Abstract
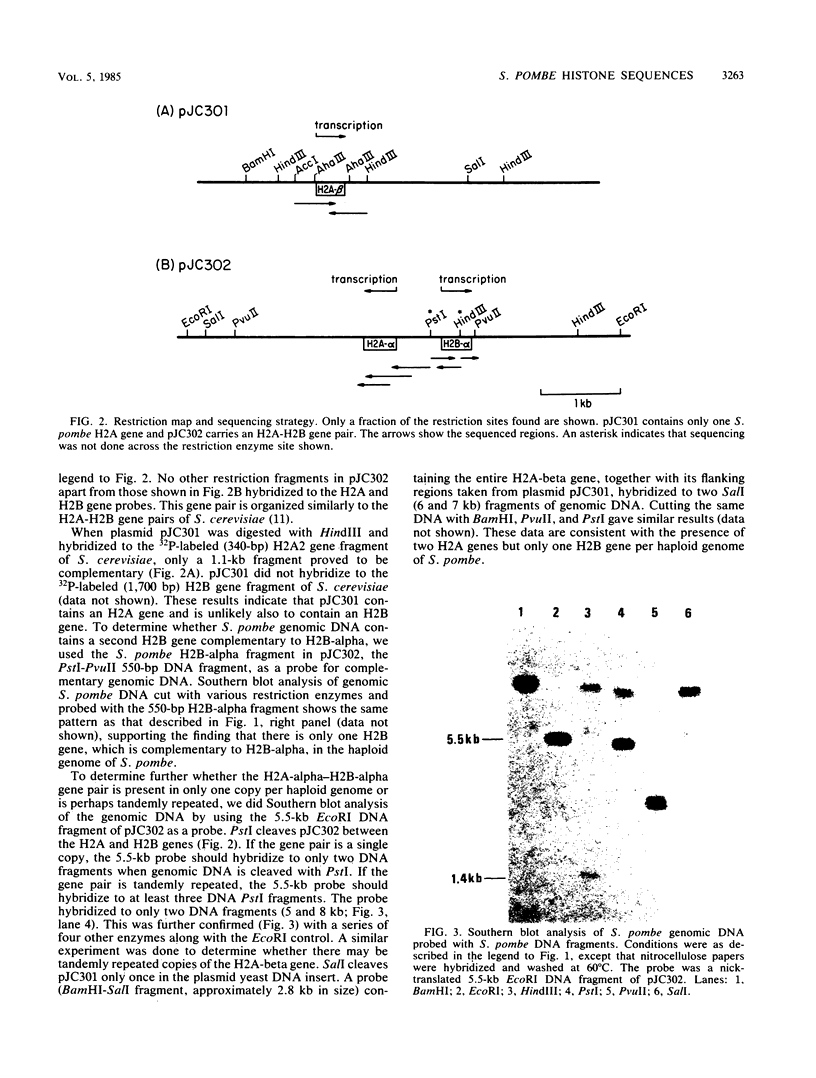
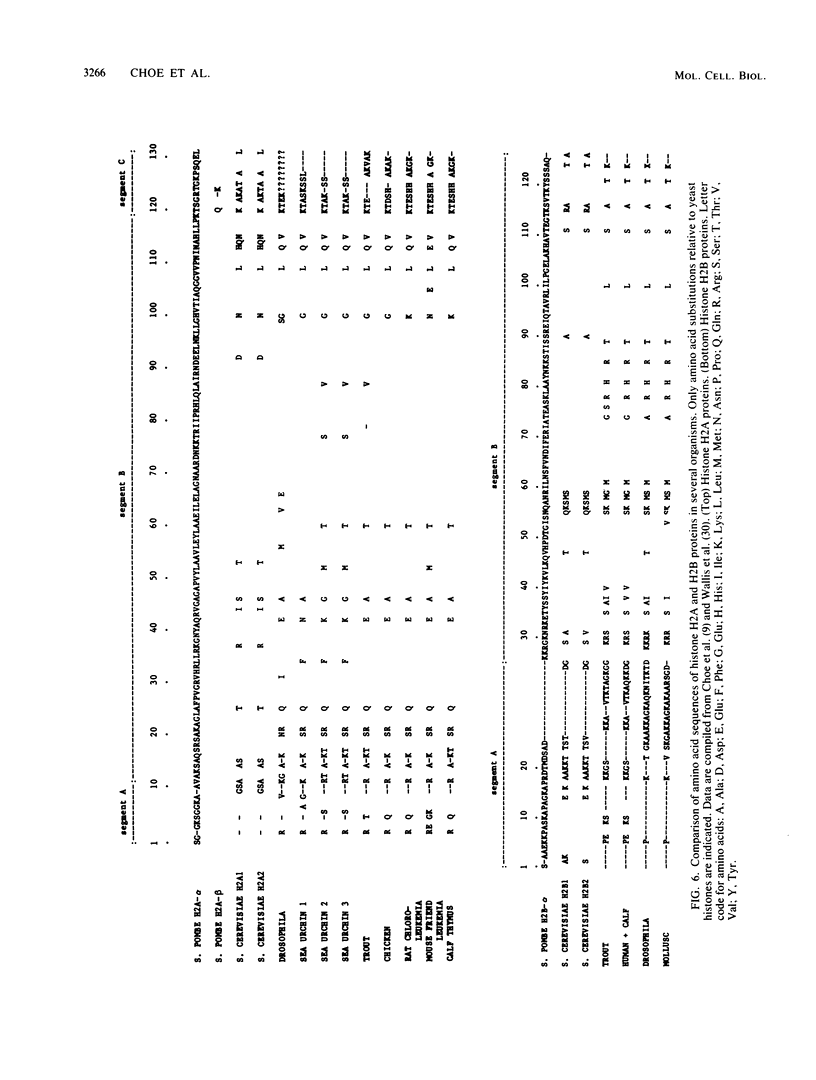
The histone H2A and H2B genes of the fission yeast Schizosaccharomyces pombe were cloned and sequenced. Southern blot and sequence analyses showed that, unlike other eucaryotes, Saccharomyces cerevisiae included, S. pombe has unequal numbers of these genes, containing two histone H2A genes (H2A-alpha and -beta) and only one H2B gene (H2B-alpha) per haploid genome. H2A- and H2B-alpha are adjacent to each other and are divergently transcribed. H2A-beta has no other histone gene in close proximity. Preceding both H2A-alpha and -beta is a highly conserved 19-base-pair sequence (5'-CATCAC/AAACCCTAACCCTG-3'). The H2A DNA sequences encode two histone H2A subtypes differing in amino acid sequence (three residues) and size (H2A-alpha, 131 residues; H2A-beta, 130 residues). H2B-alpha codes for a 125-amino-acid protein. Sequence evolution is extensive between S. pombe and S. cerevisiae and displays unique patterns of divergence. Certain N-terminal sequences normally divergent between eucaryotes are conserved between the two yeasts. In contrast, the normally conserved hydrophobic core of H2A is as divergent between the yeasts as between S. pombe and calf.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beach D. H., Klar A. J. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 1984 Mar;3(3):603–610. doi: 10.1002/j.1460-2075.1984.tb01855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bostock C. J. DNA synthesis in the fission yeast Schizosaccharomyces pombe. Exp Cell Res. 1970 Apr;60(1):16–26. doi: 10.1016/0014-4827(70)90484-2. [DOI] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Irminger J. C., Birnstiel M. L. Ubiquitous and gene-specific regulatory 5' sequences in a sea urchin histone DNA clone coding for histone protein variants. Nucleic Acids Res. 1980 Mar 11;8(5):957–977. doi: 10.1093/nar/8.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm L., Briand G., Sautière P., Crane-Robinson C. Proteolytic digestion studies of chromatin core-histone structure. Identification of limit peptides from histone H2B. Eur J Biochem. 1982 Apr 1;123(2):299–303. doi: 10.1111/j.1432-1033.1982.tb19767.x. [DOI] [PubMed] [Google Scholar]
- Böhm L., Crane-Robinson C., Sautière P. Proteolytic digestion studies of chromatin core-histone structure. Identification of a limit peptide of histone H2A. Eur J Biochem. 1980 May;106(2):525–530. doi: 10.1111/j.1432-1033.1980.tb04599.x. [DOI] [PubMed] [Google Scholar]
