Abstract
Anoikis-resistance of tumor cells is critical for anchorage-independent growth and metastasis. The inflammatory-response transcription factor NF-κB contributes to anoikis-resistance and tumor progression through mechanisms that are understood incompletely. Deleted in Breast Cancer-1 protein (KIAA1967) is over-expressed in several tumor types, and correlates with a poorer prognosis in some cases. We report here that DBC1 suppressed anoikis in normal epithelial and breast cancer cell lines. DBC1 interacted with IKK-β, stimulating its kinase activity, promoting NF-κB transcriptional activity through the phosphorylation of relA serine-536 and enhancing the expression of the NF-κB target genes, c-FLIP and bcl-xl. Our results indicate that DBC1 is an important co-factor for the control of the IKK-β-NF-κB signaling pathway that regulates anoikis.
Keywords: anoikis, DBC1, NF-κB, IKK-β
INTRODUCTION
Anoikis is defined as apoptosis that is rescued by cellular interaction with an appropriate extracellular matrix [1]. Physiologically, it is critical for cellular homeostasis and development. Anoikis-resistance is a hallmark of metastasis, because it is required for anchorage-independent growth during tumor dissemination. Identification of the factors and mechanisms that control anoikis is a high priority in cancer cell biology and developmental therapeutics. Such factors may control either the set-point, i.e., the gene expression program controlling sensitivity vs. resistance of cells (which may exist prior to detachment from matrix) or the apoptosis-triggering mechanisms that occur in detached cells, or both. Indeed, oncogenes or tumor suppressor proteins affecting both functions have been identified [2-4].
Among the signaling and transcription factor pathways involved in regulating anoikis, NF-κB is notable because it links anoikis with inflammatory signaling between and within cells [5-7]. Consistent with this, several NF-κB target genes including, c-FLIP, survivin, Bcl-2, bcl-xl, cIAP-2, xIAP, PLK1 and trkB protect tumor cells against anoikis [8-14]. Accordingly, NF-κB signaling is widely up-regulated in diverse tumor types [5,7]. This up-regulation is attributed mainly to hyperactivation of upstream signaling pathways, except in a subclass of leukemias in which activating relA mutations occur. These pathways include Akt, inflammatory cytokines, TNF and, interestingly, cell-matrix detachment of tumor cells, but not normal cells [6,14].
Phosphorylation and acetylation regulate the NF-κB activation process in several respects, including nuclear translocation, DNA binding, and potency of transcriptional activation [15,16]. The kinase IKK-β plays a particularly significant role in that it both promotes the translocation of NF-κB to the nucleus through the phosphorylation of IκB-α, as well as enhancing the ability of relA to activate transcription by phosphorylation of sites within the relA activation domain [17-20]. The mechanisms linking the regulation of NF-κB to the control of anoikis are understood incompletely, however.
Deleted in Breast Cancer (DBC1) is a nuclear protein encoded by a gene on 8p21 that was originally believed to reside within a deleted region in breast cancer, a deletion assignment that was later found to be inaccurate [21]. In fact, DBC1 over-expression has been observed in colorectal, esophageal and breast cancers, where its over-expression correlates, in some cases, with poor prognosis [22-25]. These observations suggest a potential role of DBC1 in tumor progression, although paradoxical roles as a tumor suppressor have been proposed as well [26].
In this study, we demonstrate that DBC1 suppresses anoikis by activating IKK-β through a direct interaction, increasing NF-κB activity and enhancing the expression of key anoikis-relevant cell survival genes.
MATERIALS AND METHODS
Antibodies
Antibodies used in this study were from the following sources: DBC1 (pAb, Bethyl laboratory and mAb Cell signaling); RelA (Santa cruz biotechnology); RelA acetyl-lys310 (Abcam); β-Actin (Sigma); IKK-β (rabbit mAb, Cell signaling); p-RelA S536 (Cell signaling); β-Tubulin (Santa cruz biotechnology); α-Tubulin (Millipore Mab DM1A); human c-FLIP (NF-6, Enzo life sciences, Inc); mouse c-FLIP clone DAVE-2 (Axxora), HA (Covance HA. 11 mAb (Ascites)); FLAG (Sigma mAb M2); GADPH (Sigma-Aldrich pAb G9545); Bcl-xl (rabbit pAb, Cell Signaling); cleaved caspase 3 (Cell Signaling); p-IκB-α (mouse mAb, Cell Signaling).
Reagents
Reagents were from the following sources: TNFα (R&D System); Bay 11-7082 (Sigma-Aldrich); recombinant GST-IκBα and active IKK-β (Signalchem); S-protein HRP Conjugate (Novagen).
ShRNAs and siRNAs
DBC1 siRNA duplexes from Sigma Aldrich Sense: 5′-AAACGGAGCCUACUGAACAUU-3′ Anti-Sense: 5′-AAUGUUCAGUAGGCUCCGUUU-3′. Non-targeting control siRNA (Dharmacon RNAi Technology). The siDBC1 sequences are adapted from [27]. DBC1 shRNAs in the vector pTRIPZ were from Open Biosystems: A6: 193-0178-A-6 GGTTCCACTTAACAACTA (in 5′UTR); B2: 193-0195-B-2 CGGCTCTACCTAGAGAAC (in coding sequence).
Protein Expression and Purification
Human DBC1 was expressed in E.coli BL21 using a clone in the GST vector pGEX-6P3 (generated as described below). BL21 cells were grown in Luria broth supplemented with 50 μg/mL ampicilin at 37 °C overnight. Protein expression was induced by adding 1 mM isopropyl thio-β-d-galactopyranoside (IPTG) for 2hrs. Cells were pelleted by centrifugation at 5,000 rpm and washed with 1× PBS once and resuspended in bufferA (50 mM sodium phosphate buffer (pH 7.4), 500 mM KCl, 1 mM DTT and a protease inhibitor mixture (Pierce)). The cells were lysed by French Press (twice), and the lysate was centrifuged at 100,000 rpm for 1 hr to obtain the soluble fraction of the cell extract. The supernatant was incubated with 0.3 ml of 50% slurry of gluthione-sephorose (Pharmacia-pre-equilibrated with PBS) for 2 hours at 4 degrees on rotating wheel. Beads were washed once with wash buffer (50 mM Tris-HCl, pH8, 300 mM NaCl, 1 mM DTT/PBS+protease inhibitors) and washed 3 times with cleavage buffer (50 mM Tris-HCl, pH8, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol + 0.01% Triton X-100). To elute protein, 160 L of cleavage buffer containing Protease (Pierce, 27-0843-01) mixture (12.8ul [80unit] of PreScission protease and 147.2ul of cleavage buffer) was added to 160ul of bead solution. The mixture was incubated for 5 hours at 4°C on rotating wheel, spun down for 15 seconds at full speed, and the supernatant was transferred to a new tube. The protein sample was dialyzed with kinase assay buffer (see below) overnight using Slide-A-Lyzer MINI Dialysis Units (Thermo Scientific).
qRT-PCR
The primers used for RT-PCR were as follows: CIAP-1 (BIRC2) 5-TTGAGGTGTTGGGAATCTGG-3, 5-GGCCTTTCATTCGTATCAAGAAC-3; XIAP(BIRC4) 5-GCACGGATCTTTACTTTTGGG-3, 5-GGGTCTTCACTGGGCTTC-3; Survivin 5-CAAGGAGCTGGAAGGCTG-3, 5-TTCTTGGCTCTTTCTCTGTCC-3; BCL2 5-GTGGATGACTGAGTACCTGAAC-3, 5-GCCAGGAGAAATCAAACAGAGG-3; BCLXL 5-GACATCCCAGCTCCACATC-3, 5-GTTCCCATAGAGTTCCACAAAAG-3; FLIP 5-CTTGGCCAATTTGCCTGTAT-3, 5-TCTTTGGCTTCCCTGCTAGA-3. Beta2M: 5-GGCATTCCTGAAGCTGACAG-, 5-TGGATGACGTGAGTAAACCTG-3.
For real–time quantitative RT-PCR, RNA was purified from cell lines using RNeasy Plus mini kit (Qiagen, Germantown, MD) and cDNA was synthesized with invitrogen 18080-051 Superscript III First-strand Synthesis System for RT-PCR using 1-3 μg of RNA template with oligo-dT primers. Reactions were run in total 20 μL utilizing 10ul of 2× Applied Biosystems (Foster City, CA) SYBR green PCR Master Mix, 8 μL of 10 μM primer mix and 2 L of 50× diluted cDNA.
Expression constructs
3×HA-DBC1-pCDNA3.1: DBC1 coding sequence from pCMV-SPORT6 vector (obtained from Open Biosystems) was ligated into the BglII-EcoRI site of 3×HA-pRc/CMV2 vector (D. Anderson, University of Saskatchewan). The 3×HA-DBC1 fragment is then subcloned by PCR into the HindIII-EcoRI site of pCDNA3.1.
pCDNA-IKK-β-Flag WT expression construct was obtained from Addgene (plasmid 23298).
DBC1/pGEX: Full length DBC1 coding sequence (aa1-923) amplified from DBC1-pCMV-SPORT6 vector by PCR was subcloned into pGEX-6P3 vector (GE Healthcare) that was cut with EcoRI and XhoI.
RelA-cFlag-pcDNA3 (mouse) vector was obtained from Addgene (Plasmid 20012).
RelAS536A-cFlag-pCDNA3 was generated by performing mutagenesis the Quick-change IIXL kit (Agilent).
HA-DBC1/pBabe-puro: DBC1 fragment was amplified by PCR from pCMV-Sport6 vector (Open Biosystems) using a forward primer that had an HA sequence. The fragment was ligated into the SalI site of pBabe-puro retroviral vector (addgene).
HA-FLIP-pMIG: c-FLIP coding sequence was amplified by PCR from the pcDNA3-Myc-FLIP vector (kindly provided by J. Tschopp, University of Lausanne) with inclusion of a single HA tag on the forward primers and ligated into the XhoI-EcoRI site of pMIG (Addgene).
