Abstract
In the holometabolous insect Drosophila melanogaster, genetic, physiological and anatomical aspects of olfaction are well known in the adult stage, while larval stages olfactory behavior has received some attention it has been less studied than its adult counterpart. Most of these studies focus on olfactory receptors (Or) genes that produce peripheral odor recognition. In this paper, through a loss-of-function screen using P-element inserted lines and also by means of expression analyses of larval olfaction candidate genes, we extended the uncovering of the genetic underpinnings of D. melanogaster larval olfactory behavior by demonstrating that larval olfactory behavior is, in addition to Or genes, orchestrated by numerous genes with diverse functions. Also, our results points out that the genetic architecture of olfactory behavior in D. melanogaster presents a dynamic and changing organization across environments and ontogeny.
Keywords: Drosophila melanogaster, Olfactory Behavior, Development, Genetic Architecture
Introduction
Among environmental cues that relate to differential survival and reproductive success of organisms, chemical signals appear very important (Asahina et al. 2008). The olfactory system obtains essential information on chemical cues through two major classes of stimuli: small molecules derived from food sources or the environment, and pheromones. Considerable insights into the mechanisms by which animals discriminate odors has emerged from a broad range of anatomical, physiological, biochemical and, especially, molecular studies (Dangles et al. 2009; Hallem and Carlson 2006; Matsunami and Amrein 2003; Su et al. 2009; Vosshall and Stocker 2007). In the context of studies of olfaction, Drosophila melanogaster (Diptera: Drosophilidae) has proven to be an attractive model organism because its olfactory system is relatively simple (Hallem et al. 2006; Vosshall and Stocker 2007) and olfactory behavior can be quantified by high throughput behavioral assays (Anholt and Mackay 2004; Lavagnino et al. 2008).
With respect to olfactory behavior in the larvae of D. melanogaster, the demonstration by Aceves-Piña and Quinn in 1979 that D. melanogaster larvae can perceive and discriminate different chemical stimuli (Aceves-Piña and Quinn 1979) motivated an increased interest in understanding larval olfactory behavior. Subsequent research has extended our knowledge about physiological and genetic aspects of larval olfaction using a variety of chemical stimuli (Ayyub et al. 1990; Boyle and Cobb 2005; Cobb 1996; Cobb et al. 1992; Cobb and Dannet 1994; Cobb and Domain 2000; Fishilevich et al. 2005; Ganguly et al. 2003; Kreher et al. 2005; Kreher et al. 2008; Parsons 1980). It has been proven that a subset of members of the olfactory receptors (Or) gene family are expressed in larva and participate in larval olfactory response to a large number of these stimuli (Fishilevich et al. 2005; Kreher et al. 2005; Kreher et al. 2008). Also, considerable progress has been achieved in understanding the functional organization of larval olfactory system, where events begins with stimuli interacting with olfactory receptors expressed in olfactory receptor neurons in the dorsal organ at larva anterior end. Each olfactory receptor neurons projects its axon and connect to a single glomerulus in the larval antennal lobe where projections neurons extend the olfactory signal to glomeruly in the mushroom body calyx in higher brain centres, at this point odor representation is established and translated into behavioral output (Fishilevich et al. 2005; Gerber and Stocker 2007; Kreher et al. 2005; Kreher et al. 2008; Masuda-Nakagawa et al. 2009; Vosshall and Stocker 2007).
As a holometabolous insect D. melanogaster adults and larvae stages have anatomically, physiological and behaviorally dissimilar characteristics across ontogeny. For example, in nature larval stages crawl on or inside rotten fruits in a limited space whereas adult flies move larger distances to locate food, oviposition sites and mating partners. However, the basic organization of the larval olfactory circuit is surprisingly similar to its adult counterpart but is numerically much simpler (Fishilevich et al. 2005; Gerber and Stocker 2007; Kreher et al. 2005; Kreher et al. 2008; Masuda-Nakagawa et al. 2009; Python and Stocker 2002; Vosshall and Stocker 2007). In these sense, most studies in both larva and adult stages have dealt with genes that mediate odor recognition in the periphery of the olfactory system, with focus on Or genes (Fishilevich et al. 2005; Kreher et al. 2005; Kreher et al. 2008) and odorant-binding protein (Obp) genes (Galindo and Smith 2001; Zhou et al. 2009). However, several studies on adult flies have identified others genes than Or or Obp genes to be implied in olfaction, e.g., members of the ionotropic receptors (Ir) gene family (Benton et al. 2009; Croset et al. 2010), acj6 (Ayer and Carlson 1991), scribbled (Ganguly et al. 2003), paralytic (original described as mutants named smellblind and olfD) (Aceves-Piña and Quinn 1979; Lilly and Carlson 1990; Lilly et al. 1994), CG33713 / CG33714 (Ryuda et al. 2008), a gene in the cytological region 96A2-7 uncovered by the mutant indifferent (Cobb 1996; Cobb et al. 1992; Cobb and Dannet 1994) and Vanaso which is an allele of discs large gene (Fanara et al. 2002). Thus, we can consider three possible scenarios with respect to the genetic architecture of olfaction in larval and adult: i) genes that only participate in adult olfaction, like for (Shaver et al. 1998) and Or22a (Vosshall and Stocker 2007); ii) genes only expressed in larvae, for example Or45a (Vosshall and Stocker 2007), Obp99b and Obp58c (Zhou et al. 2009); iii) genes that are involved in olfaction at both larval and adult stages, such as scribbled (Ganguly et al. 2003) and Or67b (Vosshall and Stocker 2007).
Previous studies on larval olfactory behavior were carried out by means of induced mutations with large behavioral effects (Cobb et al. 1992). More recently a mutational approach to study the genetics of olfactory behavior targeting single genes in an isogenic background identified genes that contribute to adult olfactory behavior (Sambandan et al. 2006). Here, we report the identification of genes that participate in D. melanogaster larval olfactory behavior by screening a set of co-isogenic lines that contain a single gene mutation produced by a P[GT1] gene-trap transposon insertion (Lukacsovich et al. 2001). We demonstrated that the structural components of the genetic architecture of larval olfactory behavior change between different chemical stimuli, i. e. genetic factors involved in the olfactory response to different stimuli are not the same. We also show that the genetic underpinnings that enable larval and adult olfactory behavior are distinct but present a partial overlap, i. e. different life stages share some genetic factors and others are stage-exclusive. Thus, the genetic architecture that underlies this trait undergoes rearrangement during metamorphosis from the larval to the adult stage.
Materials and Methods
Drosophila stocks and mutagenesis screening
Homozygous viable P[GT1] insertion lines constructed in a co-isogenic Canton-S B genetic background (Lukacsovich et al. 2001) were scored for larval olfactory behavioral responses. All lines were maintained by full-sib mating on cornmeal–molasses–agar medium under standard conditions of 25 ± 1° C, 70% humidity and a 12-h light: 12-h dark cycle; lights were switched on at 08:00 hours and switched off at 20:00 hours.
