Abstract
The present study aims to design hepatic targeted curcumin (CUR) nanoparticles using Gantrez (GZ) as a polymer. Three carbohydrate-based hepatocyte asialoglycoprotein receptor (ASGP-R) ligands were selected for the study, namely kappa carrageenan (KC), arabinogalactan (AG), and pullulan (P). AG and KC are galactose based while P is a glucose-based polymer. CUR-GZ nanoparticles were prepared by nanoprecipitation and anchored with the ligands by nonspecific adsorption onto preformed nanoparticles. The change in zeta potential values confirmed adsorption of the ligands. Docking simulation was evaluated as a tool to predict ligand ASGP-R interactions, using grid-based ligand docking with energies (Glide). Monomers and dimers were used as representative units of polymer for docking analysis. The binding of ASGP-R was validated using d-galactose as monomer. The interaction of the ligands with the receptor was evaluated based on Glide scores and Emodel values, both for monomers and dimers. The data of the docking study based on Glide scores and Emodel values suggested higher affinity of AG and P to the ASGP-R, compared to KC. At 1 h, following intravenous administration of the nanoparticles to rats, the in vivo hepatic accumulation in the order CUR-GZAG > CUR-GZKC > CUR-GZP correlated with the docking data based on Glide scores. However, at the end of 6 h, pullulan exhibited maximum hepatic accumulation and arabinogalactan minimum accumulation (p < 0.05). Nevertheless, as predicted by docking analysis, arabinogalactan and pullulan revealed maximum hepatic accumulation. Docking analysis using dimers as representative stereochemical units of polymers provides a good indication of ligand receptor affinity. Docking analysis provides a useful tool for the preliminary screening of ligands for hepatic targeting.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-013-9474-6) contains supplementary material, which is available to authorized users.
Key words: arabinogalactan, docking, hepatic targeting, kappa carrageenan, pullulan
INTRODUCTION
Targeted drug delivery to the liver for diseases like hepatitis, cirrhosis, fibrosis, and hepatocellular carcinoma could provide enhanced efficacy with decreased toxicity. While nanocarriers provide an important strategy for enhanced delivery to the liver, they are sequestered rapidly by hepatic Kupffer cells, thereby limiting accumulation in the hepatocytes (1,2). However, enhanced drug accumulation in hepatocytes is essential for improved therapy in such diseases. Strategies to bypass Kupffer cells coupled with receptor-based targeting to hepatocytes could provide an immense advantage. The asialoglycoprotein receptor (ASGP-R), often overexpressed in hepatic diseases, is a promising target (3). The limited presence of ASGP-R at few other sites in the body provides an additional advantage (4,5).
The ASGP-R also termed as hepatic lectin exhibits high affinity for galactose-based carbohydrates in the form of tri- and tetraantennary oligosaccharides with galactose at its terminal (6). Carbohydrate polymers such as pullulan (a glucose-based polymer) and arabinogalactan (a galactose-based polymer) have shown promise as ligands for ASGP-R-mediated targeting to liver. Enhanced hepatic accumulation of pullulan nanoparticles and pullulan drug conjugates is reported (7–14). A marked decrease in hepatic accumulation of pullulan when co-administered with arabinogalactan suggests higher affinity of the ASGP-R for galactose-based ligands. Such influence of galactose density on ligand receptor binding has been demonstrated with Tri-Gal > Di-Gal > Mono-Gal (15–19).
Receptor ligand binding is governed by the stereochemical conformation and functional groups of ligands. For the sugar monomer, the hydroxyl group position and density of sugar moieties play an important role (20). Docking simulation can provide detailed electronic properties as well as dynamic aspects of structural mechanisms to explain such ligand receptor interactions. Docking analysis could serve as a useful tool for predicting receptor–ligand interaction in vivo. Nevertheless, studies in this direction are sparse. Massarelli (21) reported alginates and ulvans as possible ASGP-R ligands by docking their monomers that constituted the building blocks for the polymer; however, this has not been confirmed in vivo.
Curcumin (CUR), a nutraceutical, has been reported to possess therapeutic benefits in hepatocellular carcinoma and other liver diseases (22,23). The present study aims to design hepatic-targeted Gantrez nanoparticles of CUR. Three carbohydrate-based ligands were selected for the study, namely kappa carrageenan, arabinogalactan, and pullulan. Arabinogalactan is a highly branched carbohydrate polymer, comprising ∼86% of galactose, with galactose/arabinose in the ratio of 6:1. Kappa carrageenan is a linear, galactose polymer comprising galactose and galactose sulfate, while pullulan is glucose-based polymer.
Evaluating docking simulation as a tool to predict ligand receptor interactions in silico and correlation with in vivo data was the objective of the study. In vivo evaluation following intravenous administration of curcumin-loaded Gantrez nanoparticles anchored with the three selected ligands was studied in healthy rats (Sprague–Dawley).
MATERIALS AND METHODS
Materials
Commercial curcumin (mixture of curcuminoids) (95% w/w) and Gantrez® AN 119 (poly(methyl vinyl ether-co-maleic anhydride)) with an average molecular weight of 200,000 were gifts from Konark Herbals & Health Care, Daman, India and ISP, Anshul Life Sciences, India, respectively. Pullulan (Hayashibara) and kappa carrageenan were gifts by Gangwal Chemicals and Lucid Colloids Ltd., India, respectively. Bisdemethoxycurcumin and demethoxycurcumin (purity >99%) were procured from Sami Chemicals, Bangalore, India. Arabinogalactan (AG) and 17β-estradiol were purchased from Sigma, Mumbai. Acetone, dextrose, methanol, orthophosphoric acid, polyethylene glycol 400, sodium ethylenediaminetetraacetate (sodium EDTA), and Tween 80 (spectroscopic or analytical grade) were acquired from SD Fine Chemicals, India. Magnesium acetate was procured from Merck, India. Sodium pertechnetate (99mTc) was freshly extracted from 99molybdenum, which was received from the Board of Radiation and Isotope Technology (Mumbai, India), by solvent extraction. Anesthetics ketamine hydrochloride injection and xylazine hydrochloride injection were procured from the local market. HPTLC plates were procured from Himedia Laboratories. Other solvents and chemicals used were analytical/spectroscopic grade. Double-distilled water filtered through a 0.45 μm-membrane filter was used throughout the experiments.