IKK-β-S-mscv-puro: Synthetic double stranded oligonucleotides (S-tag-f: CTAGC CCACC ATG AAG GAG ACCGCC GCT GCC AAG TTC GAG AGA CAG CAC ATG GAC TCC GGC GGC A; S-tag-r: agctt GCCGCCGGAGTCCATGTGCTGTCTCTCGAACTTGGCAGCGGCGGTCTCCTTCATGGTGGg) were annealed and ligated into the NheI-HindIII site of pcDNA3.1+ to generate the vector S-tag/pcDNA3.1. The NheI-EcoRI fragment from this vector was ligated together with an EcoRI-NotI fragment generated by PCR from pcDNA-IKK-β-Flag-WT vector (Addgene plasmid 23298). The ligated product was then re-amplified with primers spanning the S-tag and the IKK-β sequence, carrying XhoI and NotI sites respectively. This PCR product was subcloned into the XhoI-NotI site of pMSCV-IRES-puro.
3×Flag-IκBmutant-pBabe-hygro (super-repressor): pBabe-puro-IκBmutant was obtained from Addgene (plasmid 15291). Full length IKBmt fragment was amplified by PCR and ligated into 3×Flag-CMV-10 (Invitrogen) that was cut with NotI-BglII. The 3×Flag-IKBmt from 3×Flag-IKBmt-CMV10 vector was amplified by PCR and ligated into pBabe-hygro (Addgene) that was cut with BamHI and EcoRI.
IKK-β-pMXS-ires-puro: The IKK-β-Flag fragment from IKKb-Flag-pcDNA3 vector by PCR using NotI primers and subcloned into the Not I site of pMXS-ires-puro-FF (R. Carstens, University of Pennsylvania), creating three C-terminal Flag sequences.
NF-κB-luc reporter vector: pNF-κB-Luc contains five copies of a consensus NF-kB site in a pGL2 luciferase reporter vector (Promega).
TK-LacZ vector constructed by A. Ivanov used as an internal control for luciferase assays.
Cell lines
In this study, we used MCF10aPG2, a subclone of the human mammary epithelial cell line MCF10a developed in our lab that undergoes anoikis more rapidly than the parental line (S.H.-P. and S.M.F., unpublished data), except for acinar morphogenesis assays, where MCF10aneoT cells (F. Miller, Karmanos Cancer Center) were used. These cells were maintained in MCF10a medium (DME/F12+5% horse serum +1× penicillin-streptomycin-glutamine (PSG) + 10 μg/ml insulin, 10 ng/ml EGF, 0.5ug/ml hydrocortisone, and 0.1ug/ml cholera toxin). MDA-MB231 and MDA-MB-468 (provided by A. Ivanov) were maintained in DMEM +10% fetal bovine serum + 1× PSG. All derivative cell lines were mixed populations generated by drug selection or flow sorting after viral infection (except the HA-DBC1-expressing MCF10a cells, where high-expressing clones were selected). BT-549 cells were provided from E. Pugacheva (WVU) and maintained in RPMI + 10% fetal bovine serum + 1× PSG. Inducible DBC1 shRNA knockdown cell lines were generated by infection with the A6/pTRIPZ or B2/pTRIPZ lentiviruses (described above), followed by selection with 2 μg/ml puromycin, and after 24 hours induction with 1 μg/ml doxycycline, cells were then flow-sorted for RFP. Retroviruses were packaged by co-transfecting 60mm dishes of 293T+GP2 cells (Clontech) using Mirus LT1 reagent, with 4.5 μg of retroviral plasmid and 2.5 μg of pCMV-VSV-G. Lentiviral constructs were packaged by co-transfecting 60mm dishes of 293T cells, using Mirus LT1, with 3.3 μg of lentiviral vector, 2.2 μg of sPAX2 and 1.2 μg of CMV-VSV-G. Viral supernatants were cleared about 48 hours post-transfection by filtration through 0.45 micron cellulose acetate filters (Whatman), aliquoted and stored at −80 degrees. To improve the transfection efficiency, cells were infected by spinning down the cells with virus stock at 1,400 rpm for 1.5 hr at room temperature followed by overnight incubation at 37°. After infection, and selection in puromycin (2 μg/ml), and/or flow-sorting for GFP or RFP expression, expression of the transgenes or knockdown was verified by western blotting. To induce shRNA expression, 1 μg/ml of doxycycline was added to growth media in cells with pTRIPZ-based shRNA and incubated for 48 hours before performing any experiment.
Derivative cell lines used in this study were generated as follows:
MCF10a with inducible DBC1 shRNA knockdown: MCF10a cells were infected with the A6/pTRIPZ or B2/pTRIPZ lentiviruses (described above), selected with puromycin (2 μg/ml), and after 24 hours induction with 1 μg/ml doxycycline, cells were flow-sorted for RFP.
Breast cancer cells with inducible DBC1 shRNA knockdown: MDA-MB-231 and BT-549 cells were infected with the A6/pTRIPZ lentiviruses, and selected for puromycin (2 μg/ml). After 24 hours induction with 1 μg/ml doxycycline, cells were flow-sorted for RFP.
MCF10a+DBC1 over-expression: MCF10a cells were infected with HA-DBC1/pBabe-puro and selected for puromycin (2 μg/ml). Empty vector control cells lines were generated by infecting with pBabe-puro and selecting for puromycin.
DBC1-KO MEFs with rescued DBC1 expression: DBC1-KO mouse embryo fibroblasts [27], kindly provided by Z. Lou (Mayo Clinic College of Medicine), were immortalized with SV40 T-antigen by infection with the ZipTer retrovirus (provided by P. Soriano, Hutchinson Cancer Center). Cells were then infected with HA-DBC1/pBabe-puro vector and selected for puromycin (2 μg/ml). Empty vector control cells lines were also generated by infecting immortalized DBC1-knock out MEFs with pBabe-puro and selecting for puromycin (2μg/ml).
MCF10a over-expressing IKK-β: MCF10a cells were infected with 3×Flag-IKKβ-pMXS-puro and selected for puromycin (2 μg/ml). Empty vector control cells lines were also generated by infecting with pMXS-puro and selecting for puromycin.
MCF10a with inducible DBC1 shRNA and c-FLIP over-expression: MCF10a cells expressing DBC1 shRNA (A6/TripZ) were infected with HA-FLIP/pMIG and selected for GFP-positive cells by flow cytometry. Empty vector control cells were also generated by infecting with empty pMIG vector and selecting for GFP-positive cells by flow cytometry.
Mouse embryo fibroblasts expressing IκBmt: Immortalized DBC1-KO and WT MEFs were infected with 3×Flag-IκBmt/pBabe-hygro vector and selected for hygromycin (200 μg/ml). Empty vector control cells lines were also generated by infecting immortalized DBC1-KO and WT MEFs with pBabe-hygro and selecting for hygromycin.
MDA_MB-231 cells overexpressing S-tag-IKK-β: MDA-MB-231 cells were infected with S-tag-IKK-β/MSCV-IRES-Puro vector and selected for puromycin (2 μg/ml). Empty vector control cells lines were also generated by infecting MDA-MB231 cells with mscv-puro and selecting for puromycin.
Western blots
Novex/Invitrogen 4-12% gradient gels were transferred under standard conditions in Tris-glycine/5% methanol buffer and transfer was run overnight at 45V or at 80V for 3 hours with stirring. Secondary antibodies anti-mouse-hrp and anti-rabbit-hrp were from BioRad; Clean-Blot secondary antibody (Pierce/Thermo) was used to avoid detection of precipitated IgG on blots used for co-immunoprecipitation experiments. Blots were developed using either enzyme-linked chemiluminescence (ECL, West Pico, Pierce), or, for the more quantitative applications, using fluorescently tagged secondary antibodies, buffers and scanning instrument from LiCor; quantitation generated by the software was based on raw pixel integrations.
siRNA transfection and anoikis assays
1.5×105 cells were plated in wells of 6-well collagen-coated dishes. Two duplicate wells with target siRNA and two duplicate wells with control siRNA, using 500 μL of Opti-MEM containing 5 μl of 20 μM siRNA (or 1 μl of 100 μM DBC1 siRNA) and 5 μl of lipofectamine RNAi-max, added to a well containing 2.0 ml of Opti-MEM. After 4-6 hours, cells were re-fed with regular growth medium, and, 24 hours later, each well was split into one 60mm dish. These were further incubated forty-eight hours, trypsinized, and resuspended in 3ml of growth medium. Cells were counted and 100,000 cells were plated in a 35mm Ultralow Attachment well and incubated for specified periods of time in presence of 0.5% of methylcellulose. At each time points, including time zero, cells were spun down at 4,000 rpm for 1.5 minutes in a microfuge tube, washed in ice-cold D-PBS once and lysed in 100 uL of phosphate buffered saline (PBS) containing 0.5% Triton-×100 and 10 mM EDTA. Lysates were incubated on ice for 15 minutes and cleared at 13,200 rpm for 12 minutes and, 5-15ul were assayed in a total of 100ul of Roche Cell Death ELISA Lysis/Incubation buffer in the Roche system and read in a Perkin-Elmer Envision/Excite. Values measured at a suspension time of zero (time-zero), which were generally unaffected by the siRNAs we transfected in this study, were subtracted from the final values shown in the figures. MCF10a cells that were pre-treated with either curcumin (25 μM) for 4 hours were suspended for indicated time and collected to measure DNA fragmentation using Roche Cell Death ELISA lysis kit as described above.
Cell permeability-based cell death assays
DBC1-KO MEF (~100,000 cells) expressing either vector or HA-DBC1 vector treated with BAY 11-7082 (10 μM) for 1 hour were plated on low attachment 6-well plates containing 2 ml of growth medium. Cells were collected at indicated time points, spun down at 3,200 rpm for 15 seconds, resuspended in 100 μL of Accumax (Innovative Cell Technologies) and incubated (~5-10 minutes) at room temperature to generate single cell suspensions. Equal volumes of trypan blue staining solution (Invitrogen) were added to the cell suspension and the percentage of dead cells was counted on a hemacytometer in duplicate; time-zero cell death values were subtracted.
RelA acetylation assay
MCF10a cells expressing control shRNA or DBC1 shRNA, or mouse embryo fibroblasts (DBC1 knockout or DBC1-rescued) were plated on 6-well plates. When cells were about 80% confluent, cells were treated with TNF (20 ng/ml for MCF10a, 40 ng/ml for MEF) for 30 or 120 minutes. Cells were lysed with 5% βME/1× SDS sample buffer, boiled and analyzed on western blots using acetyl-lys310 relA antibody.
