Abstract
Compound promiscuity refers to the ability of small molecules to specifically interact with multiple targets, which represents the origin of polypharmacology. Promiscuity is thought to be a widespread characteristic of pharmaceutically relevant compounds. Yet, the degree of promiscuity among active compounds from different sources remains uncertain. Here, we report a thorough analysis of compound promiscuity on the basis of more than 1,000 PubChem confirmatory bioassays, which yields an upper-limit assessment of promiscuity among active compounds. Because most PubChem compounds have been tested in large numbers of assays, data sparseness has not been a limiting factor for the current analysis. We have determined that there is an overall likelihood of ∼50% of an active PubChem compound to interact with two or more targets. The probability to interact with more than five targets is reduced to 7.6%. On average, an active PubChem compound was found to interact with ∼2.5 targets. Moreover, if only activities consistently detected in all assays available for a given target were considered, this ratio was further reduced to ∼2.3 targets per compound. For comparison, we have also analyzed high-confidence activity data from ChEMBL, the major public repository of compounds from medicinal chemistry, and determined that an active ChEMBL compound interacted on average with only ∼1.5 targets. Taken together, our results indicate that the degree of compound promiscuity is lower than often assumed.
KEY WORDS: active compounds, activity measurements, compound promiscuity, confirmatory assays, polypharmacology, screening data, targets
INTRODUCTION
Studying interactions between bioactive compounds or drugs and biological targets, experimentally and/or computationally, has become a major topic in pharmaceutical research (1–8). Compound promiscuity is rationalized as the ability of small molecules to specifically interact with multiple targets (but does not imply non-specific binding effects). The increasing notion of compound promiscuity and ensuing polypharmacology is beginning to change specificity-centric drug discovery paradigms, at least in some therapeutic areas such as oncology (5,6). In addition, target promiscuity is also responsible for undesired (or desired) side effects of drugs (9,10). Hence, it is not surprising that much attention is currently being paid to promiscuity-related drug actions, be they positive or negative for therapy.
While it is generally accepted that polypharmacological effects often play an important role in drug efficacy and side effects, the actual degree of promiscuity among pharmaceutically relevant compounds remains unclear. Early estimates proposed that drugs might on average bind to about three to six targets. For example, when collecting activity annotations for ∼800 drugs active against ∼400 targets from drug discovery databases, a drug was found to interact with on average 2.7 targets (4). When hypothetical drug–target interactions predicted on the basis of chemical drug similarity were additionally taken into account, this target-to-drug ratio further increased to ∼6.3 (4). In addition, when all publicly available activity annotations of drugs were considered (IC50, EC50, Ki, or Kd values < 10 μM), it was found that more than 50% of current drugs interacted with more five targets and that drugs directed against G protein-coupled receptors, which was the most promiscuous drug class investigated in this study, had on average six to seven targets (8).
However, compound data selection criteria and the types of activity measurements that are considered also affect the outcome of promiscuity analysis. Depending on the source database, for example, PubChem (the major public repository of screening data) (11), ChEMBL (the major public repository for compound data from medicinal chemistry) (12), or DrugBank (13), and the data selection criteria that are applied, the number of activity/target annotations of a bioactive compound or drug might significantly vary. For example, for the drugs milrinone and buspirone, it was found that ChEMBL reported 42 and 72 potential targets, respectively (14). By contrast, in DrugBank, these drugs were only annotated with one and two targets, respectively (14). Such apparent discrepancies raise questions and might lead to inconsistent analyses. However, when compound activity data in ChEMBL were filtered and only ligand–target interactions at the highest confidence level were considered, together with equilibrium constants as activity measurements, the number of target annotations for milrinone and buspirone was reduced to one and two, respectively, consistent with DrugBank (14). Such examples are abundant and illustrate the critical role of data selection criteria for the outcome of promiscuity analysis.
It has also been observed that the amount of compounds with multiple target annotations available in ChEMBL has substantially increased over time (15). However, most of this apparent growth in compound promiscuity has resulted from assay-dependent IC50 data. By contrast, when only increasing amounts of Ki values were analyzed, there was no notable increase in across different target families over time (15). Compound promiscuity is often restricted to closely related targets. For example, in an analysis of high-confidence activity data in ChEMBL, it was shown that only 2.4% of all promiscuous compounds interacted with targets from different protein families (16). Hence, compound promiscuity across distantly or unrelated targets was a rare event.
Taken together, the findings discussed above clearly illustrate that a meaningful evaluation of compound promiscuity is more complicated than it might appear at a first glance. It remains difficult to determine the “true” degree of promiscuity among active compounds from different sources.