Method
Preparation of ligand-anchored curcumin Gantrez nanoparticles (CUR-GZ NPs)
CUR-GZ nanoparticles were prepared by nanoprecipitation. Briefly, CUR (10 mg) and Gantrez® AN 119 (10 mg) dissolved in acetone (10 mL) were added using a syringe fitted with a needle of gauge 26 1/2 (0.45 × 13 mm) to an aqueous phase (20 mL) containing Tween 80 (0.05% w/v) with continuous stirring. Aqueous magnesium acetate solution (5% w/v, 1 mL) enabled cross linking of Gantrez® AN 119 (24,25). The dispersion was stirred for approximately 3 h at 25 ± 2°C till complete removal of the organic solvent by evaporation. Centrifugation (Eltek4100 D Research Centrifuge) at 15,000 rpm for a period of 30 min was carried out to separate the CUR-GZ NPs. Following centrifugation, the supernatant was analyzed for un-entrapped CUR by UV spectrophotometry (Shimadzu, Japan) at 425 nm. Entrapment efficiency (%EE) was calculated using the equation below:
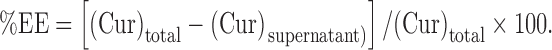 |
The resultant pellet was washed twice with water and redispersed in 2.5 mL particulate-free (filtered through 0.22 μm) water using a probe sonicator (DP120, Dakshin Mumbai, India) with 10-s pulse at 200 V, for 8 min kept in an ice bath, and freeze-dried (FreeZone 4.5, USA) using trehalose as cryoprotectant (20%). Lyophilized samples were characterized for nanoparticle size after reconstitution in particulate-free water (as mentioned in “Materials and Methods” section).
Aqueous solutions (1% w/v) of pullulan and arabinogalactan were prepared by dissolving in distilled water at room temperature (28°C). A solution of kappa carrageenan (1% w/v) in distilled water was prepared by heating at 70°C, followed by cooling to 28°C and filtration prior to use. Aliquots of these solutions to maintain a Gantrez AN 119/ligand ratio of 1:1 (w/w) were added to the CUR-GZ NP pellet obtained after centrifugation, and the dispersions were freeze dried.
Physicochemical characterization
Particle size
The mean diameter was determined by photon correlation spectroscopy, with light scattered at 90° on a N4 plus particle size instrument (Beckman Coulter, USA) at a temperature of 25°C. Dilutions of the dispersions to obtain an intensity of 5 × 104 to 1 × 106 final counts per second were carried out prior to recording the particle size.
Zeta potential
Zeta potential was determined using the Zetasizer Nano ZS (Malvern Instruments Ltd, Malvern, UK) using DTS Nano software. Nanoparticle dispersions, with the aid of a syringe, were filled into a capillary cell to record the zeta potential.
Scanning electron microscopy (SEM)
Nanodispersion was dropped on an aluminum grid and dried in vacuum. The samples were sputtered with platinum using a coater JEOL JSM 1600 and analyzed on a scanning electron microscope (JEOL JSM 6380).
Differential scanning calorimetry (DSC)
Freeze-dried samples (5 mg) were placed in aluminum pans and crimped and DSC thermograms were obtained (PerkinElmer Pyris 6 DSC). Samples were heated from 40°C to 250°C at 10°C/min. An empty aluminum pan served as reference. The samples were continuously purged with nitrogen at a flow rate of 20 mL/min.
In vitro hemolytic assay
The hemolytic potential/erythrocyte toxicity of the nanoparticles was evaluated following a reported method (26). Fresh rat blood was collected in a vial containing EDTA (anticoagulant). Erythrocytes were isolated from blood by sedimentation following centrifugation (3,500 rpm for 5 min). The erythrocytes were repeatedly washed with phosphate-buffered saline (PBS), pH 7.4, thrice by centrifugation. The erythrocyte pellet obtained after third washing was then diluted with buffer to prepare the erythrocyte stock dispersion.
Dispersions of CUR-GZ NPs with and without ligands were appropriately diluted to give 1, 2.5, 5, 10, 25, 50, 100, and 150 μg/mL of concentrations equivalent to CUR. To 1 mL of each of the solution, 100 μL of erythrocyte stock dispersion was added and the samples were incubated at 37°C for 60 min. The samples were subsequently centrifuged at 5,000 rpm for 10 min. To 100 μL of the supernatant, 2 mL of an ethanol/HCl mixture comprising l part conc. HC1 and 39 parts ethanol (99% v/v) was added. This mixture ensured solution of all the components and circumvented the precipitation of hemoglobin. The absorbance was recorded at 540 nm on a spectrophotometer using negative control as blank. PBS and double-distilled water served as negative and positive control, respectively.
Hemolysis (in percent) was calculated as follows (27):
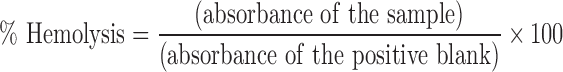 |
In vitro serum stability
The stability of CUR-GZ NPs in serum was determined by evaluating for change in particle size. Serum (10%) was diluted using phosphate-buffered saline (pH 7.4). A 4% w/v NP dispersion (1 mL) was added to 1 mL of diluted serum and incubated at 37 ± 0.5°C for 6 h respectively and evaluated for nanoparticle size for 6 h.
Docking analysis
Molecular flexible docking studies were performed using the grid-based ligand docking with energies (Glide) (28), a computational program run within Maestro (29) and a graphical user interface by Schrödinger, LLC, New York, NY, installed on an Intel Xeon workstation. ASGP-R is a hetero-oligomer comprising of two homologous subunits H1 and H2. The 3D ASGP-R structure (H1 subunit) for carbohydrate recognition domain was downloaded from Protein Data Bank (PDB code 1DV8, 2.3 Å of resolution) (http://pubchem.ncbi.nlm.nih.gov/). Before docking the ligands, the structural defects within the imported protein structure were corrected and generated using protein preparation wizard using Maestro. All the crystallographic water molecules, other than the molecules forming coordinate bonds with Ca2+ ions within the binding sites, were removed. The bond orders and hydrogen atoms were assigned to define the correct ionization states. Zero-order bonds between metal and water molecules were generated after correcting the formal charges on metal and neighboring atoms. Optimized metal binding states were generated within pH 7.0 ± 4.0. An H-bond assignment was given to orient the water molecules to optimize H-bonding. Finally, an Impref minimization using OPLS 2005 force field was performed to relieve any strain and fine tune the protein structure. The calcium ion (CA-1002) at the active site showed eight coordinate bonds which include bonding with two water molecules (HOH-11 and HOH-13).
