. When these proteins were first identified, it was initially thought that chaperones – some of which are essential for cell viability – would explain the higher efficiency of protein folding in vivo versus in vitro. More recently however, quantitative proteomics has revealed that the substrate repertoire for molecular chaperones is quite narrow under normal growth conditions [60-62], and hence cannot fully explain the higher efficiency of folding in vivo. Yet, while chaperones contribute to the productive folding of only 10-20% of the proteome, their substrates represent a much more diverse subset of proteins than current folding models (Figure 1B, Supplementary Table 3). These obligate chaperone substrates therefore provide an excellent opportunity to explore the features of folding mechanisms and native structures that lead to chaperone recruitment in vivo, versus those that do not. However, because these proteins are unlikely to refold reversibly we will need alternative approaches to study such mechanisms, for example by exploiting experiments that can quantify the kinetic partitioning between folding and aggregation, or by developing single-molecule experiments that enable a protein to fold at or near infinite dilution, thereby reducing intermolecular interactions between substrate proteins that can lead to aggregation [63, 64].
(5) Do other aspects of the cellular environment affect protein folding in vivo?
Our growing appreciation of the contributions of the cellular environment to protein folding has inspired the development of exciting new experimental approaches, including specific fluorescence and isotope labeling strategies that can explore the effects of this complex environment on protein folding thermodynamics and kinetics [65-67]. To date, many of these studies have relied on the same model proteins favored for in vitro refolding experiments. Perhaps not surprisingly, in some cases the cellular environment has minimal effects on the folding of these proteins, versus refolding in vitro; these proteins were selected as folding models in part due to their ability to fold robustly under a wide range of conditions. What is less clear is how other, more diverse types of proteins might exploit properties of the cellular environment to modulate their energy landscape for folding and increase folding efficiency. For example, the folding mechanism for a very small protein might not be altered by folding vectorially (from N to C terminus) during translation or secretion. By contrast, longer proteins take longer to synthesize – and secrete – and therefore spend more time with the N-terminal portion of the protein available for folding prior to the appearance of the C terminus. Indeed, several larger proteins are now known to have co-translational folding mechanisms in which the energy landscape for folding is significantly altered versus refolding in vitro, leading to the population of folding intermediates not detected during refolding in vitro. These differences often include significant amounts of native-like structure formation for the most N-terminal portions of the nascent protein chain even after tens or hundreds of C-terminal residues have been added to the growing polypeptide chain [41, 68, 69]. Ideally, computer simulations would enable modeling of co-translational folding processes and the formulation of experimentally testable hypotheses regarding these altered energy landscapes. Unfortunately, the complexities of both longer proteins and the cellular environment greatly increase the complexity of computer simulations, although progress is being made in these directions [70, 71].
Conclusions and future directions
Like many biological mechanisms, protein folding is a complex, multi-faceted process. In vitro refolding studies of small proteins have played an invaluable role in developing our understanding of many fundamental aspects of this process. However, the folding problem is not yet solved, and the technical constraints of any one approach will render it insufficient to fully understand such a complex process. Hence, in our opinion the field of protein folding is currently “mature” only in the sense that we have extracted much of the information available from our established methods and model systems. A comprehensive understanding of protein folding, particularly in the cell, will require addressing additional phenomena that are not amenable to detection using our current models and approaches.
For these reasons, we advise extreme caution before concluding that the folding behavior of a protein appears to be an unusual outlier. Proteomes contain thousands of proteins, and to date only a small fraction of their diversity has been explored through folding studies. Given that our selection of model proteins to date has been far from general, it remains an open question to what extent unusual “outlier” folding mechanisms contribute to the folding properties of the proteome as a whole. Studying more diverse proteins, such as those identified from proteome-wide screens that do not require reversible refolding (Figures 1B, 4), would enable us to build upon the fundamental laws of physics and chemistry that form the foundation of our current understanding of protein folding in vitro, and learn how the cellular environment exploits those laws in order to build functional cells.
We predict an exciting revolution on the horizon, as the field of protein folding develops the new experimental approaches needed to describe the folding properties of an increasingly diverse set of model proteins and explore the influence of the cellular environment on these proteins. In addition to the examples described above, we are heartened by the recent development of new techniques to study the folding features of transmembrane proteins [72-75], which represent a significant fraction of all proteomes (Figure 1) but by and large have been refractory to traditional approaches to study protein refolding in vitro. Once new methods and model systems are in place, we will be able to explore the folding of all corners of the proteome and develop a truly general understanding of how proteins fold.