We quantified larval olfactory responses in second instar larvae of each P[GT1] insertion line using the odorants benzaldehyde 1 % (v/v) (Merck Schuchardt OHG, Hohenbrunn, Germany), propionic acid (pure) (Mallinckrodt Chemical Works, New York - Los Angeles - St. Louis, U.S.A.) and nonanol (pure) (Merck Schuchardt OHG, Hohenbrunn, Germany). Larval olfactory response was quantified for 102 lines in response to propionic acid, 100 lines for benzaldehyde and 106 lines for nonanol. Of the total P-element insertion lines tested, 76 were quantified in response to the three chemical stimuli. The type of olfactory response (attractive or repulsive) of second instar D. melanogaster larvae depends on the stimulus. Propionic acid and benzaldehyde are reported to be attractive (Ayyub et al. 1990; Heimbeck et al. 1999; Oppliger et al. 2000), while a repulsive response is expected for nonanol (Boyle and Cobb 2005; Cobb et al. 1992; Cobb and Domain 2000). We employed the assay of Aceves-Piña and Quinn (1979), modified by Cobb et al. (1992) to quantify larval olfactory responses. Briefly, adult females were allowed to lay eggs for 8 h on Petri dishes filled with agar medium and yeast paste. Larvae were allowed to develop on these Petri dishes for 36 h, when they were washed from the yeast paste and the behavioral test was started. Between 10 and 30 larvae were placed at the centre of a 10-cm Petri dish filled with 10 ml of 2.5% agar. A 5 μl drop of odorant solution and a 5 μl drop of distilled water were placed on filter paper discs on opposite ends of the Petri dish. To prevent diffusion of odorants through the agar and to eliminate larval gustatory responses, the filter paper discs containing the odorant or water were placed on inverted lids cut off 1.5 ml microcentrifuge tubes. The number of individuals within a 30 mm radius from each filter disc and the larvae that remain between both 30 mm radii were counted five minutes after the introduction of the larvae. Olfactory responses tend to decline after 5 min, presumably as a result of saturation of the vapour phase (Kaiser and Cobb 2008; Rodrigues 1980). A larval response index (LRI) was calculated for each dish as:
where n designates the number of larvae and the subscripts indicate the sides of the Petri dish containing odorant, water (control) and the entire dish, respectively. This index varies between −100 (total repulsion) and +100 (total attraction). An LRI = 0 indicates indifferent behavior. Larvae respond to odorants the same when in groups as when tested individually; thus, there is no alteration of LRI due to the presence of the other individuals (Kaiser and Cobb 2008; Monte et al. 1989). Replicate measurements (5–7) were made for each line tested, distributed in different batches in which 10 to 15 lines were simultaneously assessed. In order to account for environmental variation in larval olfactory behavior between batches, 5-7 replicates of the co-isogenic transposon-free Canton-S B control line were run in parallel with each batch. Whenever P[GT1] insertion lines from different batches were included in the same statistical analysis, mean LRI of each line was corrected by subtracting mean LRI of the contemporaneous co-isogenic control. All behavioral tests were performed between 14:00 and 16:00 hours under controlled temperature (25 ± 1 °C), light (5.4 ± 0.2 × 10−5 lx) and humidity (42 ± 5%).
The magnitude of mutational variation in larval olfactory behavior was assessed by means of one-way ANOVA of replicate means expressed as deviation from the across replicate mean of the contemporaneous co-isogenic control, the model used was: Y = μ + L + ε, where μ is the overall mean, L is the random effect of the P[GT1] insertion lines and the ε is the error term. A significant L effect is interpreted as the existence of mutational genetic variation for larval olfactory behavior. Mutational broad sense heritability was computed as H2M = σ2L / (σ2L + σ2ε), where σ2L is the line effect variance component and an estimate of VG, and σ2ε is the error term variance component and an estimate of VE. To determine which lines present phenotypic differences with respect to the contemporaneous co-isogenic Canton-S B control line, a Dunnett’s test comparing mean LRI of each P[GT1] insertion line with mean LRI of the control line was performed. Lines that exhibited significant differences relative to the control were considered “smell-impaired” mutant lines (“smi lines”) (Anholt et al. 1996) and the genes disrupted by the transposon insertion as candidate larval olfactory genes. Smi lines could show an anosmic phenotype, when their LRI is different from control line and no different from 0 or a hyposmic phenotype, when their LRI is different from control line but also different from 0. To identify the transposon-tagged candidate genes, nucleotide sequences flanking the P-element insertion were aligned using BLAST with corresponding sequences of the FlyBase release FB2011_10 D. melanogaster genomic sequence (flybase.org).
Gene expression analyses
To provide further evidence for candidate genes that affect larval olfactory behavior, we quantified gene expression using two different experimental approaches. (i) 7 P[GT1] insertion lines affecting larval olfactory behavior (BG00737, BG01043, BG01324, BG01515, BG01683, BG02042, BG02081) were selected to quantify messenger RNA levels as previously described (Arya et al. 2010). Gene expression was quantified by means of Quantitative Real time PCR (qPCR) using the SYBR Green detection method (Maxima™ SYBR Green/Rox qPCR Master Mix (2X), Fermentas Life Sciences, Burlington, Ontario, Canada) and the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). These lines were selected based on aberrant behavior toward either one or multiple odorants; female sterile (1) homeotic (BG01515), easily shocked (BG02042) and Rtnl1 (BG02081) are candidate genes for larval olfactory behavior in response to propionic acid, while Gp150 (BG01043) and cricklet (BG01324) are candidate genes for larval olfactory behavior in response to nonanol. In the cases of Hsp27 (BG00737) and CG32572 (BG01683) larvae showed a hyposmic phenotype for both propionic acid and nonanol. From each line, we sampled 15 larvae from each developmental stage (first instar, second instar and third instar). Total RNA was isolated from three independent biological replicates per line and stage using the Trizol reagent (GIBCO-BRL, Gaithersburg, MD). Subsequently, cDNA was generated from 60 ng of total RNA by reverse-transcription PCR using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Primer3Plus (Untergasser et al. 2007) was used to design real-time PCR primers. For all samples, the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as an internal control. We used a mixed linear model, Y = m + L + e, where Y denotes average normalized CT value, L line (fixed), and e the error variance, to determine whether there was a significant difference between the two genotypic classes. (ii) 20 P[GT1] insertion lines affecting larval olfactory behavior were selected, using the same criteria as in messenger RNA quantification, to evaluate spatial expression using the GAL4/UAS-GFP system, where expression of a GAL4 cassette in the P[GT1] construct is driven by the endogenous promoter of the tagged gene (Lukacsovich et al. 2001). The lines were crossed with a W / +; UAS-CD8::GFP / CyO; TM3 stock and F1 larvae of these crosses were observed and photographed in vivo at approximately 36 to 42 h after hatching using an Olympus MVX10 microscope (MV PLAPO 2X lens) with a 488/30 filter coupled with a Olympus DP71 camera. F1 larvae from a cross between an isogenic transposon-free Canton-S B control line and the UAS-CD8::GFP line were used as a negative control contemporaneously to experimental genotypes crosses. All negative controls worked correctly (not shown). F1 larvae from a cross between engrailed-GAL4 line and the UAS-CD8::GFP line were used as a positive control contemporaneously to experimental genotypes crosses. These larvae showed a clear pattern of expression in the tracheal system (not shown).
All statistical analyses were performed using the STATISTICA software package (StatSoft 2001).