While kappa carrageenan comprises of repeated units of galactose, pullulan comprises of glucose and arabinogalactan comprises of galactose and arabinose. Dimers derived from these natural polymers of interest represented structural and conformational units of the polymers. With this perspective, α(1→6) d-glucose and α(1→4) d-glucose dimers of pullulan, β(1→3) d-galactose and β(1→6) d-galactose dimers of arabinogalactan, and galactose dimers of kappa carrageenan were selected as the dimer. The 3D structures of pullulan and d-galactose were downloaded from PubChem database (http://pubchem.ncbi.nlm.nih.gov/) while that for arabinogalactan and kappa carrageenan they were taken from the literature for the docking studies (30–32). The ligand structures were constructed using the 2D Sketcher in Maestro. A single low-energy 3D conformation of ligand was created with the help of LigPrep (33), using OPLS 2005 force field.
The active site of the protein was defined by taking centroid of Trp 243, Asp 241, Glu 252, Gln 239, Asp 265, and Asn 264 residues for generating the grid. The grid, i.e., a virtual box having a default length of 20 Å, was generated to restrict the docking process to take place within the grid space. Positional constraints were added to replace the oxygen atom of the Ca-coordinated water molecules (11,13) as coordinate bonding of ligand is essential for binding to ASGP-R. Docking study was performed in the extra precision (XP) mode keeping “must match at least one constraint” criterion. The other building blocks of the polysaccharides were studied using the similar grid.
Biodistribution of 99mTc-radiolabelled CUR-GZ NPs by gamma scintigraphy
Radiolabelling and stability
CUR-GZ NPs with and without ligands were radiolabelled with stannous chloride reduced 99mTc. For the reduction of stannous chloride, acidic (0.1 N HCl) solution of stannous chloride solution was mixed with 99mTc eluted in normal saline (0.1 mg of stannous chloride per millicurie of 99mTc). The nanoparticle dispersion (CUR-GZ/CUR-GZ P/CUR-GZ KC/CUR-GZ AG) was adjusted to pH 7.2 by adding citrate buffer and mixed over a shaker at 28°C and incubated for 20 min in a lead cell. The radiolabelling and binding efficiency were determined as reported earlier (27,34). The percent radiolabelling efficiency was calculated using following equation:
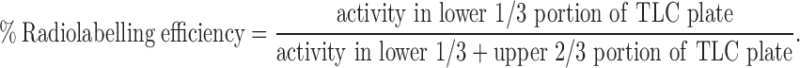 |
Radiolabelling efficiency was monitored up to 6 h to determine the stability of the radiolabelled complex.
Imaging
Healthy female Sprague–Dawley rats (BW 200 ± 20 g) were housed under standard laboratory conditions with access to pelleted feed and water ad libitum. Anesthesia was induced by injecting a cocktail solution of ketamine hydrochloride (50 mg kg−1) and xylazine hydrochloride (10 mg kg−1) intramuscularly 20 min prior to study. For acquiring the scintigraphic images through Millennium MPS Acquisition System (multipurpose single head square detector), gamma camera fitted with low energy general purpose pinhole collimator, the animals were placed prone with a distance of 95 cm maintained between the collimator to table for all acquisitions. Radiolabelled CUR-GZ, CUR-GZ P, CUR-GZ KC, and CUR-GZ AG (approx. 500 μCi) were administered to rats intravenously through the tail vein. Difference in radioactivity after dosing and before dosing was measured on a gamma counter and gave an estimate of total injected dose. The data were acquired on GENIE acquisition station and then transferred to Entegra workstation for processing. Static images were recorded at 1, 3, and 6 h post-dosing for 1 min with a resolution of ×1.33 zoom. The region of interest (liver) was demarcated using Entegra software. The percentage uptake of the radiolabelled formulation at the end of 1, 3, and 6 h was estimated in the liver and counted by background subtraction and decay correction.
At sixth hour, animals were sacrificed and organs were isolated. The organs (heart, spleen, lungs, stomach, liver, and small intestine) were washed with saline, dried, and weighed. Radioactivity in each organ was determined using a dose calibrator (Capnitech) and corrected for physical decay. The percent injected dose per gram of organ was estimated. The experimental protocol was approved by the animal ethics committee of the Bombay Veterinary College, Mumbai, India.
Statistical analysis
Data reported herein are expressed as mean ± standard deviation in the tables and mean ± standard error in the figures. Data were analyzed by one-way ANOVA with Dunnett’s test and Student’s t test. A p < 0.05 was considered for statistical significance.
RESULTS
CUR-GZ NPs
CUR-GZ NPs were successfully prepared by nanoprecipitation with very high entrapment efficiency (84.1 ± 0.3%), nanosize of 532.2 ± 15.2 nm, and a negative zeta potential. Anchoring the ligands revealed no change in particle size; however, changes in zeta potential were observed (Table I). Nanoparticles with and without ligands were successfully freeze dried with an Sf/Si ratio less than 1.3 (35). SEM imaging revealed spherical nanoparticles and particle size matched the results obtained with the photon correlation spectroscopy (Fig. 1).
Table I.
Particle, Size Zeta Potential, and S f/S i Ratios of CUR-GZ NPs (Mean ± Standard Deviation)
| NPs | Particle size (nm) | Polydispersity index | Zeta potential (mV) | S f/S i ratio |
|---|---|---|---|---|
| CUR-GZ | 532.2 ± 15.2 | 0.32 ± 0.11 | −30.5 ± 1.2 | 0.99 |
| CUR-GZ P | 539.3 ± 10.1 | 0.38 ± 0.08 | −12.1 ± 4.6 | 1.00 |
| CUR-GZ AG | 540.4 ± 8.7 | 0.31 ± 0.10 | −13.4 ± 3.8 | 1.01 |
| CUR-GZ KC | 550.1 ± 8.1 | 0.36 ± 0.09 | −24.3 ± 2.7 | 1.02 |
S f indicates particle size after freeze drying of nanoparticles; S i indicates particle size before freeze drying of nanoparticles
Fig. 1.