Reporter Assays
50,000 MCF10ap+DBC1 shRNA cells were plated on 12 well plates coated with collagen in duplicates and shRNA was induced with 1 μg/ml of doxycyclin. Next day, cells were co-transfected with 600ng NF-kB-luciferase construct, 150 ng TK-lacZ and either 250 ng of HA-DBC1-pCDNA3.1 or pCDNA3.1 vector by using Lipofectamine 2000 reagent (Invitrogen) at a ratio of 1 μg of DNA to 2 μL of Lipofectamine reagent. MEF DBC1 WT and KO expressing 3×Flag-IκB mutant construct were plated on 6 well plates coated with collagen. Next day, cells were transfected with either DBC1 or control siRNA. After 24hrs, cells were re-plated on 12 well plates coated with collagen in duplicates. Following day, cells are co-transfected with ug of NF-κB-luciferase construct and TK-LacZ (1 μg of total DNA) by using Lipofectamine 2000 reagent. After 28hrs, cells were treated with TNF (20ng/ml) for 90mins and lysed with 200ul of 1× cell culture lysis buffer (Promega). The cell lysates were centrifuged at 4C for 12mins at 13,200rpm and assayed for luciferase and beta-galactosidase activity in 100ul of luciferase assay reagent or 150ul of 1× galactosidase assay buffer (both Promega), which were analyzed in a Perkin-Elmer Envision/Excite apparatus. Light units were normalized to b-galactosidase activity to control for transfection efficiency. For MDA-MB 231 cells, 60,000 cells plated on 6-well dishes were transfected with 750 ng NF-κB-luciferase construct and 250 ng TK-LacZ. After 24 hours, cells were re-plated on low-attachment 6-well plates. Cells were harvested at time points indicated and assayed for luciferase and β-galactosidase activity. MCF10a cells plated on 6 well plates coated with collagen were transfected with either control or DBC1 siRNA. After 24 hours, cells were split and plated onto 12 well plates coated with collagen. Following day, cells were transfected with 600 ng NF-κB-luciferase construct, 150 ng TK-LacZ, and either 250 ng of pCDNA3.1 or 3×Flag-IKKβ-pCDNA3.1 vector by using Lipofectamine 2000 reagent. After 28 hours, cells were treated with TNF (20 ng/ml) for 1.5hr and assay assayed for luciferase and beta-galactosidase activity.
Effect of Bay 11-7082 on RelA localization
Cells grown on poly-L-lysine coated coverslips were treated with 10 ng/ml of Bay 11-7082 for 1 hour at 37° followed by 20 ng/ml of TNF for various times. At each time point, cells were fixed in PBS containing 4% paraformaldehyde and permeabilized with PBS containing 0.2% Triton X-100. Cells were blocked in PBS containing 10% normal goat serum, 0.1% Tween-20, and 0.1% BSA (blocking solution) for 60 minutes at room temperature. A 1:500 dilution of RelA antibody to blocking solution was then applied overnight at 4°C. Slides were washed with 0.1% PBS/Tween-20 three times and incubated for 1.5 hour with Alexa 488-conjugated goat anti-rabbit reagent (Molecular Probes) diluted 1:1000 in blocking solution. Coverslips were mounted with ProLong Gold (Molecular Probes) containing 4′, 6-Diamidino-2-phenylindole. Target proteins were detected fluorescently using microscope (Zeiss Axiovert).
Acinar Morphogenesis assay
MCF10aneoT cells expressing either control or DBC1 shRNA were assayed for morphogenesis following the protocol indicated in [50].
RelA S536 phosphorylation assay
MCF10a cells expressing either control or DBC1 shRNA were plated on 6 well plates. Cells were treated with TNF (20 ng/ml) for 1.5 hours or 3 hours and lysed with 1× SDS sample buffer +5% βME. Western blots were performed using relA phospho-S536 specific antibody or total relA antibody; buffer containing 1× TBS+0.1% Tween+ 5% BSA was used for phospho-specific antibody incubation and blocking. Three independent experiments were performed. Image J was used to measure the relative phosphorylation level against total RelA expression. The results from three different experiments were averaged and indicated in the bottom of the blots with standard deviation.
S-tag pulldown assay
MDA-MB-231 cells expressing either S-tag-IKK-β or empty vector (one 100ml dish for each sample) were pre-treated with TNF (20 ng/ml) for 1.5hr or suspended for 24 hours on low attachment plates. Cells were lysed in IP buffer (600 μL), followed by preclearing the lysates at 13,200 rpm for 10 minutes. Lysates were precipitated with 40 μL of a 50% slurry of S-protein beads (Novagen/EMD Biosciences) that had been pre-equilibrated with IP buffer containing 10 mg/ml of bovine serum albumin and washed. After overnight incubation at 4°C with rotation, beads were washed three times with IP buffer. Samples were boiled in 2× SDS sample buffer for 5 minutes and analyzed by Western blotting.
In vitro kinase assay
To each 20 μL reaction, recombinant proteins GST-IKKβ, GST-IκBα and DBC1 protein was added at the indicated amounts. The final concentrations of components in the kinase assay buffer were: 25 mM Tris-HCl, pH8, 50mM KCl, 10mM MgCl2, 1mM Na3VO4, 1mM DTT and 500 μM ATP. After 30 minute incubation at 30°, the reactions were stopped by the addition of 20 μL of 2× SDS sample buffer+βME to the tube and boiling. Samples were analyzed on western blots for IκBα phospho-S32/36.
In vivo kinase assay
MDA-MB-231 cells that express either control or DBC1 shRNA were plated on 60mm dishes. Cells (either attached or suspended for 24 hours in complete growth medium) were lysed in 600ul of standard IP buffer (25mM Tris pH8, 150mM NaCl, 0.5% Triton-×100, 10% glycerol, 1 mM EDTA, 1 mM beta-glycerophosphate, and Pierce complete protease inhibitor). After centrifugation at 13,200 rpm for 10 minutes at 4°C, supernatants were incubated with either 1 μg of IKK-β or normal IgG antibody overnight at 4°C with rotation, followed by incubation with 40 μl of pre-equilibrated protein A-sepharose for another hour. Beads were washed three times with IP buffer and washed three times with kinase assay buffer (25mM Tris-HCl pH8, 50mM KCl, 10mM MgCl2, 1mM Na3VO4, and 1mM DTT). Kinase assay buffer (40 μL) containing GST-IκBα (50 ng) fusion protein and ATP (1mM) was added to the beads, which were incubated at 30°C for 30 minutes. Following removal of the beads by centrifugation, the supernatant was diluted with 2S SDS sample buffer, heated and analyzed by western blotting and probed for phospho-IκBα, IKK-β, and DBC1.
RESULTS
DBC1 suppresses anoikis
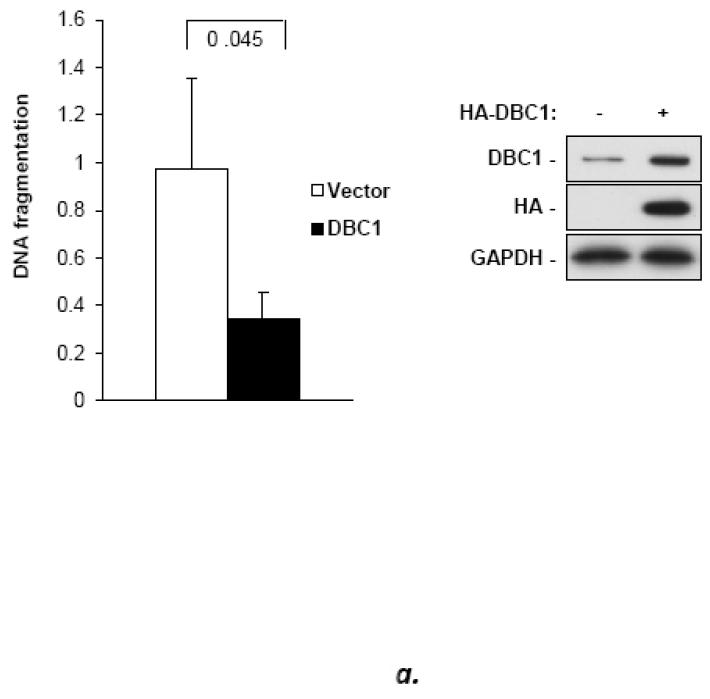
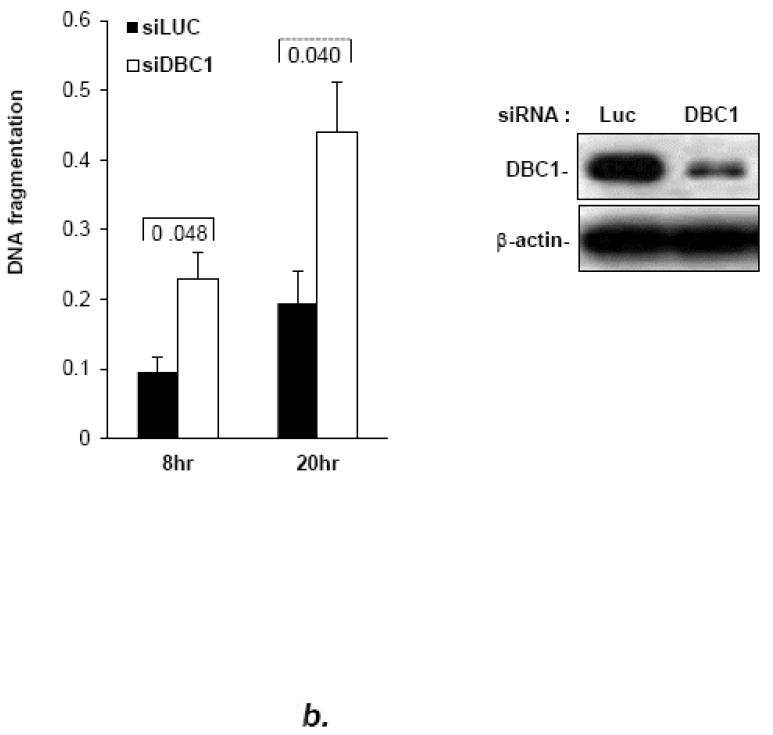
To investigate the potential role of DBC1 in anoikis, full-length DBC1 was over-expressed in the human mammary epithelial cell line MCF10a. Over-expression (~3-fold) of DBC1 conferred significant (~3-fold) resistance to anoikis after six hours of suspension, although anoikis was not permanently suppressed (Fig. 1a). Conversely, the partial depletion of DBC1 via siRNA transfection sensitized MCF10a cells to anoikis (Fig. 1b). Similar results were obtained using MCF10a cells stably transfected with DBC1 shRNA targeting a sequence distinct from the siRNA targets, ruling out off-target effects (Fig. S1). The effect of DBC1 siRNA on anoikis was further validated using a cell permeability assay for percent cell death as well as a three-dimensional acinar morphogenesis assay (Fig. S2, S3).