Results
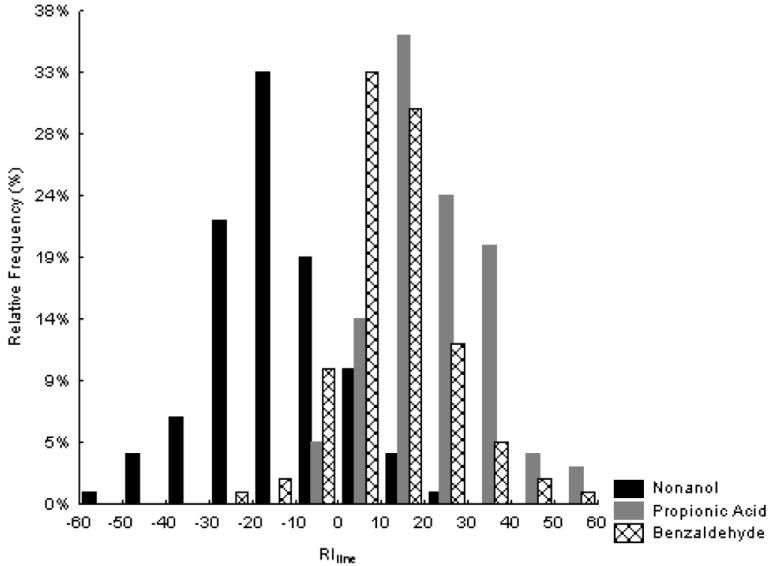
We measured larval olfactory behavioral responses of 102, 100 and 106 P-element insertion lines in response to propionic acid, benzaldehyde and nonanol, respectively. Also, in 76 of these lines the phenotypic characterization of larva olfaction was done using the three chemical stimuli. P-element insertions in the lines tested for each different chemical stimulus are evenly distributed across chromosomes. For propionic acid 36.8 % of the insertions were on chromosome II, 41.1 % on chromosome III and 22.1 % on the X chromosome. For benzaldehyde the proportions were 37.5 %, 41.5 % and 21 %, respectively. Finally, when nonanol was used as stimulus proportions were 8 %, 42 % and 20 %, respectively. The co-isogenic Canton-S B control line exhibited an attractant response to propionic acid (LRI: 27.92) and benzaldehyde (LRI: 13.12) but a repulsive response to nonanol (LRI:−24.67) as expected from previous reports (Ayyub et al. 1990; Boyle and Cobb 2005; Cobb et al. 1992; Cobb and Domain 2000; Heimbeck et al. 1999; Oppliger et al. 2000). The effect of P-element insertions on larval olfactory response to propionic acid, benzaldehyde and nonanol showed substantial phenotypic variation among lines (Figure 1) and mutational variance was highly significant for each stimulus (Table I). The broad-sense mutational heritability for larval olfactory behavior was the largest for propionic acid (H2M = 0.324), followed by nonanol (H2M = 0.246) and benzaldehyde (H2M = 0.171) (Table I).
Figure 1.
Frequency distribution of mean LRI among P[GT1] insertion lines. Black bars represent LRI scores of lines tested in response to nonanol, gray bars in response to propionic acid and striped bars in response to benzaldehyde.
Table I.
Parameters of the Line factor for the ANOVA of avoidance scores of P[GT1] insertion lines for each chemical used as stimulus.
| Stimulus | d. f. | F | P | H 2 M |
|---|---|---|---|---|
| Propionic Acid | 101 | 3.97 | < 1×10−6 | 0.324 |
| Benzaldehyde | 99 | 2.265 | < 1×10−6 | 0.171 |
| Nonanol | 105 | 3.032 | < 1×10−6 | 0.246 |
d.f.: degrees of freedom. H2M = σ2L / (σ2L + σ2ε), mutational broad sense heritability.
Identification of smell impaired co-isogenic P[GT1] insertion lines for larval olfactory behavior
Comparisons between P[GT1] insertion lines and the co-isogenic Canton-S B control line allowed us to identify lines that showed a significant reduction in larval olfactory responses. Of the total 308 lines analysed 17 showed a significant reduction in olfactory response to propionic acid, 3 to benzaldehyde and 8 to nonanol (Table II). These lines are considered smell impaired (smi) lines and the genes disrupted by the insertion of the transposon are candidate genes that contribute to larval olfactory behavior. All smi lines presented a hyposmic phenotype, since their LRI is different not only from the control line but also different from 0. The proportion of smi lines represents 16.7%, 3% and 7.5% of the total lines screened when the odorant used was propionic acid, benzaldehyde and nonanol, respectively. The proportion of smi lines detected in our screen using benzaldehyde as odorant was similar to the ~ 4% and 6% reported in previous mutant screens for adult olfaction (Anholt et al. 1996; Sambandan et al. 2006). Candidate genes that contribute to larval olfactory behavior present a heterogeneous set of gene ontology categories including other behavioral and physiological functions as: defense response, response to heat, inter-male aggressive behavior, immune response, adult learning and memory, mechanosensory behavior, response to hypoxia and male mating behaviour; and developmental functions like: determination of lifespan, nervous system development, open tracheal system development, mesoderm development, mushroom body development (gene ontology categories of candidate genes were obtained from FlyBase). In previous studies several of the smi lines were analysed and considered as mutants for other traits like: starvation resistance (Harbison et al. 2004), developmental time (Mensch et al. 2008), startle-induced locomotion (Yamamoto et al. 2008), body size (Carreira et al. 2009). It is evident the pleiotropic nature of genes that contribute to larval olfactory behavior. Finally, 8 of the larval olfactory behavior candidate genes are reported as adult olfactory genes in previous studies (Table 2). Only 4 lines exhibited a smi phenotype for more than one odorant, while none presented a hyposmic phenotype for all three stimuli. Lines where the P-element is inserted in Heat shock protein 27 (BG00737), CG32572 (BG01683) and located 2.7 kb upstream from CG6175 (BG01733) presented a different olfactory phenotype with respect to the control line for propionic acid (Dunnett’s test; BG00737: p = 0.034; BG01683: p < 0.001; BG01733: p < 0.001) and nonanol (Dunnett’s test; BG00737: p = 0.021; BG01683: p = 0.0208; BG01733: p = 0.0107). Two insertions near jing exhibited significant differences with respect to the control for propionic acid (Dunnett’s test: p = 0.004) and benzaldehyde (Dunnett’s test: p = 0.044) for lines BG01257 and BG01897, respectively. The fact that different insertions in this gene affected larval olfactory behavior differently suggests that the precise location of the P[GT1] insertion determines the phenotypic effect, as observed previously for others traits (Carreira et al. 2009; Rollmann et al. 2006).
Table II.