- Certa U., Colavito-Shepanski M., Grunstein M. Yeast may not contain histone H1: the only known 'histone H1-like' protein in Saccharomyces cerevisiae is a mitochondrial protein. Nucleic Acids Res. 1984 Nov 12;12(21):7975–7985. doi: 10.1093/nar/12.21.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J., Kolodrubetz D., Grunstein M. The two yeast histone H2A genes encode similar protein subtypes. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1484–1487. doi: 10.1073/pnas.79.5.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Wallis J. Colony hybridization. Methods Enzymol. 1979;68:379–389. doi: 10.1016/0076-6879(79)68027-8. [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr, Rosbash M., Kaback D. B. Isolation of yeast histone genes H2A and H2B. Cell. 1979 Dec;18(4):1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- Kolodrubetz D., Rykowski M. C., Grunstein M. Histone H2A subtypes associate interchangeably in vivo with histone H2B subtypes. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7814–7818. doi: 10.1073/pnas.79.24.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Plasmids carrying the yeast OMP decarboxylase structural and regulatory genes: transcription regulation in a foreign environment. Cell. 1983 Feb;32(2):371–377. doi: 10.1016/0092-8674(83)90456-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robinow C. F. The Number of Chromosomes in SCHIZOSACCHAROMYCES POMBE: Light Microscopy of Stained Preparations. Genetics. 1977 Nov;87(3):491–497. doi: 10.1093/genetics/87.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. Structure of the Schizosaccharomyces pombe cytochrome c gene. Mol Cell Biol. 1982 Feb;2(2):106–116. doi: 10.1128/mcb.2.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. R., Hall B. D. The primary structure of the alcohol dehydrogenase gene from the fission yeast Schizosaccharomyces pombe. J Biol Chem. 1983 Jan 10;258(1):143–149. [PubMed] [Google Scholar]
- Rykowski M. C., Wallis J. W., Choe J., Grunstein M. Histone H2B subtypes are dispensable during the yeast cell cycle. Cell. 1981 Aug;25(2):477–487. doi: 10.1016/0092-8674(81)90066-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. M., Andrésson O. S. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J Mol Biol. 1983 Sep 25;169(3):663–690. doi: 10.1016/s0022-2836(83)80164-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Toda T., Adachi Y., Hiraoka Y., Yanagida M. Identification of the pleiotropic cell division cycle gene NDA2 as one of two different alpha-tubulin genes in Schizosaccharomyces pombe. Cell. 1984 May;37(1):233–242. doi: 10.1016/0092-8674(84)90319-2. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Hereford L., Grunstein M. Histone H2B genes of yeast encode two different proteins. Cell. 1980 Dec;22(3):799–805. doi: 10.1016/0092-8674(80)90556-5. [DOI] [PubMed] [Google Scholar]
- Wallis J. W., Rykowski M., Grunstein M. Yeast histone H2B containing large amino terminus deletions can function in vivo. Cell. 1983 Dec;35(3 Pt 2):711–719. doi: 10.1016/0092-8674(83)90104-6. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]