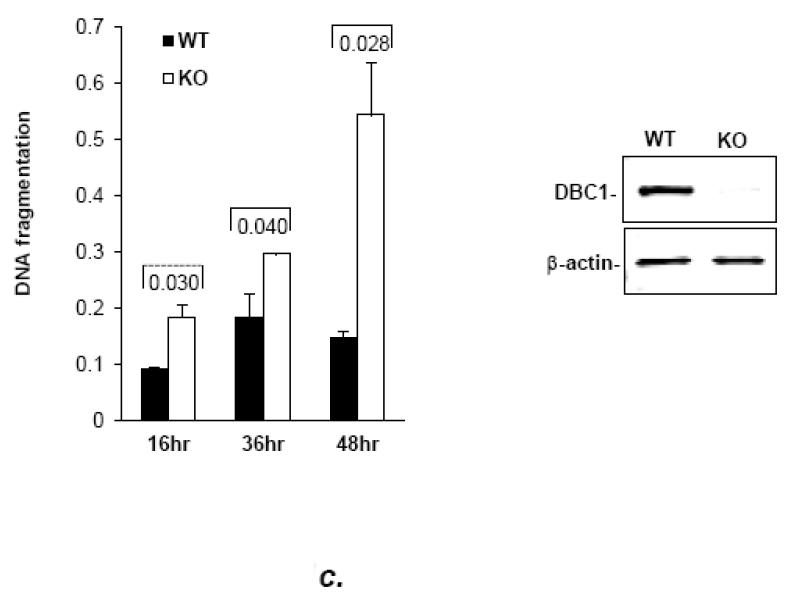
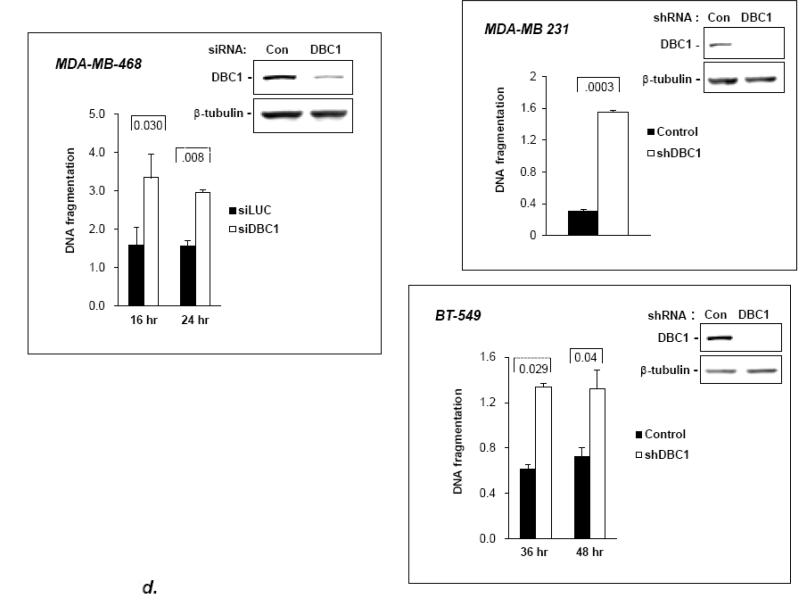
Fig. 1. DBC1 suppresses anoikis.
(a) DBC1 over-expression suppresses anoikis in MCF10a cells. MCF10a infected with DBC1 retroviral expression vector or control vector were suspended for six hours and assayed for anoikis using a cell death ELISA assay. (b) DBC1 suppresses anoikis in MCF10a cells: DBC1 knockdown approach. MCF10a transfected with DBC1 siRNA were assayed for anoikis using DNA fragmentation ELISA. Western blotting confirmed the knock down of DBC1. (c) DBC1 knockout promotes anoikis. Mouse embryo fibroblasts with wild-type or DBC1-knockout genotypes were assayed for anoikis by cell death ELISA. Expression level of DBC1 was confirmed by western blotting.(d) DBC1 suppresses anoikis in breast cancer cell lines. The indicated breast cancer cell lines were infected with either control or DBC1 shRNA vectors and assayed for anoikis using DNA fragmentation ELISA. DBC1 knock down was confirmed by western blotting. Samples were assayed in duplicate and the data shown are representative of three experiments
The generality of the protective effect of DBC1 on anoikis was then addressed in several diverse cell lines. First, mouse embryo fibroblasts (MEF) derived from DBC1−/− mice [27] were found to be more sensitive to anoikis than DBC1+/+ MEF (Fig. 1c). To exclude potential artifacts due to spurious differences between MEF cell lines, DBC1−/− MEFs were rescued with DBC1 retroviral expression vector, which reverted them to the wild-type level of anoikis-sensitivity (Fig. S4). The anoikis-sensitization effect of DBC1 depletion was also observed in three breast cancer cell lines: MDA-MB-231, BT-549 and MDA-MB-468 (Fig. 1d). These data indicate that DBC1 functions as a generalized anoikis suppressor.
DBC1 affects the expression of NF-κB target genes
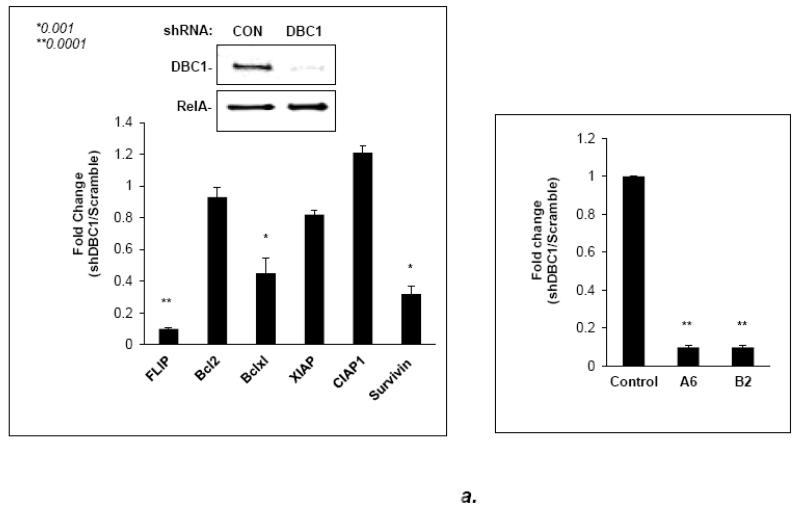
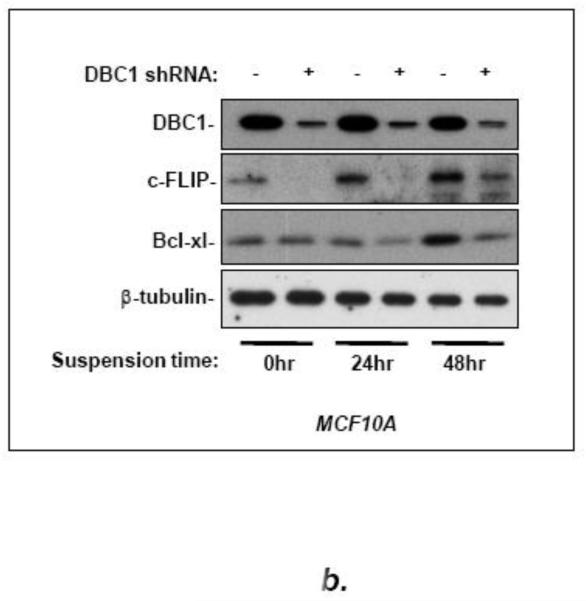
We hypothesized that DBC1 might affect NF-κB, in light of the documented importance of this factor for anoikis regulation [8,12,14]. To test this, we initially probed for several NF-κB target genes in cells with or without DBC1 expression. MCF10a cells with DBC1 depletion were probed by qRT-PCR, showing significant decreases in the levels of the NF-κB target genes c-FLIP, Bcl-x-l, and survivin; other target genes, Bcl-2, xIAP, and cIAP, were unaffected (Fig. 2a). Positive regulation of both c-FLIP and bcl-xl proteins by DBC1 was observed in both attached and detached MCF10a cells (Fig. 2b), as well as in the comparison of wild-type vs. DBC1 knockout MEFs (Fig. S5).
Fig. 2. DBC1 affects expression of NF-kB target genes.
(a) DBC1 induces expression level of NF-kB target genes in MCF10A cells. (Left panel): MCF10a cells expressing DBC1 or control shRNAs were analyzed by real time PCR for the expression level of the indicated NF-kB target genes; DBC1 knockdown was confirmed by western blotting; RelA was used as a loading control as DBC1 did not affect its level (Right panel): MCF10a cells expressing two different DBC1 shRNAs were assayed for c-FLIP, to rule out off-target effects. (b) DBC1 knockdown reduces c-FLIP and Bcl-xl expression in MCF10A. (Left panel): Expression levels of c-FLIP and Bcl-xl in attached or suspended MCF10A cells were assayed by western blotting. Samples were assayed in triplicate and the data shown are representative of two experiments
These data indicated that DBC1 protein enhanced the expression of some NF-κB target genes, under attached or detached conditions.