Smell impaired P[GT1] insertion lines.
|
P-element line |
Disrupted locus | P[GT1] insertion site | Mutational effect (/ΔLRI/) |
Smell impaired for adult OB? | |
|---|---|---|---|---|---|
| Propionic acid |
|||||
| BG00737 | Hsp27 | In 5′-UTR and exon 1 | 24.44* | NO | |
| BG01011 | Spinophilin (Spn) /misshapen (msn) | 55 bp upstream 5′ region /1443 bp downstream 3′ region |
16** | YES (Sambandan et al. 2006) | |
| BG01179 | defense repressor 1 (dnr1) | In intron | 39.69*** | NO | |
| BG01223 | Glutamine synthetase 2 (Gs2) | In intron | 6.5* | NO | |
| BG01228 | derailed (drl) | 22,8 kbp downstream 3′ region | 12.24* | YES (Moreau-Fauvarque et al. 2002) | |
| BG01257 | jing | 290 bp downstream 5′ region | 28.54** | YES (Sambandan et al. 2006) | |
| BG01330 | CG11883 | In intron | 11 4*** | NO | |
| BG01376 | Chd64 | In 5′-UTR | 9.4** | NO | |
| BG01380 | Oseg4 / draper (drpr) | In exon 1 / In 5′-UTR | 9.42** | NO / YES (MacDonald et al. 2006) | |
| BG01399 | CG17646 | In intron | 7.5* | NO | |
| BG01404 | bicoid-interacting protein 3 (bin3) | In intron | 31.1** | YES (Sambandan et al. 2006) | |
| BG01515 | female sterile (1) homeotic (fs(1)h) | In exons 19 and 20, also in introns | 28.97** | NO | |
| BG01683 | CG32572 | In intron | 39.2*** | NO | |
| BG01733 | CG6175 | 2725 bp upstream 5′ region | 38.7*** | NO | |
| BG02042 | easily shocked (eas) | In exon 2 | 32.5* | NO | |
| BG02081 | Rtnl1 | In 5′-UTR, exon 10 and intron | 27.3* | YES (Sambandan et al. 2006) | |
| BG02823 | scylla (scyl) | 130 bp downstream 5′ region | 35.7* | NO | |
|
| |||||
| Benzaldehyde | |||||
| BG01735 | CG13130 / big brain (bib) | In exon 2 / In 5′-UTR and exon 1 | 20.25* | NO / NO | |
| BG01897 | jing | 143 bp upstream 5′ region | 21* | YES (Sambandan et al. 2006) | |
| BG02102 | Quasimodo † | 2,15 kbp upstream 5′ region | 20.8* | NO | |
|
| |||||
| Nonanol | |||||
| BG00737 | Hsp27 | In 5′-UTR and exon 1 | 29.3* | NO | |
| BG01043 | Gp150 | In 5′-UTR and exons 1, 7 and 8 | 20.9* | NO | |
| BG01173 | CG5361 | 11,8 kbp upstream 5′ region | 32** | NO | |
| BG01315 | couch potato (cpo) | In intron | 23.8* | YES (Sambandan et al. 2006) | |
| BG01324 | cricklet (clt) | In 5′-UTR and exon 4 | 33.2** | NO | |
| BG01600 |
Chronologically inappropriate
morphogenesis (chinmo) |
1244 bp upstream 5′ region | 38.6* | NO | |
| BG01683 | CG32572 | In intron | 25.4* | NO | |
| BG01733 | CG6175 | 2725 bp upstream 5′ region | 31.9* | NO | |
Lines and/or genes that showed a smell impaired mutant phenotype in response to more than one odorant are in bold. Candidate genes used for further characterization of expression patterns are underlined. Mutational effects were calculated as the absolute values of the difference between the LRI of the smell impaired P[GT1] line versus the isogenic control line. Significant deviations from the control line using a Dunnett’s test are indicated by asterisks.
: p < 0.05
: p < 0.01
: p < 0.001.
Information on P[GT1] insertion site and gene name was obtained from FlyBase (flybase.org). Genes categorization as “smell impaired for adult OB?” was based on the previous works cited in each case.
also known as l(2)05510.
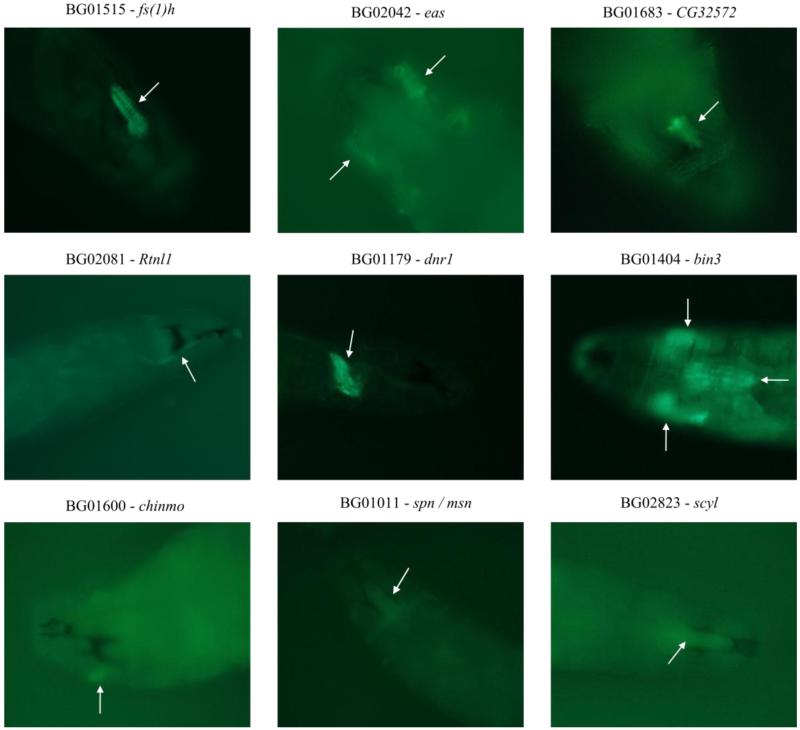
Since the P[GT1] construct can enable endogenous promoters to drive expression of GAL4, we could utilize the GAL4/UAS-GFP system (Lukacsovich et al. 2001) to localize expression of the candidate gene in 9 smi lines (BG01011, BG01179, BG01404, BG01515, BG01600, BG01683, BG02042, BG02081, BG02823; Figure 2). Results show that female sterile (1) homeotic (fs(1)h), defense repressor 1 (dnr1), bicoid-interacting protein 3 (bin3), spinophilin (spn) / misshapen (msn) and scylla (scyl) are expressed in larval brain hemispheres and/or in the cone-shaped ventral nerve cord. CG32572 is expressed in a region near or in a ganglion located below larval sense organs and Rtnl1 is expressed in what appears to be a set of neurons or interneurons that project to and from the anterior part of the larvae. Finally, candidate genes easily shocked (eas), scyl and chronologically inappropriate morphogenesis (chinmo) are expressed in the tracheal system (Figure 2). We did not detect GFP expression in the remaining lines, presumably because the location of the transposon insertion prevented GAL4 expression from being driven by the promoter or the insertion disrupted the promoter itself. Lines BG01173, BG01228, BG01257, BG01315 and BG01897 could not be analyzed.
Figure 2.
Expression of candidate genes affecting larval olfactory behavior in second instar larvae. Images of GFP expression of whole F1 larvae from a cross between P-element insertion lines and UAS-CD8::GFP line are shown. fs(1)h: ventral view of the head, expression in one of the brain hemispheres and in the cone-shaped ventral nerve cord (indicated by the white arrow). eas: ventral view of the head, expression in the anterior segment of the tracheal system (white arrows). CG32572: dorsal view of the head, expression in the region corresponding to ganglia located below the larval sense organs (arrow). Rtnl1: dorsal view of the head, expression in a longitudinal nerve (white arrow) innervating peripheral sensory organs. dnr1: lateral view, expression in the cone-shaped ventral nerve cord (white arrow). bin3: dorsal view of the head, expression in brain hemispheres and in the cone-shaped ventral nerve cord (horizontal white arrow) and the anterior segment of the tracheal system (vertical white arrows). chinmo: dorsal view of the head, expression in the anterior segment of the tracheal system (white arrow). spn / msn: ventral view of the head, expression in the brain hemispheres and the cone-shaped ventral nerve cord (white arrow). scyl: dorsal view of the head, expression in the brain hemispheres and the cone-shaped ventral nerve cord (white arrow). Contemporaneously to experimental genotypes, positive and negative controls were run; and all controls worked correctly (not shown).