Supplementary Material
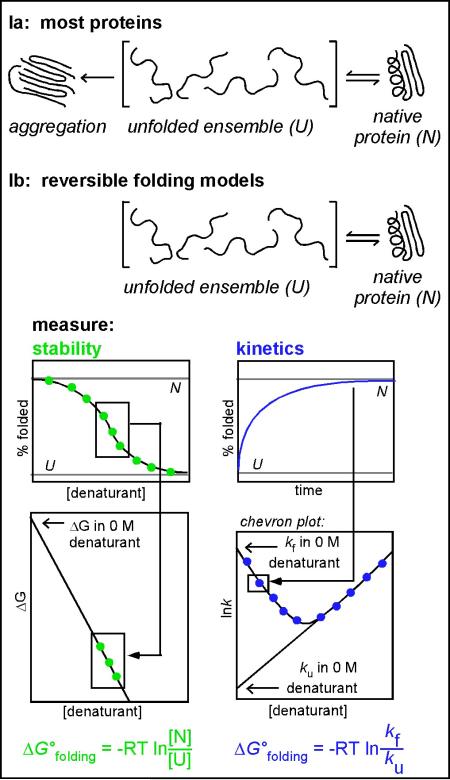
Box 1. The standard set of experiments used to characterize protein folding in vitro.
This approach is applicable only to proteins that reversibly unfold and refold in a two-state reaction in vitro. Conditions must be identified that eliminate aggregation (Figure Ia), a common side reaction. Protein folding in reversible systems (Figure Ib) is often monitored using optical methods including tryptophan fluorescence emission and circular dichroism spectroscopy. Thermodynamic stability (ΔGofolding) is measured by equilibrating the protein in various concentrations of a chemical denaturant and measuring the conformation spectroscopically at each denaturant concentration (Figure Ib, green). The linear transition region in the sigmoidal unfolding/refolding titration can be extrapolated to calculate ΔGofolding in 0 M denaturant. The same spectroscopic tools can be used to monitor refolding and unfolding kinetics (Figure II, blue). The rate constants for folding (kf) and unfolding (ku) are dependent on the residual denaturant concentration. By systematically varying the final denaturant concentration, one can construct a V-shaped chevron plot (bottom right) where the folding and unfolding rate constants are plotted versus the final concentration of denaturant [76]. Each “arm” of the V-shaped plot can be extrapolated to obtain the unfolding and refolding rate constants at 0 M denaturant. For proteins that fold via a simple one-step process, these rate constants can also be used to calculate ΔGofolding.
Figure Ia: For most proteins, dilution of the ensemble of unfolded conformations (U) out of denaturant results in a kinetic competition between correct folding (N) versus aggregation. This results in <100% reversibility of the refolding reaction, meaning that such proteins cannot be used for the thermodynamic and kinetic analyses described below.
Figure Ib: For proteins that do refold reversibly, there are widespread assays available to study protein stability and folding kinetics. For such assays, conditions must be identified where folding is reversible: upon dilution, the ensemble of unfolded conformations (U) is converted exclusively to the native structure (N) with no detectable aggregation or hysteresis. Under these conditions, the free energy of folding (ΔGofolding) can be determined either from equilibrium denaturation measurements (left; green) or from kinetic measurements of rate constants (kf and ku) (right; blue). R = gas constant; T = temperature.
Highlights.
Proteins must fold successfully in order to function.
General conclusions are emerging for folding of small reversibly refolding proteins.
Proteomes are more complex and diverse than most proteins used as folding models.
Valuable insights can be gleaned from the folding of unconventional model proteins.
New approaches are needed to understand the folding properties of an entire proteome.