DBC1 regulates anoikis through NF-kB
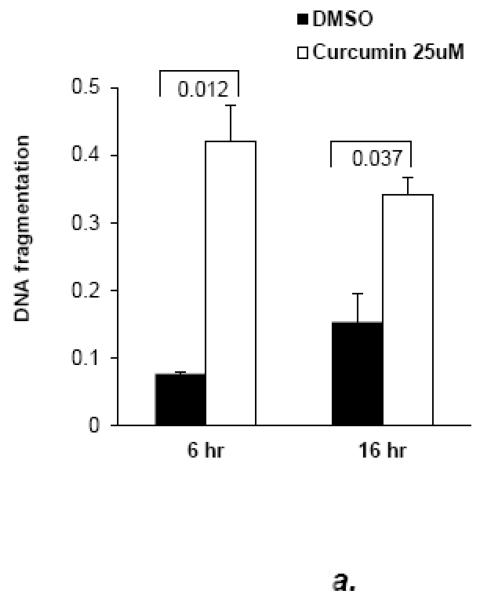
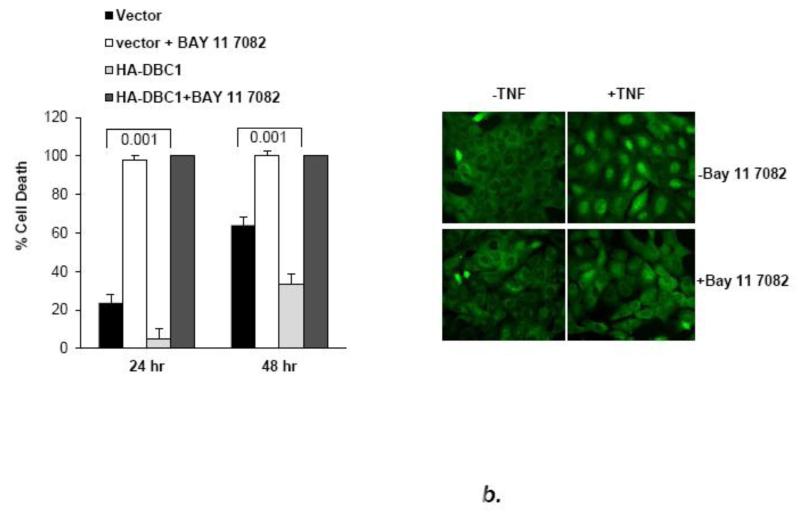
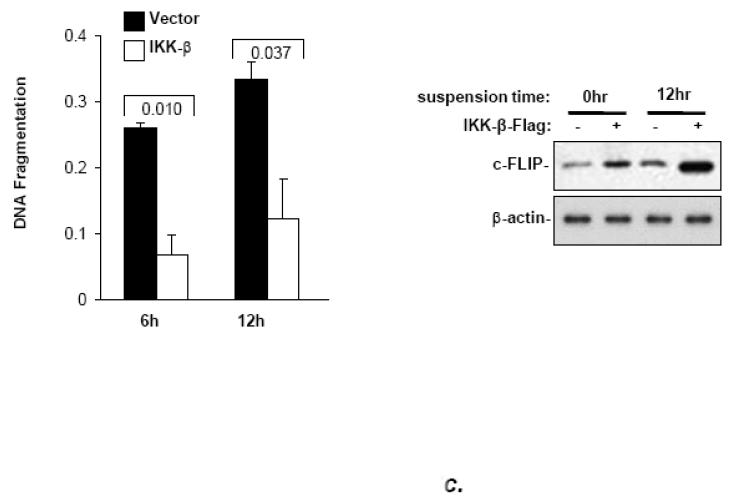
Initially, the role of NF-κB in anoikis of MCF10a cells was tested by the use of the dietary, non-specific inhibitor, curcumin and a highly specific pharmacologic inhibitor of IKK-β activity, Bay 11-7082 [28,29]. Both compounds, used at similar concentrations to those previously reported for cell lines, sensitized MCF10a cells and mouse embryo fibroblasts to anoikis (Fig. 3a, b). The anoikis-hypersensitive state in BAY 11-7082-treated mouse embryo fibroblasts was not reversed by DBC1 expression, indicating that DBC1 protects primarily through the NF-κB pathway (Fig. 3b). Further supporting the role of NF-κB in anoikis, over-expression of IKK-β which has been shown previously to activate the NF-κB pathway [30], protected MCF10a against anoikis efficiently and induced higher levels of c-FLIP expression, both in attached and, especially, detached cells (Fig. 3c).
Fig. 3. DBC1 regulates anoikis through NF-κB.
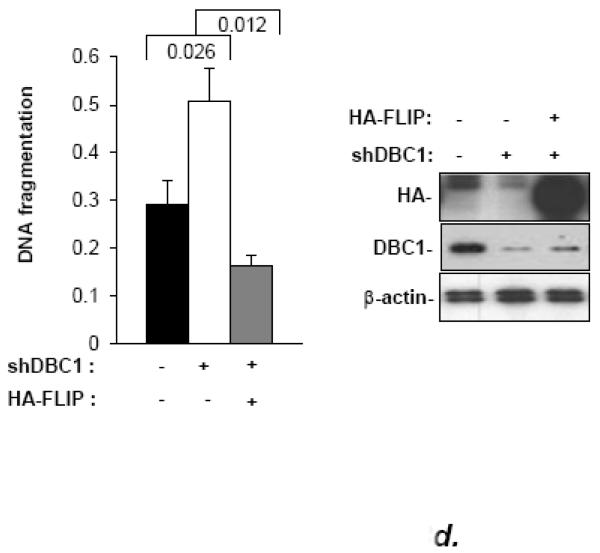
(a) Curcumin enhances the anoikis response in MCF10a cells (DNA fragmentation ELISA). (b) The specific NF-kB inhibitor BAY11-7082 enhances the anoikis response in mouse embryo fibroblasts, which is not rescued by DBC1. (Left panel): DBC1-knockout MEFs that were stably infected with empty vector or retroviral DBC1 expression vector were assayed for anoikis in the presence or absence of Bay 11 7082 (cell permeability/trypan blue assay). (Right panel): Confirmation of inhibition of relA nuclear translocation by BAY11-7082 (immunofluorescence). (c) IKK-β over-expression suppresses anoikis and induces c-FLIP expression in MCF10a cells. (Left panel): MCF10a cells over-expressing Flag-IKK-β were assayed for anoikis (DNA fragmentation ELISA). (Right panel): Expression of c-FLIP in MCF10a cells upon detachment was analyzed by western blotting. (d) DBC1 regulates anoikis in part through c-FLIP. MCF10a+DBC1 shRNA cells expressing either control or HA-FLIP retroviral vector were assayed for anoikis (cell death ELISA). Over-expression of HA-FLIP was confirmed by western blotting. Samples were assayed in duplicate and the data shown are representative of two to three experiments
Among NF-κB target genes, c-FLIP is particularly noted for its ability to regulate anoikis [11]. To test the role of c-FLIP in the suppression of anoikis by DBC1, MCF10a cells that express inducible DBC1 shRNA were infected with a constitutive c-FLIP retroviral construct. This was found to reverse the effect of DBC1 knockdown (i.e., confer resistance to anoikis; Fig. 3d). These data indicate that DBC1 promoted the expression of multiple NF-κB target genes, including c-FLIP, suppressing anoikis.
DBC1 activates NF-kB-mediated transcription
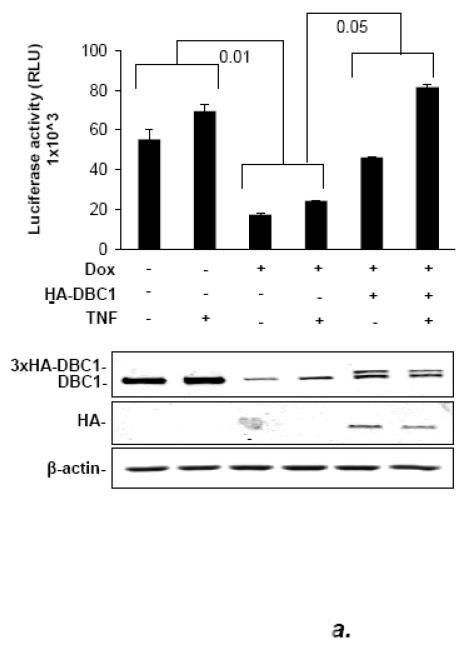
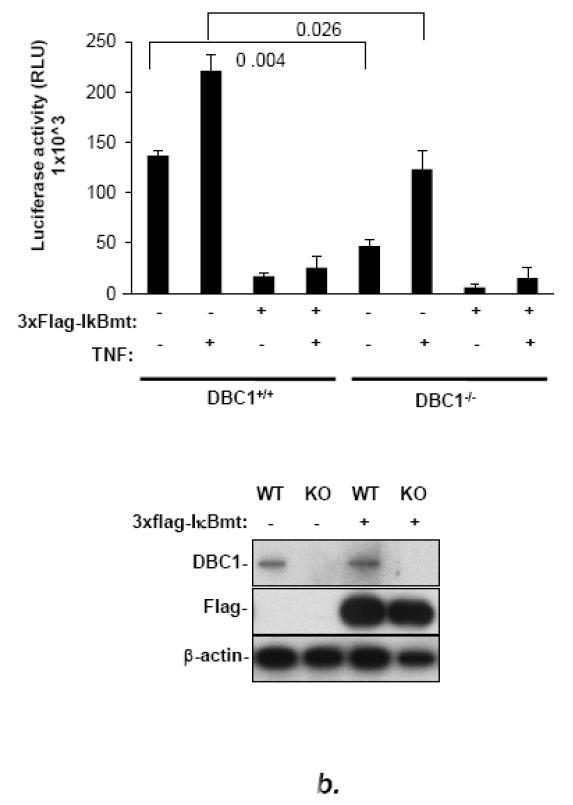
The results described above suggested that DBC1 activated the transcriptional activity of NF-κB. To test this, an NF-κB-responsive reporter construct was co-transfected into MCF10a cells stably bearing a doxycycline-inducible DBC1 shRNA construct. The depletion of DBC1 inhibited NF-κB-driven luciferase expression significantly (Fig. 4a). As a control, cells were co-transfected with DBC1 cDNA expression vector to rescue DBC1 expression (because the shRNA was targeted to the 3′ UTR), which rescued the NF-κB activity, ruling out off-target effects. The effect of DBC1 on NF-κB activity was observed in cells with or without stimulation by TNF-α. The effect of DBC1 was confirmed by additional experiments in DBC1 knockout vs. DBC1 wild-type MEFs, which were co-transfected with the NF-κB-luc reporter vector, in the presence or absence of a dominant-negative mutant of Iκ B-α (“Iκ Bmt”) that constitutively inhibits relA translocation regardless of stimulatory signals [31]. DBC1 enhanced NF-κB reporter activity in this system, which was inhibited to below baseline level by the Iκ Bmt construct, indicating the dependence of DBC1-stimulated NF-κB activity on the canonical kinase pathway (Fig. 4b).