Effects of P[GT1] insertions on transcript levels of candidate genes implicated in larval olfactory behavior
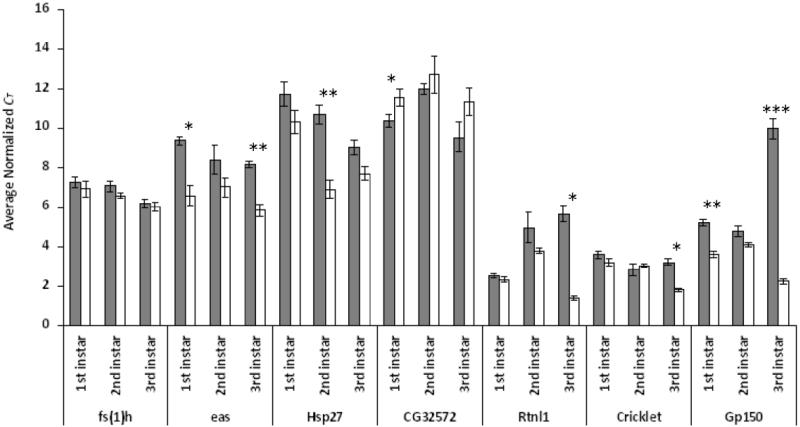
We also quantified messenger RNA levels in first, second and third instar larvae of seven P[GT1] insertion lines. These mutants showed aberrant olfactory responses either to one odorant (fs(1)h (BG01515), eas (BG02042), Rtnl1 (BG02081), clt (BG01324) and Gp150 (BG01043)), or to more than one odorant (Hsp27 (BG00737) and CG32572 (BG01683)). All of these smi lines, except BG01515 (fs(1)h), showed significant alterations in transcript abundance in one or more larval stages, confirming that the P-element insertion affected the expression of the tagged genes (Figure 3). In four smi lines the transposon tagged genes (Hsp27, Rtnl1, clt, Gp150) were associated with increased transcript levels. In these lines the P-element is inserted in the 5′ UTR. In eas, increased transcript level is associated with transposon insertion in an exon, a pattern that could be caused by altered expression of splice variants. Interruption of an intron in CG32572 is associated with a decrease in the level of transcript in first instar larvae. Most lines showed altered transcript abundance in only one larval stage, with the exception of eas and Gp150 where expression was augmented in two larval stages, the first and third instars (Figure 3).
Figure 3.
Expression of candidate gene transcripts for D. melanogaster larval olfactory behavior. Levels of mRNA for candidate genes in P-element insertion lines (grey bars) and control Canton S-B lines (white bars) are shown. mRNA levels were measured in first, second and third instar larvae. CT values were normalized to an internal control (Gapdh gene). Standard errors were obtained from the normalized CT value of three independent biological replicates per line and stage. Significant differences in gene expression level are indicated by asterisks (ANOVA, *: p < 0.05, **: p < 0.01, ***: p < 0.001).
Discussion
A considerable amount of information has been accumulated about functional organization (Benton et al. 2009; Keene and Waddell 2007; Laissue and Vosshall 2008; Vosshall 2000; Vosshall and Stocker 2007) and genetic networks (Anholt et al. 2003; Sambandan et al. 2006; Zhou et al. 2009) underlying olfaction in D. melanogaster. However, most of these studies focused on the adult olfactory system while larval olfaction has received less attention, although there are several studies on the genetics of larval olfactory behavior (Boyle and Cobb 2005; Cobb et al. 1992; Cobb and Dannet 1994; Cobb and Domain 2000; Gerber and Stocker 2007; Lavagnino et al. 2008; Monte et al. 1989; Parsons 1980; Vosshall and Stocker 2007; Zhou et al. 2009). Adding to these previous investigations, our results reveal that larval olfactory behavior is a complex trait orchestrated by ensembles of pleiotropic genes. In fact, 9% of P-element mutated lines screened in our study exhibited a hyposmic larval phenotype. This percentage of mutant lines is lower than reported in similar screens on different traits using the same set of P[GT1] insertion lines: Norga et al. (2003) showed that about 20% of P[GT1] insertions have a significant effect on adult sensory bristle number, Yamamoto et al. (2008) found that 37 % of the P[GT1] insertion lines affected startle-induced locomotion, Carreira et al. (2009) and Mensch et al. (2008) reported that 60 % of mutants affected body size and developmental time, respectively; and, Harbison et al. (2004) observed that 40% of these P-element lines affected starvation resistance. However, the percentage of mutant lines detected in our screen is similar to the 4% and 6% reported in previous screenings for adult olfaction (Anholt et al. 1996; Sambandan et al. 2006).
Candidate genes identified in this study were not previously reported to be involved in D. melanogaster larval olfactory behavior (Table II). These candidate genes present a heterogeneous array of functions, none of them being an olfactory receptor. These results points that the genetic underpinnings of D. melanogaster larval olfactory behavior is, in addition to olfactory receptor genes, orchestrated by numerous genes with diverse functions. Receptors may play a major role in olfactory response by sensing odors in the periphery of olfactory system, but other genes may also play non trivial roles not only in making olfactory response possible but also in defining the characteristic of the behavioral response. Three of the lines that showed a hyposmic larval phenotype deserves a more detailed discussion: (i) in BG01011 the P[GT1]-element is inserted 55 bp upstream of the Spinophilin gene and 1443 bp downstream of the misshapen gene. Even though the insertion is located closer to Spinophilin than misshapen, an effect on misshapen cannot be ruled out because the distance to the insertion point is within the reported range of action of the P[GT1] transposable element (Bellen et al. 2004), (ii) more complex is the transposon insertion in BG01380 since it simultaneously affects two genes, Oseg4 and draper (drpr), as it is located in a ~ 100 bp region that overlaps between these two genes; and (iii) line BG01735 also showed the same effect since the insertion affects the two closely located genes CG13130 and big brain (bib). Gene families that are clustered in the genome are notably refractory to P-element insertion; therefore, we did not identify transposons in or near Or, Obp and Ir genes (Sambandan et al. 2006). Also, given that the total number of lines screened does not cover the whole genome and that chemoreceptor genes make up about only 2% of the genome, this lack of transposon hit to Or, Obp or Ir genes could be due to chance.