SEM of a CUR-GZ, b CUR-GZ P, c CUR-GZ KC, and d CUR-GZ AG
The DSC thermograms of the various samples are depicted in Fig. 2. Crystalline CUR showed a thermal event at 177.63°C (ΔH = 90.64 J/g) corresponding to its melting point. The trehalose thermogram showed an endotherm at 100°C attributed to dehydration of the compound (36), which was evident in the thermograms of the freeze-dried ligand-anchored nanoparticles. CUR-GZ NPs showed a mild thermal event at 173°C due to the melting of CUR. While a small peak for CUR exhibited for CUR-GZ suggesting partial amorphization, ligand-anchored nanoparticles exhibited no peak for CUR.
Fig. 2.

Comparative DSC thermograms
Nanoparticles with/without ligands revealed low hemolysis of less than 10% even at high concentration of 150 μg/mL of CUR (Fig. 3a) suggesting adequate safety (37,38). No marked difference in particle size (Fig. 3b) confirmed the serum stability of the nanoparticles for 6 h.
Fig. 3.
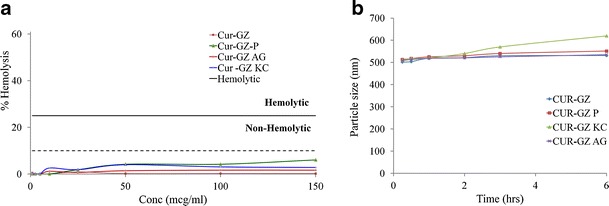
a In vitro hemolysis and b in vitro serum stability of CUR-GZ NPs (mean ± standard error, n = 3)
Docking analysis
The active binding site for ASGP-R could be easily located on the surface of the H1 subunit. Figure 4a shows optimized docked pose of d-galactose at the active binding site and the interaction diagram of d-galactose with ASGP-R. Following docking into the active site, the ligands were ranked on the basis of their Glide score (or G score) and Emodel scores.
Fig. 4.

Glide docking of a d-galactose, b β(1→3) d-galactose dimer of arabinogalactan, and c β(1→6) d-galactose dimer of arabinogalactan at the active binding site of ASGP-R and its ligand interaction diagrams
The Glide score is an experimental scoring function that calculates the ligand binding free energy (39). It includes contributing terms such as electrostatic and van der Waals interactions and terms that influence ligand binding as seen in formula below:
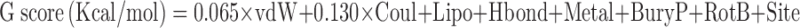 |
where vdW is van der Waals energy, Coul is Coulomb energy, Lipo is lipophilic contact term, Hbond is hydrogen bonding term, Metal is metal binding term, BuryP is penalty for buried polar groups, RotB is penalty for freezing rotatable bonds, and Site is polar interactions in the active site, and the coefficients of vdW and Coul are a = 0.065 and b = 0.130.
Model energy score (Emodel) is a combination of the energy grid score, the binding affinity, and the internal strain energy (in flexible docking) of the model. Therefore, the Emodel is a more significant function in selecting best-docked pose for each ligand (pose selection), which is then ranked accordingly based on Glide score. The Glide score and Glide Emodel scores of the various ligands are shown in Table II. All the dimeric ligands showed good dock score compared to the known ligand d-galactose and formed similar type of interactions with the active sites.
Table II.
Docking Results of Various Ligands in the Original ASGP-R Crystal Structure Using Glide-XP
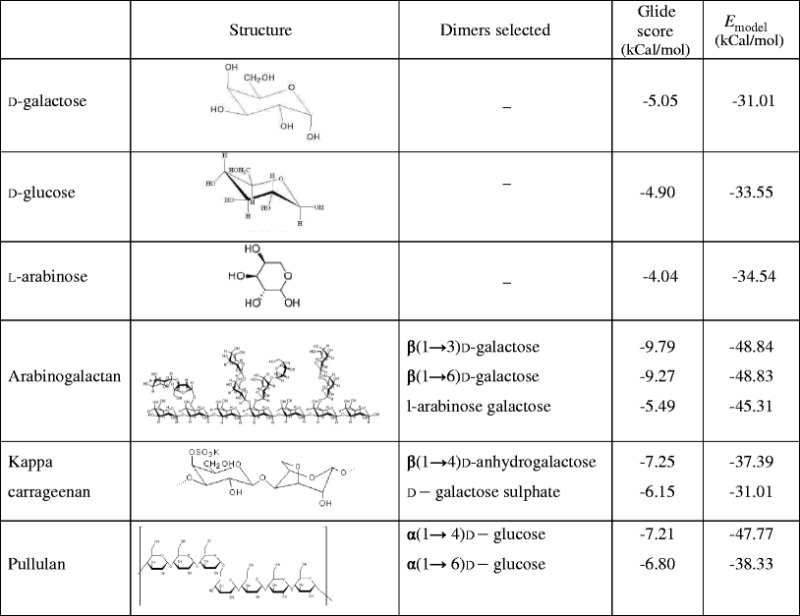
All the ligands maintained the hydrophobic interaction of Trp 243 with either of C3, C4, C5, or C6 atoms of monomer shown in Supplementary Fig. 1. d-galactose exhibited four hydrogen bonds. Similarly, kappa carrageenan, a 100% galactose ligand, also exhibited four hydrogen bonds, comparable to d-galactose. However, arabinogalactan exhibited five, six, and seven bonds attributed to the dimers, arabinose–galactose dimer, β(1→6), d-galactose dimer (Fig. 4c), and β(1→3) d-galactose dimer, respectively (Fig. 4b). Pullulan exhibited a higher number of hydrogen bonds of 7 and 8 attributed to α(1→6) d-glucose dimer and α(1→4) d-glucose dimer, respectively. A comparison of the different G score suggests that the ligands would show an order of binding AG > KC > P > d-galactose > d-glucose > l-arabinose.
Biodistribution in rats by gamma scintigraphy
The radiolabelled complexes were stable up to 80% at the end of 6 h (Supplementary Fig. 2) indicating adequate stability and their suitability for in vivo studies. In vivo stability of the 99mTc-labelled complex was confirmed by the absence of radioactivity in the thyroid.