Fig. 4. DBC1 stimulates NF-κB transcriptional activity.
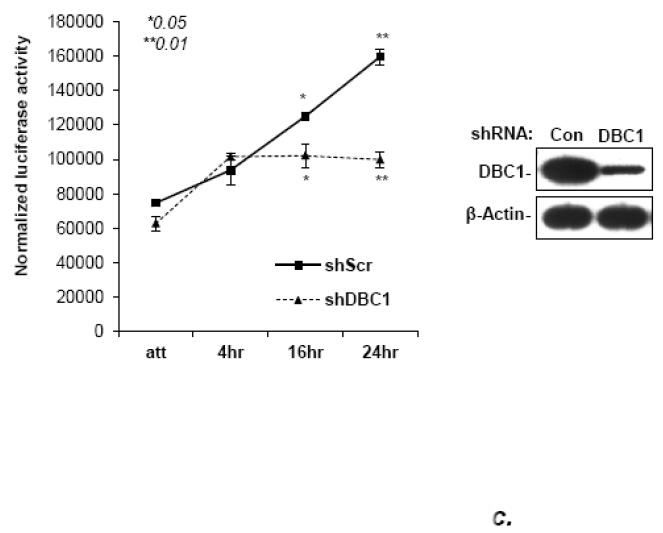
(a) DBC1 promotes NF-κB activation in MCF10a cells. MCF10a+shDBC1 cells were transfected with 3×-NF-κB-luc, and either empty pCDNA3.1 or 3×HA-DBC1-pCDNA3.1. The cells were then assayed for luciferase activity following stimulation with TNF in the indicated samples. Knockdown and re-expression of (shRNA-resistant) DBC1 was confirmed by western blotting. (b) DBC1 promotes NF-κB activation in mouse embryo fibroblasts, which is sensitive to inhibition by IκBmt. Wild-type or DBC1-knockout MEFs were transfected with 3×HA-DBC1 expression vector or empty vector (pcDNA3.1), 3×-NF-κB-Luc, and Flag-I Bmt. Cells were assayed for luciferase activity following TNF treatment (in indicated samples). DBC1 knockdown and transfected Flag-IκBmt expression were confirmed by western blot. (c) DBC1 suppresses NF-kB activity induced upon detachment. MDA-MB-231 cells expressing either control or DBC1 shRNA were transfected with 3×-NF-kB-luc and assayed for luciferase activity following the indicated times of detachment. Samples were assayed in duplicate and the data shown are representative of two to three experiments
Breast cancer cell lines, but not normal mammary epithelial cells, respond to detachment conditions by activating NF-κB activity, contributing to anoikis-resistance [14]. To determine whether this stimulation was DBC1-dependent, MDA-MB-231 cells with or without DBC1 shRNA expression were transfected with the NF-κB reporter construct and assayed for NF-κB activity following detachment. Control cells activated NF-κB almost linearly over a 24-hour period, as reported previously [14]. By contrast, cells with DBC1 knockdown showed significantly reduced NF-κB activity, especially at time points after four hours (Fig. 4c). This indicated that the detachment-induced increase in NF-κB activity in a breast cancer cell line was partially dependent upon DBC1 protein.
DBC1 interacts with IKK-β and stimulates its activity
By immunofluorescence, we confirmed previous reports that DBC1 was a nuclear protein [27], and, consistent with this, the translocation of relA to the nucleus upon stimulation with TNF was not affected by DBC1 (Fig. S6). This excluded mechanisms involving the canonical activity of the IKK complex in promoting relA translocation from cytoplasm to nucleus. We also addressed the possibility that DBC1 – an endogenous inhibitor of SIRT1 deacetylase activity [32,33]—activated the transcriptional activation domain of relA through acetylation, but relA K310 acetylation was not detectable in MCF10a cells (data not shown), and there was no significant effect of DBC1 acetylation in mouse embryo fibroblasts (Fig. S7).
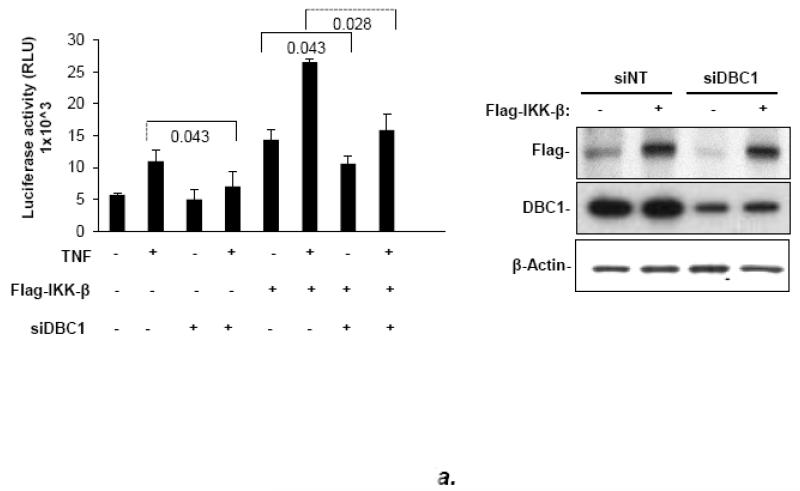
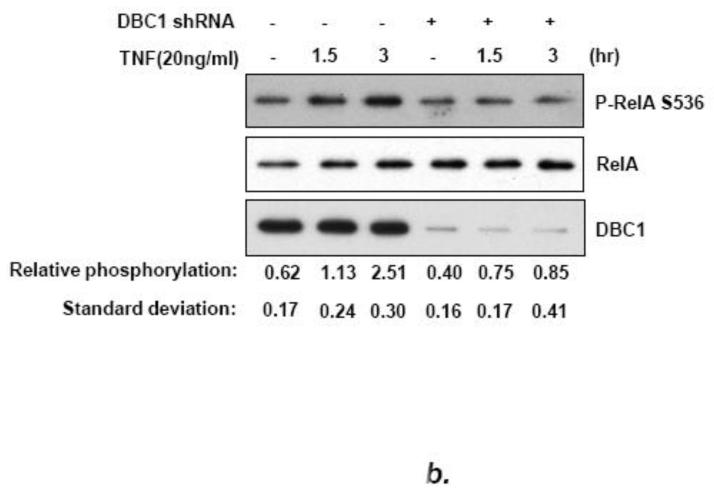
Interestingly, DBC1 stimulated the activation of NF-κB activity by co-transfected IKK-β in reporter assays (Fig. 5a). Moreover, the major phosphorylation site for IKK-β on relA, S536, potentiated transactivation by relA using a co-transfection assay (Fig. S8), as reported previously in other systems [17-20]. We reasoned that perhaps DBC1 stimulated the kinase activity of IKK-β on relA S536. Consistent with this, the phosphorylation of relA S536 was suppressed by shRNA-mediated depletion of DBC1 (Fig. 5b).
Fig. 5. DBC1 interacts with IKK-b and stimulates its activity.
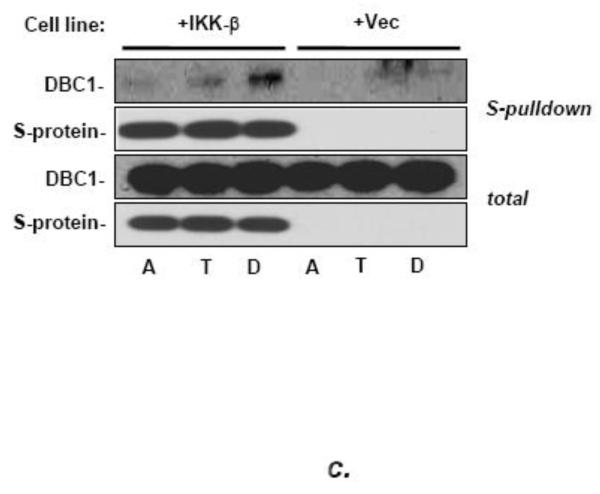
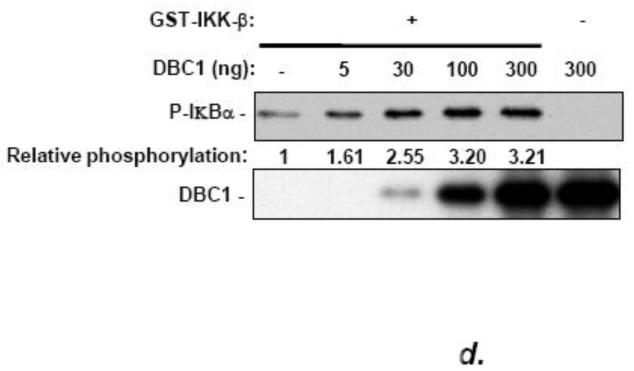
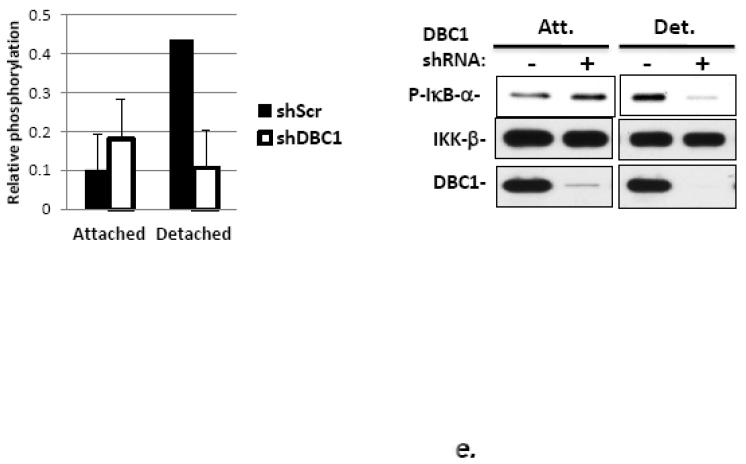
(a) DBC1 promotes IKK-βb induced NF-κB activation. MCF10a cells that were pretreated with either control or DBC1 siRNA were transfected with Flag-IKK-β and 3×-NF-κB-luc and assayed for luciferase activity. Knockdown of DBC1 and expression of Flag-IKK-β were confirmed by western blotting. Samples were assayed in duplicate and the data shown are representative of two experiments. (b) DBC1 promotes the phosphorylation of RelA S536. (Left panel): MCF10A stably expressing control or DBC1 shRNA were analyzed by western blotting for phospho-RelA S536 and total relA upon TNF stimulation. The numbers on the bottom represents the average of three independent experiments. (c) DBC1 interacts with IKK-β. (Top panel): Lysates from MDA-MB-231 cells that expressed S-tag-IKK-β or empty vector), stimulated with either TNF (T) or detachment (D) were precipitated with S-protein agarose beads and analyzed for DBC1 and S-tag-IKK-β protein by western blotting. (Lower panel): Lysates from MDA-MB-231 cells treated as above were immunoprecipitated with IKK-β antibody and interaction with DBC1 was determined by western blotting. The data shown are representative of three experiments. (d) DBC1 stimulates IKK-β kinase activity (in vitro). Kinase activity of recombinant IKK-β was assayed on recombinant GST-IκB-αsubstrate in the presence of the indicated amounts of recombinant DBC1 protein. Kinase activity was detected using a phospho-IκB-α antibody. The data shown are representative of three experiments. (e) DBC1 stimulates IKK-β kinase activity (in vivo). MDA-MB-231 cells with control or DBC1 shRNAs (see figure 4c) were assayed for IKK-β under attached or suspended conditions; phosphorylation of substrate was normalized to total IKK-β to generate the values shown, which were averaged together over two experiments.