We performed two experimental approaches to provide additional support for the involvement of the candidate genes in olfactory behavior. First, we analysed spatial patterns of expression to determine whether candidate genes for larval olfactory behavior are expressed in olfactory organs or the central nervous system. Among the candidate genes that showed a clear expression signal (fs(1)h, dnr1, bin3, spn / msn and scyl) most are expressed in larval brain hemispheres and/or in the cone-shaped ventral nerve cord (Figure 2). The fact that these candidate genes are expressed in the larval central nervous system is consistent with a role in olfactory information processing. CG32572 is expressed in ganglia located below the larval sense organs, whereas Rtnl1 is expressed in a region consistent with neurons or interneurons that project to and from the anterior part of the larvae, where the first contact with odor molecules takes place in the dorsal organ (Figure 2). In cases where gene expression was observed in the tracheal system (eas, scyll and chinmo) (Figure 2) it is difficult to explain the relationship with larval olfactory behavior; anyhow, is not entirely surprising that candidate genes for larval olfaction are expressed in tracheal system since these genes have been proved to be involved in other traits, confirming its pleiotropic nature. Thus, these genes may play a role in organization and function of larval nervous system related to olfaction and also in the development of tracheal system. Second, we quantified levels of messenger RNA in candidate genes to establish whether the P-element insertion affected the expression of these genes at different stages of D. melanogaster ontogeny. Six of the seven smi lines tested, with the exception of BG01515 (fs(1)h), were associated with alterations in transcript abundance in one or more larval stages (Figure 3), confirming that P-element insertion affect the expression of the tagged genes. In four of the seven smi lines whose tagged genes (Hsp27, Rtnl1, clt, Gp150) were associated with increased transcript levels the P-element insertion affects 5′ UTR region, the only exception being eas where insertion affects an exon. The fact that the P-element was inserted where generally regulatory sequences are located within the genome could explain these results. It is known that gene expression regulatory sequences and trans-acting factors binding to them present a heterogeneous array of types (transcription factors, micro RNAs, small RNAs, long non coding RNAs, etc.) but functionally all can be grouped in two categories: enhancers and silencers or suppressors. Whereas transcription factors can regulate transcription positively or negatively; micro RNAs appear to regulate gene expression mostly, while not always, through repression (Hobert 2008). If so, the possibility that gene expression regulatory sequences located in 5′ UTR region are involved in down regulation of gene expression becomes very plausible. Hence, when these regions are disrupted in smi lines, gene expression increases with respect to wild-type. In this sense, it is worth saying that in most studies that use P-element inserted lines there is always a proportion of mutants with RNA over-expression (Edwards et al. 2009; Harbison et al. 2005; Sambandan et al. 2006). Also, a notable result is that the effects of single P-element insertions on gene expression depend on developmental stage (Figure 3). This phenomenon was reported previously in investigations carried out with the same lines and messenger RNA levels quantification technique (Edwards et al. 2009; Rollmann et al. 2007; Sambandan et al. 2006). As stated in those papers, differential disruption by the transposon of distinct promoter elements that are active at different developmental stages is the cause of the observed results. The biological interpretation of these results in the case of olfaction is that in cases were expression is variable in previous stages than second instar larvae (like in eas, CG32572 and Gp150) the adverse effects on olfactory behavior are likely the consequence of early disruptions of gene function that has consequences in posterior manifestations of the trait. In particular for D. melanogaster, disruption of early developmental genes have proved to contribute to olfaction in later stages (Rollmann et al. 2007; Sambandan et al. 2006), the same is true for another behavioral trait like aggressive behavior (Edwards et al. 2009). The different chemical stimuli used in the assays performed in our screen (benzaldehyde, propionic acid and nonanol) can be considered as different “chemical environments” and our results show that most candidate genes participate in the genetic architecture of larval olfactory behavior exclusively in one “chemical environment”, proving that structural components of genetic architecture change between different chemical stimuli, i. e. genetic factors involved in the olfactory response to different stimuli are not the same. This result showing a dynamic genetic architecture of larval olfactory behavior across “chemical environments” should be contextualized in an ecological natural scenario, where the genetic nature of this behavior make sense taking into consideration the spatial and temporal heterogeneity of D. melanogaster larvae feeding sites (Carson 1971). Thereby, this variation of structural components of larva olfaction genetic architecture, the substantial phenotypic and mutational variation found in this study (Table 1, Figure 1) and the large amount of variation found for larval olfaction in natural populations (Lavagnino et al. 2008; Lavagnino and Fanara 2011) are in agreement with a ecological context of highly variable substrates and climates for the development of behavioral patterns in D. melanogaster. Moreover, this kind of relation has been proposed for other larval behaviors, such as locomotion (Del Pino et al. 2012), dispersal and prepupation behavior (Medina-Muñoz and Godoy-Herrera 2005).
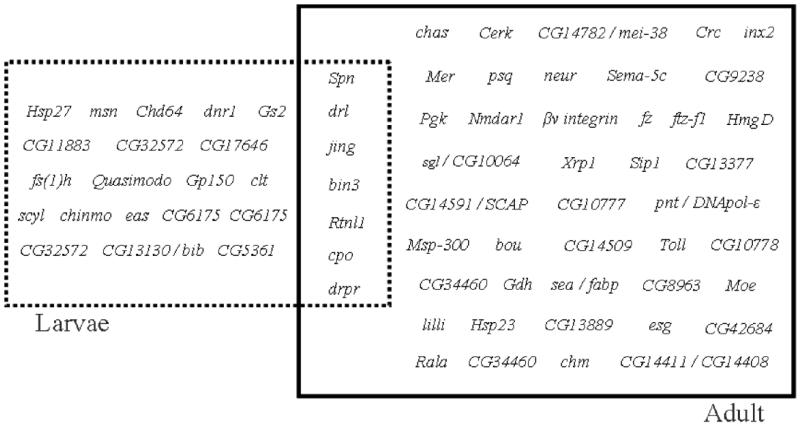
Also, D. melanogaster experiences different environments during ontogeny, especially between larvae stages and adult, where chemosensory inputs are likely to be specific for each stage of the life cycle (Zhou et al. 2009). Regarding the genetic bases of olfaction, Obp and Or genes expressed only in larvae olfaction, only in adult olfaction or at both stages have been identified (Gerber and Stocker 2007; Vosshall and Stocker 2007; Zhou et al. 2009) and, although it has not been confirmed, is quite plausible that Ir genes also present this pattern. Adding to these contributions, we did an analysis of the structural genetic components of olfactory behavior across development by comparing candidate genes for larval olfactory behavior (Table 2) with its adult counterpart identified in previous studies (MacDonald et al. 2006; Moreau-Fauvarque et al. 2002; Sambandan et al. 2006). Figure 4 shows that many candidate genes found to participate in olfaction for a determined life cycle are exclusive, but there is a considerable number of genes involved in both stages, a similar pattern from the one found in genes that orchestrate the first contact with odors (Obp and Or genes) (Gerber and Stocker 2007; Vosshall and Stocker 2007; Zhou et al. 2009). The consistency of the results between our study and the ones about Obp and Or genes demonstrate that the genetic architecture for olfactory behavior is distinct with partial overlap for larval and adult stages.
Figure 4.
Comparisons between genes implicated in olfactory behavior of larvae and adult stages of D. melanogaster life cycle. Candidate genes implicated in larval olfactory behavior are bordered by the dashed line box while genes implicated in adult olfactory behavior according to Sambandan et al. (2009) are shown in the solid line box. The intersection of both boxes shows candidate genes implicated in both larval and adult olfactory behavior, these genes were identified in our study and by Sambandan et al. (2009), Moreau-Fauvarque et al. (2002) and MacDonald et al. (2006). Only genes tested for both larval and adult olfactory behavior are included in the diagram.