At 6 h, 99mTc activities were mainly found in RES (liver, lung, and spleen), whereas radioactivity in bone, muscle, stomach, and rest of the body was minimal (Fig. 5). Maximum hepatic accumulation was seen with CUR-GZ P followed by CUR-GZ KC, CUR-GZ, and CUR-GZ AG, respectively. CUR-GZ P and CUR-GZ AG revealed a significantly lower (p < 0.05) lung and heart accumulation compared to CUR-GZ, while CUR-GZ P showed significantly higher blood concentration suggesting the stealth property of pullulan.
Fig. 5.

Comparative biodistribution profile of 99mTc-labelled CUR-GZ, CUR-GZ AG, CUR-GZ KC, and CUR-GZ P 6 h post-intravenous administration (mean ± standard error, n = 4, ***p < 0.01 between CUR-GZ and different CUR-GZ NPs, **p < 0.05 between CUR-GZ and different CUR-GZ NPs)
The hepatic accumulation of ligand anchored nanoparticles over 6 h is depicted in Fig. 6. CUR-GZ, CUR-GZ P, and CUR-GZ KC revealed an increase in hepatic accumulation over a period of 6 h while CUR-GZ AG revealed maximum concentration at 1 h followed by a decrease in concentration over time. These observations are confirmed from the scintigraphic images. The scintigraphic images of radiolabelled CUR-GZ NPs after 1, 3, and 6 h post-intravenous administration is shown in Fig. 7.
Fig. 6.

Hepatic accumulation of 99mTc-labelled CUR-GZ, CUR-GZ AG, CUR-GZ KC, and CUR-GZ P 1, 3, and 6 h post-intravenous administration (mean ± standard error, n = 4, ***p < 0.01 between CUR-GZ and different CUR-GZ NPs)
Fig. 7.
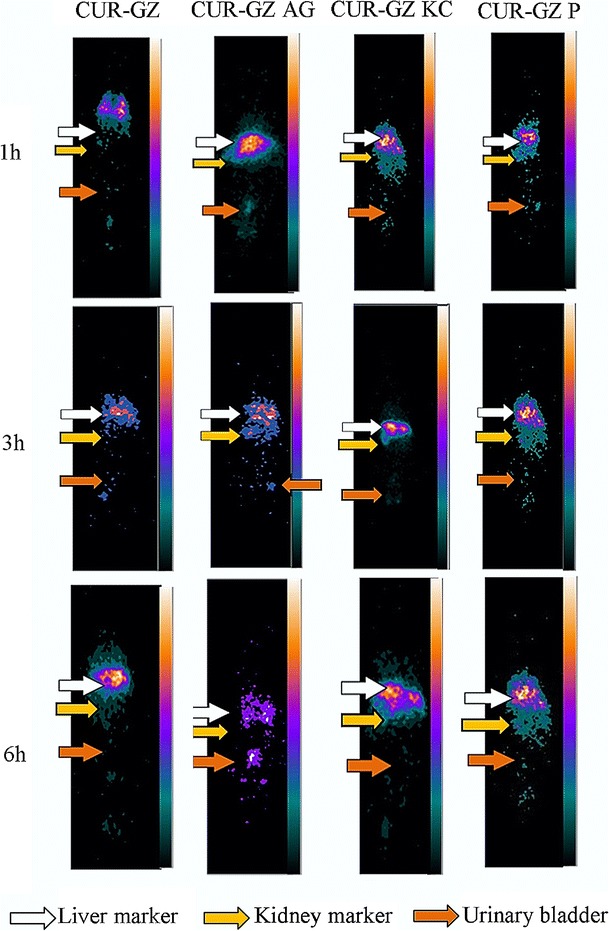
Gamma scintigraphic images a 1 h, b 3 h, c and 6 h post-intravenous administration of 99mTc-labelled CUR-GZ, CUR-GZ AG, CUR-GZ KC, and CUR-GZ P
DISCUSSION
Gantrez is a high molecular weight anhydride polymer which rapidly hydrolyses in aqueous medium to release a number of anionic groups (−COOH). The negative zeta potential of the CUR-GZ NPs is attributed to this property of Gantrez. The increase in zeta potential, significant with arabinogalactan and pullulan as ligands and less significant with kappa carrageenan, is attributed to the negative charge on kappa carrageenan due to galactose sulfate moieties. Moreover, the alteration in zeta potential confirmed the adsorption of ligands on the nanoparticles. This method of functionalization by nonspecific adsorption of ligands onto preformed nanoparticles provides the advantage of being simple (40–43). The negative charge of nanoparticles suggests the possibility of longer circulation and hence a higher probability of bypass of the Kupffer cells to reach hepatocytes (44). High serum stability and low in vitro hemolysis suggested suitability of nanoparticles for intravenous administration.
Docking of polymers is generally a subject of debate due to the myriad conformations a polymer can acquire in solutions. Massarelli (21) reported docking of the monomeric carbohydrates, d-glucuronic acid, mannuronic acid, iduronic acid, sulfated l-rhamnose, and d-xylose, the basic structural units for alginates and ulvans, to explain affinity of these polymers with ASGP-R. In our study, we evaluated both monomers as dimers. While monomers represent chemical units, dimers could function as representatives of stereochemical polymer units, due to their greater constraints, and hence could better mimic polymer conformation and interactions in vivo compared to monomers.
Human ASGP-R (human lectin) is a calcium dependent, C-type receptor on hepatocytes, which recognizes asialoglycoproteins with terminal nonreducing galactose for endocytosis. The binding of ASGP-R was validated using d-galactose as monomer. Binding of galactose to the receptor was stabilized by four hydrogen bonds and hydrophobic bonding of Try 243 of active site of receptor with C3, C4, C5, and C6 atoms of d-galactose. The two and three −OH groups of galactose which formed coordinate bonds with the calcium ion of the receptor coincided exactly with the positions of water (water molecule nos. 11 and 13). The interatomic distance between the coordinate bond of Ca and O2 was 2.8 Å (reported—3.0 Å) (21) and that between Ca and O3 was 2.6 Å (reported—2.9 Å) (21) validated the ASGP-R binding grid. The Ecoulombic energy determined by Glide (−20.71) as opposed to the reported value of −28.84 by Massarelli I (21) using DelPhi is attributed to the PDB structure.