Acknowledgements
We gratefully acknowledge helpful conversations with David Agard, Igor Drobnak, Adrian Elcock, Jonathan King, Jeff Peng and Susan Marqusee during the preparation of this manuscript. This work was supported by NIH grants GM074807 and GM097573 (P.L.C.). E.B. was supported by NIH training grant GM075762. J.L.C. was supported by a Clare Boothe Luce Graduate Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 3.Plaxco KW, et al. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 4.Jackson SE. How do small single-domain proteins fold? Fold. Des. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 5.Galzitskaya OV, et al. Chain length is the main determinant of the folding rate for proteins with three-state folding kinetics. Proteins. 2003;51:162–166. doi: 10.1002/prot.10343. [DOI] [PubMed] [Google Scholar]
- 6.Ouyang Z, Liang J. Predicting protein folding rates from geometric contact and amino acid sequence. Protein Sci. 2008;17:1256–1263. doi: 10.1110/ps.034660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naganathan AN, Munoz V. Insights into protein folding mechanisms from large scale analysis of mutational effects. Proc. Natl. Acad. Sci. USA. 2010;107:8611–8616. doi: 10.1073/pnas.1000988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tartaglia GG, Vendruscolo M. Proteome-level interplay between folding and aggregation propensities of proteins. J. Mol. Biol. 2010;402:919–928. doi: 10.1016/j.jmb.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Sawle L, Ghosh K. How do thermophilic proteins and proteomes withstand high temperature? Biophys. J. 2011;101:217–227. doi: 10.1016/j.bpj.2011.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzpatrick AW, et al. Inversion of the balance between hydrophobic and hydrogen bonding interactions in protein folding and aggregation. PLoS Comput. Biol. 2011;7:e1002169. doi: 10.1371/journal.pcbi.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramshini H, et al. Large proteins have a great tendency to aggregate but a low propensity to form amyloid fibrils. PloS ONE. 2011;6:e16075. doi: 10.1371/journal.pone.0016075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vendruscolo M. Proteome folding and aggregation. Curr. Opin. Struct. Biol. 2012;22:138–143. doi: 10.1016/j.sbi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 13.King J, et al. Thermolabile folding intermediates: Inclusion body precursors and chaperonin substrates. FASEB J. 1996;10:57–66. doi: 10.1096/fasebj.10.1.8566549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 15.Lindorff-Larsen K, et al. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 16.Lane TJ, Pande VS. A simple model predicts experimental folding rates and a hub-like topology. J Phys Chem B. 2012;116:6764–6774. doi: 10.1021/jp212332c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman GR, et al. Taming the complexity of protein folding. Curr. Opin. Struct. Biol. 2011;21:4–11. doi: 10.1016/j.sbi.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sosnick TR, Barrick D. The folding of single domain proteins--have we reached a consensus? Curr. Opin. Struct. Biol. 2011;21:12–24. doi: 10.1016/j.sbi.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins: Struct. Funct. Genet. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- 20.Ptitsyn OB. Molten globule and protein folding. Adv. Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 21.Service RF. Problem solved* (*sort of). Science. 2008;321:784–786. doi: 10.1126/science.321.5890.784. [DOI] [PubMed] [Google Scholar]
- 22.Wolynes PG, et al. Chemical physics of protein folding. Proc. Natl. Acad. Sci. USA. 2012;109:17770–17771. doi: 10.1073/pnas.1215733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dill KA, et al. Physical limits of cells and proteomes. Proc. Natl. Acad. Sci. USA. 2011;108:17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piana S, et al. Atomic-level description of ubiquitin folding. Proc. Natl. Acad. Sci. USA. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hecht MH, et al. De novo design, expression, and characterization of Felix: A 4-helix bundle protein of native-like sequence. Science. 1990;249:884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 26.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 27.Johnson SM, et al. The transthyretin amyloidoses: from delineating the molecular mechanism of aggregation linked to pathology to a regulatory-agency-approved drug. J. Mol. Biol. 2012;421:185–203. doi: 10.1016/j.jmb.2011.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koga N, et al. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willis MS, et al. Investigation of protein refolding using a fractional factorial screen: a study of reagent effects and interactions. Protein Sci. 2005;14:1818–1826. doi: 10.1110/ps.051433205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowieson NP, et al. An automatable screen for the rapid identification of proteins amenable to refolding. Proteomics. 2006;6:1750–1757. doi: 10.1002/pmic.200500056. [DOI] [PubMed] [Google Scholar]
- 31.Clark PL. Protein folding in the cell: Reshaping the folding funnel. Trends Biochem. Sci. 2004;29:527–534. doi: 10.1016/j.tibs.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Gershenson A, Gierasch LM. Protein folding in the cell: challenges and progress. Curr. Opin. Struct. Biol. 2011;21:32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randles LG, et al. Understanding pathogenic single-nucleotide polymorphisms in multidomain proteins--studies of isolated domains are not enough. FEBS J. 2013;280:1018–1027. doi: 10.1111/febs.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarney ER, et al. Is there or isn't there? The case for (and against) residual structure in chemically denatured proteins. Crit. Rev. Biochem. Mol. Biol. 2005;40:181–189. doi: 10.1080/10409230591008143. [DOI] [PubMed] [Google Scholar]