By high-throughput proteomics analysis of the NF-κB pathway, DBC1 was identified previously as a candidate interactor of IKK-β [34]. To demonstrate an interaction of DBC1 with IKK-β, S-tagged IKK-β expressed in MDA-MB-231 cells was precipitated with S-protein agarose and probed for DBC1. Interactions clearly above a background control (i.e., a sample of MDA-MB-231 cells without the S-tagged construct) were observed, with highest efficiency in cells that were detached for twenty-four hours prior to lysis (Fig. 5c). A fully endogenous co-immunoprecipitation of IKK-β with DBC1 was observed as well, but these results were not as consistently reproducible as those using the S-tagged construct, perhaps reflecting the low abundance of nuclear IKK-β expression [35](data not shown).
DBC1 stimulated NF-κB transcriptional activity and the phosphorylation of relA S536 and interacted with IKK-β as well. To test the effect of DBC1 on IKK-β kinase activity, recombinant proteins were assayed in vitro. The results showed a dose-dependent stimulation of IKK-β activity by DBC1 protein (Fig. 5d), with maximal stimulation at a DBC1: IKK-β molar ratio of approximately 1.0-1.5. Correspondingly, the knockdown of DBC1 decreased the enzymatic activity of IKK-β that was immunoprecipitated from detached MDA-MB-231 cells and assayed (figure 5e). These observations confirmed that DBC1 interacted with and stimulated the activity of IKK-β resulting in enhanced NF-κB activity and protection from anoikis.
DISCUSSION
The key finding of this study is that DBC1 protein contributes to the protection of normal and transformed cells against anoikis. Notably, DBC1 is not invariably anti-apoptotic. For example, the interaction of DBC1 with SIRT1 is enhanced by genotoxic and/or oxidative stress, through ATM-mediated DBC1 phosphorylation, enhancing p53 acetylation – a pro-apoptotic effect of DBC1 with respect to genotoxic/oxidative stress-induced apoptotic responses, in contrast with its role in anoikis [21].
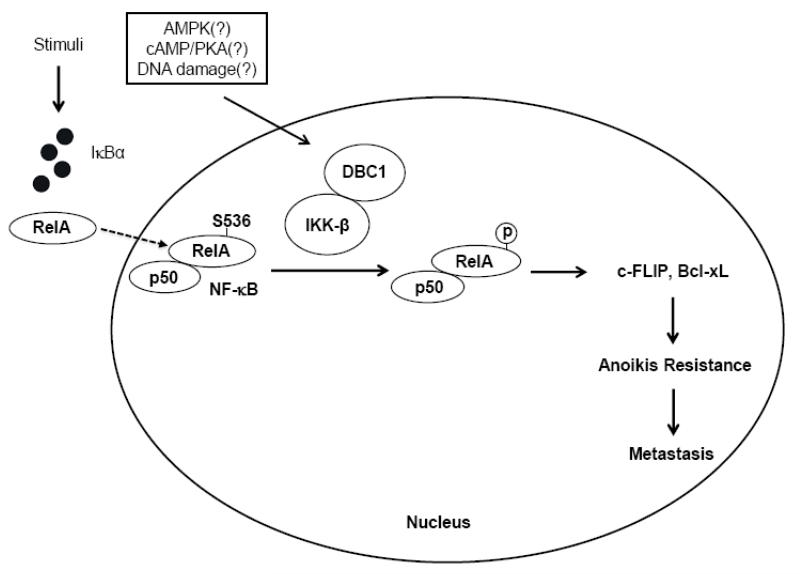
The novel pathway by which DBC1 protein promotes NF-κB activity and anoikis-resistance is depicted in figure 6. DBC1 may be conceptualized as a co-factor for IKK-β that stimulates its kinase activity on relA (S536), promoting the transcriptional activation of NF-κB target genes that enhance anoikis-resistance. Among the most robustly DBC1-sensitive genes were c-FLIP and bcl-xl, both of which have significant roles in anoikis-resistance of tumor cells, documented both here and elsewhere [11,36]. In this connection, novel drugs that act by destabilizing c-FLIP expression are being developed with the goal of sensitizing tumor cells to anoikis [37].
Fig. 6.
Notably, IKK complexes can promote cell survival by additional, NF-κB-independent mechanisms as well, for example, through the phosphorylation of Foxo3A, p53, and mTORC (the latter resulting in survivin up-regulation) [5]. Although our data indicate a dominant role of NF-κB in the DBC1 cell survival effect, we cannot exclude that other IKK-βdependent, NF-κB-independent effects might play additional roles. In this connection, IKK-βI/IKK-β appear to be responsible for increased relA S536 phosphorylation in some tumor cells [38]; the potential of DBC1 to affect additional IKK complexes will be interesting to explore. Note also that while IKK-β clearly is important also for the phosphorylation of IκB-αDBC1 did not affect the phosphorylation of this substrate in vivo, consistent with the primarily nuclear localization of DBC1, the stimulus-responsive nuclear and cytoplasmic localization of IKK-β and the constitutive cytoplasmic localization of IκB-α[27,39].
Recent studies indicate that increased relA S536 phosphorylation and DBC1 over-expression each correlate with worse prognosis in colorectal cancer, a cancer type that is influenced dramatically by inflammatory stimuli that activate NF-κB [25,40]. Correlation of more aggressive disease with DBC1 over-expression has been reported in breast cancer also [22,23], and our data here show that DBC1 is important for the detachment-induced NF-κB activation that is observed in breast cancer cell lines, reported by the Richer laboratory [14]. These observations suggest that DBC1 over-expression, which has been demonstrated in esophageal cancer as well [24], promotes tumor progression through the pathway that we have elucidated here.
Biochemically, DBC1 was previously shown to regulate several transcription factors or regulators of transcription factors. As an endogenous inhibitor of SIRT1 deacetylase activity [32,33], DBC1 enhanced the acetylation and stability of the SIRT1 substrate p53, potentially enhancing apoptosis in certain contexts. However, a recent study in liver cancer cells showed an effect of DBC1 neither on SIRT1 activity nor p53 acetylation, complicating the interpretation of this phenomenon [41]. We did not observe an effect of DBC1 on the acetylation of relA in mouse embryo fibroblasts, and failed to detect significant relA acetylation in MCF10a cells, diminishing the relevance of this as a potential mechanism for DBC1 to regulate NF-κB, at least in our cell lines. DBC1 regulates several other factors as well, including the SUV39H1 histone methyltransferase, histone deacetylase-3, BRCA1, estrogen receptor and androgen receptor, thus having multiple pleiotropic effects on transcription [42-47]. In fact, DBC1 may also modulate alternative splicing, as it is a component of the DBIRD complex that integrates mRNA splicing with RNA polymerase II-mediated transcription [48]. Thus, it is not only possible but likely that DBC1 affords anoikis-resistance by mechanisms additional to the NF-κB mediated one described here, which remain to be identified. In this connection, though, microarray analysis identified c-FLIP plus a surprisingly short list of other DBC1-regulated genes, suggesting that NF-κB and/or additional c-FLIP regulatory proteins may be the most significant DBC1 targets (data not shown).
The interaction of DBC1 with IKK-β signifies that NF-κB is positioned for activation by novel mechanisms that have not yet been suggested. For example, the interaction between SIRT1 and DBC1 is dissociated by cAMP/PKA, acting through AMP-dependent protein kinase (AMPK), suggesting a novel mechanism for the metabolic control of SIRT1 [49]. Although it is not yet clear how this dissociation is achieved, it is intriguing to note that NF-κB is inhibited by AMPK. This raises the possibility that the interaction of DBC1 with IKK-β may also be regulated by AMPK or other signal transducers, providing novel metabolic control mechanisms for NF-κB and anoikis.