Acknowledgements
The authors wish to thank T.F.C. Mackay (North Carolina State University, Raleigh, N.C., U.S.A.) for providing P[GT1] insertion lines and R. R. H. Anholt (North Carolina State University, Raleigh, N.C., U.S.A.) for comments on the manuscript. We also are grateful to F. Ceriani and G. Bernabó (Fundación Instituto Leloir, Buenos Aires, Argentina) for assistance with the GAL4/UAS-GFP system experiments. This work was supported by grants from Universidad de Buenos Aires, CONICET and NIH-Fogarty grant TW 007070. NJL is a recipient of a postdoctoral scholarship and JJF is a member of Carrera del Investigador Científico of CONICET (Argentina).
Bibliography
- Aceves-Piña EO, Quinn WG. Learning in normal and mutant Drosophila larvae. Science. 1979;206(4414):93–96. doi: 10.1126/science.206.4414.93. [DOI] [PubMed] [Google Scholar]
- Anholt RR, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, Ganguly I, Rollmann SM, Kamdar KP, Mackay TF. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 2003;35(2):180–184. doi: 10.1038/ng1240. [DOI] [PubMed] [Google Scholar]
- Anholt RR, Lyman RF, Mackay TF. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster. Genetics. 1996;143(1):293–301. doi: 10.1093/genetics/143.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt RR, Mackay TF. Quantitative genetic analyses of complex behaviours in Drosophila. Nat Rev Genet. 2004;5(11):838–849. doi: 10.1038/nrg1472. [DOI] [PubMed] [Google Scholar]
- Arya GH, Weber AL, Wang P, Magwire MM, Negron YL, Mackay TF, Anholt RR. Natural variation, functional pleiotropy and transcriptional contexts of odorant binding protein genes in Drosophila melanogaster. Genetics. 2010;186(4):1475–1485. doi: 10.1534/genetics.110.123166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahina K, Pavlenkovich V, Vosshall LB. The survival advantage of olfaction in a competitive environment. Curr Biol. 2008;18(15):1153–1155. doi: 10.1016/j.cub.2008.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer RK, Jr., Carlson J. acj6: a gene affecting olfactory physiology and behavior in Drosophila. Proc Natl Acad Sci U S A. 1991;88(12):5467–5471. doi: 10.1073/pnas.88.12.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyub C, Paranjape J, Rodrigues V, Siddiqi O. Genetics of olfactory behavior in Drosophila melanogaster. J Neurogenet. 1990;6(4):243–262. doi: 10.3109/01677069009107114. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167(2):761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136(1):149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle J, Cobb M. Olfactory coding in Drosophila larvae investigated by cross-adaptation. J Exp Biol. 2005;208(18):3483–3491. doi: 10.1242/jeb.01810. [DOI] [PubMed] [Google Scholar]
- Carreira VP, Mensch J, Fanara JJ. Body size in Drosophila: genetic architecture, allometries and sexual dimorphism. Heredity. 2009;102(3):246–256. doi: 10.1038/hdy.2008.117. [DOI] [PubMed] [Google Scholar]
- Carson HL. The ecology of Drosophila breeding sites. University of Hawaii Foundation Lyon Arboretum Fund; 1971. p. 32. [Google Scholar]
- Cobb M. Genotypic and phenotypic characterization of the Drosophila melanogaster olfactory mutation Indifferent. Genetics. 1996;144(4):1577–1587. doi: 10.1093/genetics/144.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb M, Bruneau S, Jallon JM. Genetic and developmental factors in the olfactory response of Drosophila melanogaster larvae to alcohols. Proc Biol Sci. 1992;248(1322):103–109. doi: 10.1098/rspb.1992.0048. [DOI] [PubMed] [Google Scholar]
- Cobb M, Dannet F. Multiple genetic control of acetate-induced olfactory responses in Drosophila melanogaster larvae. Heredity. 1994;73:444–455. doi: 10.1038/hdy.1994.192. [DOI] [PubMed] [Google Scholar]
- Cobb M, Domain I. Olfactory coding in a simple system: adaptation in Drosophila larvae. Proc Biol Sci. 2000;267(1457):2119–2125. doi: 10.1098/rspb.2000.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6(8):e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangles O, Irschick D, Chittka L, Casas J. Variability in sensory ecology: expanding the bridge between physiology and evolutionary biology. Q Rev Biol. 2009;84(1):51–74. doi: 10.1086/596463. [DOI] [PubMed] [Google Scholar]
- Del Pino F, Salgado E, Godoy-Herrera R. Plasticity and genotype x environment interactions for locomotion in Drosophila melanogaster larvae. Behav Genet. 2012;42(1):162–169. doi: 10.1007/s10519-011-9490-1. [DOI] [PubMed] [Google Scholar]
- Edwards AC, Zwarts L, Yamamoto A, Callaerts P, Mackay TF. Mutations in many genes affect aggressive behavior in Drosophila melanogaster. BMC Biol. 2009;7:29. doi: 10.1186/1741-7007-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanara JJ, Robinson KO, Rollmann SM, Anholt RR, Mackay TF. Vanaso is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics. 2002;162(3):1321–1328. doi: 10.1093/genetics/162.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich E, Domingos AI, Asahina K, Naef F, Vosshall LB, Louis M. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr Biol. 2005;15(23):2086–2096. doi: 10.1016/j.cub.2005.11.016. [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001;159(3):1059–1072. doi: 10.1093/genetics/159.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly I, Mackay TF, Anholt RR. Scribble is essential for olfactory behavior in Drosophila melanogaster. Genetics. 2003;164(4):1447–1457. doi: 10.1093/genetics/164.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber B, Stocker RF. The Drosophila larva as a model for studying chemosensation and chemosensory learning: a review. Chem Senses. 2007;32(1):65–89. doi: 10.1093/chemse/bjl030. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125(1):143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Dahanukar A, Carlson JR. Insect odor and taste receptors. Annu Rev Entomol. 2006;51:113–135. doi: 10.1146/annurev.ento.51.051705.113646. [DOI] [PubMed] [Google Scholar]
- Harbison ST, Chang S, Kamdar KP, Mackay TF. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biol. 2005;6(4):R36. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Yamamoto AH, Fanara JJ, Norga KK, Mackay TF. Quantitative trait loci affecting starvation resistance in Drosophila melanogaster. Genetics. 2004;166(4):1807–1823. doi: 10.1534/genetics.166.4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimbeck G, Bugnon V, Gendre N, Haberlin C, Stocker RF. Smell and taste perception in Drosophila melanogaster larva: toxin expression studies in chemosensory neurons. J Neurosci. 1999;19(15):6599–6609. doi: 10.1523/JNEUROSCI.19-15-06599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319(5871):1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Cobb M. The behaviour of Drosophila melanogaster maggots is affected by social, physiological and temporal factors. Animal Behaviour. 2008;75:1619–1628. [Google Scholar]