- 35.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 36.Lai Z, et al. Guanidine hydrochloride-induced denaturation and refolding of transthyretin exhibits a marked hysteresis: Equilibria with high kinetic barriers. Biochemistry. 1997;36:10230–10239. doi: 10.1021/bi963195p. [DOI] [PubMed] [Google Scholar]
- 37.Fuchs A, et al. In vitro folding pathway of phage P22 tailspike protein. Biochemistry. 1991;30:6598–6604. doi: 10.1021/bi00240a032. [DOI] [PubMed] [Google Scholar]
- 38.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 39.Hammarstrom P, et al. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 40.Mitraki A, et al. Global suppression of protein folding defects and inclusion body formation. Science. 1991;253:54–58. doi: 10.1126/science.1648264. [DOI] [PubMed] [Google Scholar]
- 41.Ugrinov KG, Clark PL. Cotranslational folding increases GFP folding yield. Biophys. J. 2010;98:1312–1320. doi: 10.1016/j.bpj.2009.12.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia K, et al. Identifying the subproteome of kinetically stable proteins via diagonal 2D SDS/PAGE. Proc. Natl. Acad. Sci. USA. 2007;104:17329–17334. doi: 10.1073/pnas.0705417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park C, et al. Energetics-based protein profiling on a proteomic scale: identification of proteins resistant to proteolysis. J. Mol. Biol. 2007;368:1426–1437. doi: 10.1016/j.jmb.2007.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han JH, et al. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 45.Young TA, et al. Comparison of proteolytic susceptibility in phosphoglycerate kinases from yeast and E. coli: modulation of conformational ensembles without altering structure or stability. J. Mol. Biol. 2007;368:1438–1447. doi: 10.1016/j.jmb.2007.02.077. [DOI] [PubMed] [Google Scholar]
- 46.Borgia MB, et al. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474:662–665. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stigler J, et al. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 48.Kovacs D, et al. Intrinsically disordered proteins undergo and assist folding transitions in the proteome. Arch. Biochem. Biophys. 2013;531:80–89. doi: 10.1016/j.abb.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Wright PE, Dyson HJ. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elcock AH. Models of macromolecular crowding effects and the need for quantitative comparisons with experiment. Curr. Opin. Struct. Biol. 2010;20:196–206. doi: 10.1016/j.sbi.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker D, Agard DA. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 52.Bryan PN, Orban J. Proteins that switch folds. Curr. Opin. Struct. Biol. 2010;20:482–488. doi: 10.1016/j.sbi.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaswal SS, et al. Energetic landscape of alpha-lytic protease optimizes longevity through kinetic stability. Nature. 2002;415:343–346. doi: 10.1038/415343a. [DOI] [PubMed] [Google Scholar]
- 54.Luo X, et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 55.Burmann BM, et al. An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuinstra RL, et al. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc. Natl. Acad. Sci. USA. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Ann. Rev. Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 58.Hartl FU, et al. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 59.Bukau B, et al. Getting newly synthesized proteins into shape. Cell. 2000;101:119–122. doi: 10.1016/S0092-8674(00)80806-5. [DOI] [PubMed] [Google Scholar]
- 60.Fujiwara K, et al. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010;29:1552–1564. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calloni G, et al. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Thulasiraman V, et al. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cecconi C, et al. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 64.Shank EA, et al. The folding cooperativity of a protein is controlled by its chain topology. Nature. 2010;465:637–640. doi: 10.1038/nature09021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghaemmaghami S, Oas TG. Quantitative protein stability measurement in vivo. Nat. Struct. Biol. 2001;8:879–882. doi: 10.1038/nsb1001-879. [DOI] [PubMed] [Google Scholar]
- 66.Ignatova Z, Gierasch LM. Monitoring protein stability and aggregation in vivo by real-time fluorescent labeling. Proc. Natl Acad. Sci. USA. 2004;101:523–8. doi: 10.1073/pnas.0304533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ebbinghaus S, et al. Protein folding stability and dynamics imaged in a living cell. Nat. Methods. 2010;7:319–323. doi: 10.1038/nmeth.1435. [DOI] [PubMed] [Google Scholar]
- 68.Frydman J, et al. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat. Struct. Biol. 1999;6:697–705. doi: 10.1038/10754. [DOI] [PubMed] [Google Scholar]
- 69.Evans MS, et al. Cotranslational folding promotes β-helix formation and avoids aggregation in vivo. J. Mol. Biol. 2008;383:683–692. doi: 10.1016/j.jmb.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput. Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elcock AH. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. PLoS Comput. Biol. 2006;2:e98. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlebach JP, et al. Probing membrane protein unfolding with pulse proteolysis. J. Mol. Biol. 2011;406:545–551. doi: 10.1016/j.jmb.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burgess NK, et al. Beta-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Findlay HE, et al. Unfolding free energy of a two-domain transmembrane sugar transport protein. Proc. Natl. Acad. Sci. USA. 2010;107:18451–18456. doi: 10.1073/pnas.1005729107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Booth PJ, Curnow P. Folding scene investigation: membrane proteins. Curr. Opin. Struct. Biol. 2009;19:8–13. doi: 10.1016/j.sbi.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan HS, Dill KA. Protein folding in the landscape perspective: Chevron plots and non-Arrhenius kinetics. Proteins Struct. Funct. Genet. 1998;30:2–33. doi: 10.1002/(sici)1097-0134(19980101)30:1<2::aid-prot2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.