The coordinate bonds, hydrophobic bonds, and hydrogen bonds play a critical role in ligand receptor interaction. Stabilization of ligand with ASGP-R is achieved by coordinate bonds, hydrophobic bonds, and hydrogen bonds. Higher hydrogen bonding is exhibited by the branched carbohydrates (arabinogalactan and pullulan) which could assume conformations that lead to a favorable interaction with the lectin-binding sites, not observed in linear conformations (kappa carrageenan). This correlates with the Glide scores. Coordination of the receptor calcium ion with at least two oxygen atoms of the sugars (3,45) is considered an important parameter for ASGP-R binding. Preferable oxygen atoms are the 3-hydroxyl and 4-hydroxyl. The Ca bond although seen with arabinogalactan and pullulan was absent with kappa carrageenan. This suggests that despite comparable Glide scores, kappa carrageenan would probably exhibit lower affinity to the receptor, which is reflected in the Emodel values (Table II).
At 1 h, the in vivo hepatic accumulation in the order CUR-GZ AG > CUR-GZ KC > CUR-GZ P correlated with the docking data based on Glide scores and is confirmed by the linear and high correlation (Fig. 8) of Glide scores against the hepatic accumulation.
Fig. 8.
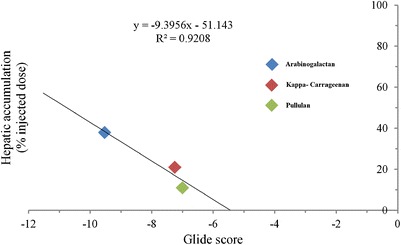
In vivo docking analysis correlation after 1 h of intravenous administration of CUR-GZ NP
Nevertheless, at the end of 6 h, the in vivo data revealed a complete reversal in ligand affinity with pullulan exhibiting maximum hepatic accumulation and arabinogalactan minimum accumulation (p < 0.05) (Figs. 6 and 7). The decrease in CUR-GZ AG is attributed to rapid clearance due to the low molecular weight of arabinogalactan (46). Intense radioactivity seen in the kidney at 3 h and in the bladder at 6 h with arabinogalactan (Fig. 7) confirmed the same, in contrast to pullulan and kappa carrageenan with a higher molecular weight (200 kDa) (47,48), which revealed high hepatic accumulation at 6 h in the order CUR-GZ P > CUR-GZ KC. The higher hepatic accumulation of CUR-GZ P is attributed to the stealth property (27) and hydrophilicity (49).
The data of the docking study based on Glide scores and Emodel values suggest higher affinity of arabinogalactan and pullulan to the ASGP-R, compared to kappa carrageenan and was confirmed from the in vivo data. This suggests arabinogalactan and pullulan as ligands with greater affinity for liver targeting via the ASGP-R.
Docking analysis provides a good indication of ligand receptor affinity based on physicochemical parameters. The properties that influence the ultimate disposition of ligands in vivo are however not accounted for in docking analysis. Nevertheless, the benefits of docking as a tool to predict ligand receptor interaction are not to be undermined. Docking could definitely help in the preliminary screening of ligands for hepatic targeting.
CONCLUSION
The present study reveals that arabinogalactan as a ligand could enable rapid and high uptake of curcumin nanoparticles, while pullulan as ligand facilitated prolonged hepatic retention of the nanoparticles. More importantly, arabinogalactan and pullulan revealed significantly lower lung and heart accumulation suggesting their relevance as targeting ligands, in the design of nanoparticles with improved cardio and pulmonary toxicity profiles.
Electronic supplementary material
Interactions of (a) β(1→3) d-galactose dimer (b) β(1→6) d-galactose dimer (c) arabinose–galactose dimer (d) α(1→4) d-glucose dimer (e) α(1→6) d-glucose dimer (f) dimer of kappa carrageenan with the binding site of ASGP-R (JPEG 154 kb)
Radiolabelling efficiency of CUR-GZ, CUR-GZ AG, CUR-GZ KC and CUR-GZ P (mean ± standard error, n = 3) (JPEG 24 kb)
Acknowledgments
Anisha A. D’Souza and Puneet Jain are thankful to the Institute of Chemical Technology, Mumbai and University Grants Commission, India for fellowship.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- 1.Moghimi SM, Hunter AC, Murray JC. Long-circulating and target-specific nanoparticles: theory to practice. Pharmacol Rev. 2001;53:283–318. [PubMed] [Google Scholar]
- 2.Sadauskas E, Wallin H, Stoltenberg M, Vogel U, Doering P, Larsen A, Danscher G. Kupffer cells are central in the removal of nanoparticles from the organism. Part Fibre Toxicol. 2007;4:10. doi: 10.1186/1743-8977-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, Nantz MH, Zern MA. Targeting hepatocytes for drug and gene delivery: emerging novel approaches and applications. Front Biosci. 2002;7:717–25. doi: 10.2741/wu2. [DOI] [PubMed] [Google Scholar]
- 4.Harvey HA, Porat N, Campbell CA, Jennings M, Gibson BW, Phillips NJ, Apicella MA, Blake MS. Gonococcal lipooligosaccharide is a ligand for the asialoglycoprotein receptor on human sperm. Mol Microbiol. 2000;36(5):1059–70. doi: 10.1046/j.1365-2958.2000.01938.x. [DOI] [PubMed] [Google Scholar]
- 5.Park JH, Kim KL, Cho EW. Detection of surface asialoglycoprotein receptor expression in hepatic and extra-hepatic cells using a novel monoclonal antibody. Biotechnol Lett. 2006;28(14):1061–9. doi: 10.1007/s10529-006-9064-0. [DOI] [PubMed] [Google Scholar]
- 6.Ramadugu SK, Chung YH, Fuentes EJ, Rice KG, Margulis CJ. In silico prediction of the 3D structure of trimeric asialoglycoprotein receptor bound to triantennary oligosaccharide. J Am Chem Soc. 2010;132:9087–95. doi: 10.1021/ja1021766. [DOI] [PubMed] [Google Scholar]