- Keene AC, Waddell S. Drosophila olfactory memory: single genes to complex neural circuits. Nat Rev Neurosci. 2007;8(5):341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- Kreher SA, Kwon JY, Carlson JR. The molecular basis of odor coding in the Drosophila larva. Neuron. 2005;46(3):445–456. doi: 10.1016/j.neuron.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Kreher SA, Mathew D, Kim J, Carlson JR. Translation of sensory input into behavioral output via an olfactory system. Neuron. 2008;59(1):110–124. doi: 10.1016/j.neuron.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue PP, Vosshall LB. The olfactory sensory map in Drosophila. Adv Exp Med Biol. 2008;628:102–114. doi: 10.1007/978-0-387-78261-4_7. [DOI] [PubMed] [Google Scholar]
- Lavagnino NJ, Anholt RR, Fanara JJ. Variation in genetic architecture of olfactory behaviour among wild-derived populations of Drosophila melanogaster. J Evol Biol. 2008;21(4):988–996. doi: 10.1111/j.1420-9101.2008.01546.x. [DOI] [PubMed] [Google Scholar]
- Lavagnino NJ, Fanara JJ. Phenotypic plasticity for Drosophila melanogaster (Diptera: Drosophilidae) larval olfactory behaviour in response to whole fruit olfactory stimuli. Revista de la Sociedad Entomológica Argentina. 2011;70:369–372. [Google Scholar]
- Lilly M, Carlson J. smellblind: a gene required for Drosophila olfaction. Genetics. 1990;124(2):293–302. doi: 10.1093/genetics/124.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M, Kreber R, Ganetzky B, Carlson JR. Evidence that the Drosophila olfactory mutant smellblind defines a novel class of sodium channel mutation. Genetics. 1994;136(3):1087–1096. doi: 10.1093/genetics/136.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacsovich T, Asztalos Z, Awano W, Baba K, Kondo S, Niwa S, Yamamoto D. Dual-tagging gene trap of novel genes in Drosophila melanogaster. Genetics. 2001;157(2):727–742. doi: 10.1093/genetics/157.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50(6):869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Masuda-Nakagawa LM, Gendre N, O’Kane CJ, Stocker RF. Localized olfactory representation in mushroom bodies of Drosophila larvae. Proc Natl Acad Sci U S A. 2009;106(25):10314–10319. doi: 10.1073/pnas.0900178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Amrein H. Taste and pheromone perception in mammals and flies. Genome Biol. 2003;4(7):220. doi: 10.1186/gb-2003-4-7-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Muñoz MC, Godoy-Herrera R. Dispersal and prepupation behavior of Chilean sympatric Drosophila species that breed in the same site in nature. Behavioral Ecology. 2005;16(1):316–322. [Google Scholar]
- Mensch J, Lavagnino N, Carreira VP, Massaldi A, Hasson E, Fanara JJ. Identifying candidate genes affecting developmental time in Drosophila melanogaster: pervasive pleiotropy and gene-by-environment interaction. BMC Dev Biol. 2008;8:78. doi: 10.1186/1471-213X-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte P, Woodard C, Ayer R, Lilly M, Sun H, Carlson J. Characterization of the larval olfactory response in Drosophila and its genetic basis. Behav Genet. 1989;19(2):267–283. doi: 10.1007/BF01065910. [DOI] [PubMed] [Google Scholar]
- Moreau-Fauvarque C, Taillebourg E, Preat T, Dura JM. Mutation of linotte causes behavioral defects independently of pigeon in Drosophila. Neuroreport. 2002;13(17):2309–2312. doi: 10.1097/00001756-200212030-00028. [DOI] [PubMed] [Google Scholar]
- Norga KK, Gurganus MC, Dilda CL, Yamamoto A, Lyman RF, Patel PH, Rubin GM, Hoskins RA, Mackay TF, Bellen HJ. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr Biol. 2003;13(16):1388–1396. doi: 10.1016/s0960-9822(03)00546-3. [DOI] [PubMed] [Google Scholar]
- Oppliger FY, Guerin PM, Vlimant M. Neurophysiological and behavioural evidence for an olfactory function for the dorsal organ and a gustatory one for the terminal organ in Drosophila melanogaster larvae. J Insect Physiol. 2000;46(2):135–144. doi: 10.1016/s0022-1910(99)00109-2. [DOI] [PubMed] [Google Scholar]
- Parsons PA. Larval responses to environmental ethanol in Drosophila melanogaster: variation within and among populations. Behav Genet. 1980;10(2):183–190. doi: 10.1007/BF01066268. [DOI] [PubMed] [Google Scholar]
- Python F, Stocker RF. Adult-like complexity of the larval antennal lobe of D. melanogaster despite markedly low numbers of odorant receptor neurons. J Comp Neurol. 2002;445(4):374–387. doi: 10.1002/cne.10188. [DOI] [PubMed] [Google Scholar]
- Rodrigues V. Olfactory behavior of Drosophila melanogaster. In: Siddiqi O, Bvabu P, Hall LM, Hall JC, editors. Development and Neurobiology of Drosophila. Plenum Press; London: 1980. pp. 361–372. [Google Scholar]
- Rollmann SM, Magwire MM, Morgan TJ, Ozsoy ED, Yamamoto A, Mackay TF, Anholt RR. Pleiotropic fitness effects of the Tre1-Gr5a region in Drosophila melanogaster. Nat Genet. 2006;38(7):824–829. doi: 10.1038/ng1823. [DOI] [PubMed] [Google Scholar]
- Rollmann SM, Yamamoto A, Goossens T, Zwarts L, Callaerts-Vegh Z, Callaerts P, Norga K, Mackay TF, Anholt RR. The early developmental gene Semaphorin 5c contributes to olfactory behavior in adult Drosophila. Genetics. 2007;176(2):947–956. doi: 10.1534/genetics.106.069781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryuda M, Tsuzuki S, Tanimura T, Tojo S, Hayakawa Y. A gene involved in the food preferences of larval Drosophila melanogaster. J Insect Physiol. 2008;54(10-11):1440–1445. doi: 10.1016/j.jinsphys.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Sambandan D, Yamamoto A, Fanara JJ, Mackay TF, Anholt RR. Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster. Genetics. 2006;174(3):1349–1363. doi: 10.1534/genetics.106.060574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver SA, Varnam CJ, Hilliker AJ, Sokolowski MB. The foraging gene affects adult but not larval olfactory-related behavior in Drosophila melanogaster. Behav Brain Res. 1998;95(1):23–29. doi: 10.1016/s0166-4328(97)00206-4. [DOI] [PubMed] [Google Scholar]
- StatSoft I. A Data Analysis Software System. Release 6.0. Tulsa, OK: 2001. STATISTICA. [Google Scholar]
- Su CY, Menuz K, Carlson JR. Olfactory perception: receptors, cells, and circuits. Cell. 2009;139(1):45–59. doi: 10.1016/j.cell.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–74. doi: 10.1093/nar/gkm306. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB. Olfaction in Drosophila. Curr Opin Neurobiol. 2000;10(4):498–503. doi: 10.1016/s0959-4388(00)00111-2. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Zwarts L, Callaerts P, Norga K, Mackay TF, Anholt RR. Neurogenetic networks for startle-induced locomotion in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2008;105(34):12393–12398. doi: 10.1073/pnas.0804889105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Stone EA, Mackay TF, Anholt RR. Plasticity of the chemoreceptor repertoire in Drosophila melanogaster. PLoS Genet. 2009;5(10):e1000681. doi: 10.1371/journal.pgen.1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]