Supplementary Material
Acknowledgements
S.M. Frisch was supported by NIH grant R01CA123359. The flow cytometry core facility (Mary Babb Randolph Cancer Center) was supported by NIH grants RR020866 and P20 RR16440 and we thank Kathy Brundage for performing the flow-sorting. We also wish to thank Zenkun Lou, Eduardo Chini for the DBC1-knockout cells, Alexey Ivanov, Elena Pugacheva, Jurg Tschopp, Russ Carstens, Yon Rojanasakul and Sierra Talbott for constructs; we also thank the Biochemistry Protein Core for assistance with recombinant protein production.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
REFERENCES
- 1.Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guadamillas MC, Cerezo A, Del Pozo MA. Overcoming anoikis--pathways to anchorage-independent growth in cancer. J Cell Sci. 124:3189–3197. doi: 10.1242/jcs.072165. [DOI] [PubMed] [Google Scholar]
- 3.Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;xxx:xxx. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett. 2008;272:177–185. doi: 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin AS. Regulation of cell death and autophagy by IKK and NF-kappaB: critical mechanisms in immune function and cancer. Immunol Rev. 246:327–345. doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal BB, Sung B. NF-kappaB in cancer: a matter of life and death. Cancer Discov. 1:469–471. doi: 10.1158/2159-8290.CD-11-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunol Rev. 246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 8.Toruner M, Fernandez-Zapico M, Sha JJ, Pham L, Urrutia R, et al. Antianoikis effect of nuclear factor-kappaB through up-regulated expression of osteoprotegerin, BCL-2, and IAP-1. J Biol Chem. 2006;281:8686–8696. doi: 10.1074/jbc.M512178200. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Li H, Wu X, Yoo BH, Yan SR, et al. Detachment-induced upregulation of XIAP and cIAP2 delays anoikis of intestinal epithelial cells. Oncogene. 2006;25:7680–7690. doi: 10.1038/sj.onc.1209753. [DOI] [PubMed] [Google Scholar]
- 10.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, et al. IAP regulation of metastasis. Cancer Cell. 17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mawji IA, Simpson CD, Hurren R, Gronda M, Williams MA, et al. Critical role for Fas-associated death domain-like interleukin-1-converting enzyme-like inhibitory protein in anoikis resistance and distant tumor formation. J Natl Cancer Inst. 2007;99:811–822. doi: 10.1093/jnci/djk182. [DOI] [PubMed] [Google Scholar]
- 12.Yan SR, Joseph RR, Rosen K, Reginato MJ, Jackson A, et al. Activation of NF-kappaB following detachment delays apoptosis in intestinal epithelial cells. Oncogene. 2005;24:6482–6491. doi: 10.1038/sj.onc.1208810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin DC, Zhang Y, Pan QJ, Yang H, Shi ZZ, et al. PLK1 Is transcriptionally activated by NF-kappaB during cell detachment and enhances anoikis resistance through inhibiting beta-catenin degradation in esophageal squamous cell carcinoma. Clin Cancer Res. 17:4285–4295. doi: 10.1158/1078-0432.CCR-10-3236. [DOI] [PubMed] [Google Scholar]
- 14.Howe EN, Cochrane DR, Cittelly DM, Richer JK. miR-200c Targets a NF-kappaB Up-Regulated TrkB/NTF3 Autocrine Signaling Loop to Enhance Anoikis Sensitivity in Triple Negative Breast Cancer. PLoS One. 7:e49987. doi: 10.1371/journal.pone.0049987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calao M, Burny A, Quivy V, Dekoninck A, Van Lint C. A pervasive role of histone acetyltransferases and deacetylases in an NF-kappaB-signaling code. Trends Biochem Sci. 2008;33:339–349. doi: 10.1016/j.tibs.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 17.Jeong SJ, Pise-Masison CA, Radonovich MF, Park HU, Brady JN. A novel NF-kappaB pathway involving IKKbeta and p65/RelA Ser-536 phosphorylation results in p53 Inhibition in the absence of NF-kappaB transcriptional activity. J Biol Chem. 2005;280:10326–10332. doi: 10.1074/jbc.M412643200. [DOI] [PubMed] [Google Scholar]
- 18.Sakurai H, Chiba H, Miyoshi H, Sugita T, Toriumi W. IkappaB kinases phosphorylate NF-kappaB p65 subunit on serine 536 in the transactivation domain. J Biol Chem. 1999;274:30353–30356. doi: 10.1074/jbc.274.43.30353. [DOI] [PubMed] [Google Scholar]
- 19.Yang F, Tang E, Guan K, Wang CY. IKK beta plays an essential role in the phosphorylation of RelA/p65 on serine 536 induced by lipopolysaccharide. J Immunol. 2003;170:5630–5635. doi: 10.4049/jimmunol.170.11.5630. [DOI] [PubMed] [Google Scholar]
- 20.Adli M, Merkhofer E, Cogswell P, Baldwin AS. IKKalpha and IKKbeta each function to regulate NF-kappaB activation in the TNF-induced/canonical pathway. PLoS One. 5:e9428. doi: 10.1371/journal.pone.0009428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zannini L, Buscemi G, Kim JE, Fontanella E, Delia D. DBC1 phosphorylation by ATM/ATR inhibits SIRT1 deacetylase in response to DNA damage. J Mol Cell Biol. 4:294–303. doi: 10.1093/jmcb/mjs035. [DOI] [PubMed] [Google Scholar]
- 22.Hiraike H, Wada-Hiraike O, Nakagawa S, Saji S, Maeda D, et al. Expression of DBC1 is associated with nuclear grade and HER2 expression in breast cancer. Exp Ther Med. 2011;2:1105–1109. doi: 10.3892/etm.2011.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H, Kim KR, Noh SJ, Park HS, Kwon KS, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum Pathol. 42:204–213. doi: 10.1016/j.humpath.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Kim SH, Kim JH, Yu EJ, Lee KW, Park CK. The overexpression of DBC1 in esophageal squamous cell carcinoma correlates with poor prognosis. Histol Histopathol. 2012;27:49–58. doi: 10.14670/HH-27.49. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Gu Y, Sha S, Kong X, Zhu H, et al. DBC1 is over-expressed and associated with poor prognosis in colorectal cancer. Int J Clin Oncol. doi: 10.1007/s10147-012-0506-5. [DOI] [PubMed] [Google Scholar]
- 26.Kim JE, Chen J, Lou Z. p30 DBC is a potential regulator of tumorigenesis. Cell Cycle. 2009;8 doi: 10.4161/cc.8.18.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 120:545–558. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shehzad A, Khan S, Sup Lee Y. Curcumin molecular targets in obesity and obesity-related cancers. Future Oncol. 8:179–190. doi: 10.2217/fon.11.145. [DOI] [PubMed] [Google Scholar]
- 29.Mori N, Yamada Y, Ikeda S, Yamasaki Y, Tsukasaki K, et al. Bay 11-7082 inhibits transcription factor NF-kappaB and induces apoptosis of HTLV-I-infected T-cell lines and primary adult T-cell leukemia cells. Blood. 2002;100:1828–1834. doi: 10.1182/blood-2002-01-0151. [DOI] [PubMed] [Google Scholar]
- 30.Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 31.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 34.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto K, Hikiba Y, Nakagawa H, Hirata Y, Hayakawa Y, et al. Promotion of DNA repair by nuclear IKKbeta phosphorylation of ATM in response to genotoxic stimuli. Oncogene. 2012 doi: 10.1038/onc.2012.192. [DOI] [PubMed] [Google Scholar]
- 36.Alfano D, Iaccarino I, Stoppelli MP. Urokinase signaling through its receptor protects against anoikis by increasing BCL-xL expression levels. J Biol Chem. 2006;281:17758–17767. doi: 10.1074/jbc.M601812200. [DOI] [PubMed] [Google Scholar]
- 37.Mawji IA, Simpson CD, Gronda M, Williams MA, Hurren R, et al. A chemical screen identifies anisomycin as an anoikis sensitizer that functions by decreasing FLIP protein synthesis. Cancer Res. 2007;67:8307–8315. doi: 10.1158/0008-5472.CAN-07-1687. [DOI] [PubMed] [Google Scholar]
- 38.Adli M, Baldwin AS. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J Biol Chem. 2006;281:26976–26984. doi: 10.1074/jbc.M603133200. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchiya Y, Asano T, Nakayama K, Kato T, Jr., Karin M, et al. Nuclear IKKbeta is an adaptor protein for IkappaBalpha ubiquitination and degradation in UV-induced NF-kappaB activation. Mol Cell. 39:570–582. doi: 10.1016/j.molcel.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 40.Lewander A, Gao J, Carstensen J, Arbman G, Zhang H, et al. NF-kappaB p65 phosphorylated at serine-536 is an independent prognostic factor in Swedish colorectal cancer patients. Int J Colorectal Dis. 27:447–452. doi: 10.1007/s00384-011-1356-8. [DOI] [PubMed] [Google Scholar]
- 41.Bae HJ, Chang YG, Noh JH, Kim JK, Eun JW, et al. DBC1 does not function as a negative regulator of SIRT1 in liver cancer. Oncol Lett. 4:873–877. doi: 10.3892/ol.2012.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu EJ, Kim SH, Heo K, Ou CY, Stallcup MR, et al. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor alpha activity and co-activator synergy. Nucleic Acids Res. 39:6932–6943. doi: 10.1093/nar/gkr347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Chen L, Kabra N, Wang C, Fang J, et al. Inhibition of SUV39H1 methyltransferase activity by DBC1. J Biol Chem. 2009;284:10361–10366. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chini CC, Escande C, Nin V, Chini EN. HDAC3 is negatively regulated by the nuclear protein DBC1. J Biol Chem. 2010;285:40830–40837. doi: 10.1074/jbc.M110.153270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiraike H, Wada-Hiraike O, Nakagawa S, Koyama S, Miyamoto Y, et al. Identification of DBC1 as a transcriptional repressor for BRCA1. Br J Cancer. 2010;102:1061–1067. doi: 10.1038/sj.bjc.6605577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trauernicht AM, Kim SJ, Kim NH, Boyer TG. Modulation of estrogen receptor alpha protein level and survival function by DBC-1. Mol Endocrinol. 2007;21:1526–1536. doi: 10.1210/me.2007-0064. [DOI] [PubMed] [Google Scholar]
- 47.Fu J, Jiang J, Li J, Wang S, Shi G, et al. Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J Biol Chem. 2009;284:6832–6840. doi: 10.1074/jbc.M808988200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Close P, East P, Dirac-Svejstrup AB, Hartmann H, Heron M, et al. DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature. 484:386–389. doi: 10.1038/nature10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nin V, Escande C, Chini CC, Giri S, Camacho-Pereira J, et al. Role of deleted in breast cancer 1 (DBC1) protein in SIRT1 deacetylase activation induced by protein kinase A and AMP-activated protein kinase. J Biol Chem. 287:23489–23501. doi: 10.1074/jbc.M112.365874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.