- 7.Akiyoshi K, Kobayashi S, Shichibe S, Mix D, Baudys M, Kim SW, Sunamoto J. Self-assembled hydrogel nanoparticle of cholesterol-bearing pullulan as a carrier of protein drugs: complexation and stabilization of insulin. J Contr Release. 1998;54(3):313–20. doi: 10.1016/S0168-3659(98)00017-0. [DOI] [PubMed] [Google Scholar]
- 8.Hosseinkhani H, Aoyama T, Ogawa O, Tabata Y. Liver targeting of plasmid DNA by pullulan conjugation based on metal coordination. J Contr Release. 2002;83(2):287–302. doi: 10.1016/S0168-3659(02)00201-8. [DOI] [PubMed] [Google Scholar]
- 9.Jo J, Ikai T, Okazaki A, Yamamoto M, Hirano Y, Tabata Y. Expression profile of plasmid DNA by spermine derivatives of pullulan with different extents of spermine introduced. J Contr Release. 2007;118(3):389–98. doi: 10.1016/j.jconrel.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Kaneo Y, Tanaka T, Nakano T, Yamaguchi Y. Evidence for receptor-mediated hepatic uptake of pullulan in rats. J Contr Release. 2001;70:365–73. doi: 10.1016/S0168-3659(00)00368-0. [DOI] [PubMed] [Google Scholar]
- 11.Kim H, Na K. Evaluation of succinylated pullulan for long-term protein delivery in poly(lactide-co-glycolide) microsphere. Macromol Res. 2010;18(8):812–9. doi: 10.1007/s13233-010-0814-4. [DOI] [Google Scholar]
- 12.Kun N, Bae YH. Self-assembled hydrogel nanoparticles responsive to tumor extracellular pH from pullulan derivative/sulfonamide conjugate: characterization, aggregation, and adriamycin release in vitro. Pharm Res. 2002;19(5):681–8. doi: 10.1023/A:1015370532543. [DOI] [PubMed] [Google Scholar]
- 13.Na K, Bae YH. Self-assembled hydrogel nanoparticles responsive to tumor extracellular pH from pullulan derivative/sulfonamide conjugate: characterization, aggregation, and adriamycin release in vitro. Pharm Res. 2002;19(5):681–8. doi: 10.1023/A:1015370532543. [DOI] [PubMed] [Google Scholar]
- 14.Rekha MR, Sharma CP. Pullulan as a promising biomaterial for biomedical applications: a perspective. Trends Biomater Artif Organs. 2007;20(2):116–21. [Google Scholar]
- 15.Ashwell G, Harford J. Carbohydrate-specific receptors of the liver. Annu Rev Biochem. 1982;51:531–4. doi: 10.1146/annurev.bi.51.070182.002531. [DOI] [PubMed] [Google Scholar]
- 16.Hashida M, Nishikawa M, Yamashita F, Takakura Y. Cell-specific delivery of genes with glycosylated carriers. Adv Drug Deliv Rev. 2001;52(3):187–96. doi: 10.1016/S0169-409X(01)00209-5. [DOI] [PubMed] [Google Scholar]
- 17.Niidome T, Urakawa M, Sato H, Takahara Y, Anai T, Hatakayama T, Wada A, Hirayama T, Aoyagi H. Gene transfer into hepatoma cells mediated by galactose-modified α-helical peptides. Biomaterials. 2000;21:1811–9. doi: 10.1016/S0142-9612(00)00076-4. [DOI] [PubMed] [Google Scholar]
- 18.Nishikawa M, Takemura S, Yamashita F, Takakura Y, Meijer DKF, Hashida M, Swart P. Pharmacokinetics and in vivo gene transfer of plasmid DNA complexed with mannosylated poly(L-lysine) in mice. J Drug Target. 2000;8:29. doi: 10.3109/10611860009009207. [DOI] [PubMed] [Google Scholar]
- 19.Ren T, Zhang G, Liu D. Synthesis of galactosyl compounds for targeted gene delivery. Bioorg Med Chem. 2001;9:2969–78. doi: 10.1016/S0968-0896(01)00203-6. [DOI] [PubMed] [Google Scholar]
- 20.Meier M, Bider MD, Malashkevich VN, Spiess M, Burkhard P. Crystal structure of the carbohydrate recognition domain of the H1 subunit of the asialoglycoprotein receptor. J Mol Biol. 2000;300(4):857–65. doi: 10.1006/jmbi.2000.3853. [DOI] [PubMed] [Google Scholar]
- 21.Massarelli I, Murgia L, Bianucci AM, Chiellini F, Chiellini E. Understanding the selectivity mechanism of the human asialoglycoprotein receptor (ASGP-R) toward Gal- and Man- type ligands for predicting interactions with exogenous sugars. Int J Mol Sci. 2007;8:13–28. doi: 10.3390/i8010013. [DOI] [Google Scholar]
- 22.Anand P, Thomas SG, Kunnumakkara AB, Sundaram C, Harikumar KB, Sung B, Tharakan ST, Misra K, Priyadarsini IK, Rajasekharan KN, Aggarwal BB. Biological activities of curcumin and its analogues (congeners) made by man and mother nature. Biochem Pharmacol. 2008;76:1590–611. doi: 10.1016/j.bcp.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Yoysungnoen P, Wirachwong P, Changtam C, Suksamrarn A, Patumraj S. Anti-cancer and anti-angiogenic effects of curcumin and tetrahydrocurcumin on implanted hepatocellular carcinoma in nude mice. World J Gastroenterol. 2008;14:2003–9. doi: 10.3748/wjg.14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Date PV, Samad A, Devarajan PV. Freeze thaw: a simple approach for prediction of optimal cryoprotectant for freeze drying. AAPS PharmSciTech. 2010;11(1):304–13. doi: 10.1208/s12249-010-9382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devarajan PV, Sonavane GS, Doble M. Computer-aided molecular modeling: a predictive approach in the design of nanoparticulate drug delivery system. J Biomed Nanotechnol. 2005;1:1–9. doi: 10.1166/jbn.2005.051. [DOI] [Google Scholar]
- 26.Bock TK, Muller BW. A novel assay to determine the hemolytic activity of drugs incorporated in colloidal carrier systems. Pharm Res. 1994;11(4):589. doi: 10.1023/A:1018987120738. [DOI] [PubMed] [Google Scholar]
- 27.Guhagarkar SA, Gaikwad RV, Samad A, Malshe VC, Devarajan PV. Polyethylene sebacate-doxorubicin nanoparticles for hepatic targeting. Int J Pharm. 2010;401(1–2):113–22. doi: 10.1016/j.ijpharm.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Glide, version 5.8, Schrödinger, LLC, New York, NY, 2012.
- 29.Maestro, version 9.3, Schrödinger, LLC, New York, NY, 2012.
- 30.Rick P, Millane RP, Chandrasekaran R, Arnott S. The molecular structure of kappa-carrageenan and comparison with iota-carrageenan. Carbohydr Res. 1988;182(1):1–17. doi: 10.1016/0008-6215(88)84087-4. [DOI] [Google Scholar]
- 31.Redgwell RJ, Curti D, Fischer M, Nicolas P, Fay LB. Coffee bean arabinogalactans: acidic polymers covalently linked to protein. Carbohydr Res. 2002;337:239–53. doi: 10.1016/S0008-6215(01)00316-0. [DOI] [PubMed] [Google Scholar]
- 32.Tryfona T, Liang HC, Kotake T, Kaneko S, Marsh J, Ichinose H, Lovegrove A, Tsumuraya Y, Shewry PR, Stephens E, Dupree P. Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr Res. 2010;345(18):2648–56. doi: 10.1016/j.carres.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 33.LigPrep, version 2.5, Schrödinger, LLC, New York, NY, 2012.
- 34.Reddy L, Sharma R, Chuttani K, Mishra A, Murthy R. Etoposide-incorporated tripalmitin nanoparticles with different surface charge: formulation, characterization, radiolabeling, and biodistribution studies. AAPS J. 2004;6(3):55–64. doi: 10.1208/aapsj060323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saez A, Guzma’n M, Molpeceres J, Aberturas MR. Freeze drying of polycaprolactone and poly(d,l-lactic-glycolic) nanoparticles induce minor particle size changes affecting the oral pharmacokinetics of loaded drugs. Eur J Pharm Biopharm. 2000;50:379–87. doi: 10.1016/S0939-6411(00)00125-9. [DOI] [PubMed] [Google Scholar]
- 36.Ogain ON, Li J, Tajber L, Corrigan OI, Healy AM. Particle engineering of materials for oral inhalation by dry powder inhalers. I-Particles of sugar excipients (trehalose and raffinose) for protein delivery. Int J Pharm. 2011;405(1–2):23–35. doi: 10.1016/j.ijpharm.2010.11.039. [DOI] [PubMed] [Google Scholar]
- 37.Amin K, Dannenfelser RM. In vitro hemolysis: guidance for the pharmaceutical scientist. J Pharm Sci. 2006;95(6):1173–6. doi: 10.1002/jps.20627. [DOI] [PubMed] [Google Scholar]
- 38.Dobrovolskaia MA, Clogston JD, Neun BW, Hall JB, Patri AK, McNeil SE. Method for analysis of nanoparticle hemolytic properties in vitro. Nano Lett. 2008;8(8):2180–7. doi: 10.1021/nl0805615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 40.Iden DL, Allen TM. In vitro and in vivo comparison of immunoliposomes made by conventional coupling techniques with those made by a new post-insertion approach. Biochim Biophys Acta. 2001;1513:207–16. doi: 10.1016/S0005-2736(01)00357-1. [DOI] [PubMed] [Google Scholar]
- 41.Illum L, Jones PDE, Kreuter J, Baldwin RW, Davis SS. Adsorption of monoclonal antibodies to polyhexylcyanoacrylate nanoparticles and subsequent immunospecific binding to tumour cells in vitro. Int J Pharm. 1983;17(1):65–76. doi: 10.1016/0378-5173(83)90019-4. [DOI] [Google Scholar]
- 42.Zhang L, Yu F, Cole AJ, Chertok B, David AE, Wang J, Yang VC. Gum arabic coated magnetic nanoparticles for potential application in simultaneous magnetic targeting and tumor imaging. AAPS J. 2009;11(4):693–9. doi: 10.1208/s12248-009-9151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexis F, Pridgen E, Linda K, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2005;5(4):505–15. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichikawa Y, Lee RT, Lee YC. Synthesis of N-acetylglucosamine derivatives as probes for specificity of chicken hepatic lectin. Glycoconj J. 1990;7(4):335–48. doi: 10.1007/BF01073377. [DOI] [PubMed] [Google Scholar]
- 45.Lee YC. Biochemistry of carbohydrate-protein interaction. FASEB J. 1992;6:3193–200. doi: 10.1096/fasebj.6.13.1397841. [DOI] [PubMed] [Google Scholar]
- 46.Kaneo Y, Ueno T, Tanaka T, Iwase H, Yamaguchi Y, Uemura T. Pharmacokinetics and biodisposition of fluorescein-labeled arabinogalactan in rats. Int J Pharm. 2000;201:59–69. doi: 10.1016/S0378-5173(00)00405-1. [DOI] [PubMed] [Google Scholar]
- 47.Dexter LB. Hayashibara International Inc. Pullulan GRAS Claim. 2002. http://www.accessdata.fda.gov/scripts/fcn/gras_notices/215492e.pdf.
- 48.Weiner ML, Nuber D, Blakemore WR, Harriman JF, Cohen SM. A 90-day dietary study on kappa carrageenan with emphasis on the gastrointestinal tract. Food Chem Toxicol. 2007;45(1):98–106. doi: 10.1016/j.fct.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 49.Hasuda H, Kwon OH, Kang IK, Ito Y. Synthesis of photoreactive pullulan for surface modification. Biomaterials. 2005;26(15):2401–6. doi: 10.1016/j.biomaterials.2004.07.065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Interactions of (a) β(1→3) d-galactose dimer (b) β(1→6) d-galactose dimer (c) arabinose–galactose dimer (d) α(1→4) d-glucose dimer (e) α(1→6) d-glucose dimer (f) dimer of kappa carrageenan with the binding site of ASGP-R (JPEG 154 kb)
Radiolabelling efficiency of CUR-GZ, CUR-GZ AG, CUR-GZ KC and CUR-GZ P (mean ± standard error, n = 3) (JPEG 24 kb)