Herein, we present an analysis designed to further refine our view of compound promiscuity that focuses on PubChem confirmatory bioassays (11), which confirm the activity of hits from biological screens and eliminate false positives. In confirmatory bioassays, compound activity is assessed on the basis of titration curves and IC50 value determination. An intrinsic advantage of focusing on confirmatory screening data is that PubChem compounds have typically been tested in many different assays, which works against the often quoted underestimation of compound promiscuity due to data sparseness (i.e., many active compounds might have only been tested against one or a few targets) (4). Thus, given the availability of many assay readouts for confirmed active compounds and their screening hit character, one would anticipate that the analysis of these data should yield an upper-level estimate of promiscuity among bioactive compounds. To put the results into perspective, a comparative promiscuity analysis of PubChem confirmatory screening data and compound activity data from ChEMBL was also carried out. Taken together, our results reveal a lower degree of promiscuity among active compounds than expected.
MATERIALS AND METHODS
Data Collection
From the PubChem BioAssay database (11), confirmatory assays with dose–response data and activity against a single protein target were assembled. For each assay, only compounds explicitly designated as “active” or “inactive” were further considered. Compounds annotated as “inconclusive” or “unspecified” were omitted. In addition, qualifying assays were further grouped according to their targets. From all compounds, scaffolds were extracted by removing all R-groups from ring systems and ring-connecting substructures (17). Hence, these scaffolds represent compound core structures.
Promiscuity Assessment
For all compounds, the number of confirmatory assays in which they were active and the number of targets they were active against were determined. For compounds active in multiple assays against the same target, the consistency of the activity across different assays was recorded.
As a control, promiscuity was also analyzed for bioactive compounds assembled from ChEMBL, release 14 (12). Compounds with reported direct interactions against human targets (with highest ChEMBL confidence score of 9) (12) and explicit Ki or IC50 values were collected. These two types of potency measurements were separately considered. Therefore, two subsets of compounds with Ki or IC50 values were generated.
For a compound, an assay promiscuity index (API) was calculated by dividing the number of assays with reported activity by the number of assays that the compound was tested in.
All data mining calculations were carried out with in-house generated programs.
RESULTS AND DISCUSSION
Analysis Concept
By systematically analyzing confirmed activity data of compounds from more than 1,000 assays against more than 400 targets, we were able to assess the general tendency of compounds to be promiscuous. Confirmatory follow-up assays, the source of our analysis, relied on titration experiments yielding IC50 values and hence provided a consistent data structure for our analysis. Because most compounds were tested in large numbers of assays and no potency threshold was applied to confirmed active compounds, the analysis was expected to provide an upper-level estimate of compound promiscuity. This was further assessed by carrying out a comparative promiscuity analysis for ChEMBL compounds.
Assay, Target, and Compound Distribution
Table I provides a summary of the assay and compound data we analyzed. Active compounds were extracted from a total of 1,085 qualifying confirmatory assays that involved 439 different targets. Thus, for individual targets, multiple assays were often available. Table II lists a subset of targets with multiple assays. PubChem screening targets were diverse and involved many different enzymes and receptors. From confirmatory bioassays, a total of 437,288 compounds were obtained, 140,112 of which were active in one or more assays. In addition, 297,176 compounds were found to be consistently inactive in all assays they were tested in. A subset of 179,189 consistently inactive compounds was tested in more than 50 different confirmatory assays.
Table I.
Confirmatory Assays, Targets, and Compounds
| Number of | PubChem BioAssay | |
|---|---|---|
| Confirmatory assays | 1,085 | |
| Targets | 439 | |
| Compounds | Total | 437,288 |
| Active | 140,112 | |
| Compound-assay combinations | Total | 25,996,196 |
| Active | 358,655 | |
| Inactive | 25,637,541 | |
The number of qualifying confirmatory assays taken from the PubChem BioAssay database and the corresponding number of targets are reported. In addition, the total number of compounds in qualifying confirmatory assays and number of compounds that were active in one or more assays are given. Furthermore, compound-assay combinations are provided. “Active” and “Inactive” report the number of compound-assay pairings involving active or inactive compounds, respectively
Table II.
Targets with Multiple Assays
| Target name | Number of | |
|---|---|---|
| Assays | Active compounds | |
| Glucocerebrosidase | 17 | 413 |
| Opioid receptor, kappa 1 | 16 | 71 |
| Apelin receptor | 13 | 258 |
| Alkaline phosphatase, intestinal | 12 | 289 |
| Intestinal alkaline phosphatase | 11 | 373 |
| ATAD5 protein | 11 | 4169 |
| NADPH oxidase 1 | 11 | 55 |
Seven targets associated with more than 10 confirmatory assays are reported. For each target, the number of assays and the number of active compounds are provided.
ATAD5 ATPase family AAA domain-containing protein 5, NADPH nicotinamide adenine dinucleotide phosphate
From all active and inactive compounds, a total of 101,876 unique scaffolds were extracted. Of these, 48,299 scaffolds were exclusively represented by consistently inactive compounds. Furthermore, assays and tested compounds yielded nearly 26 million compound-assay pairings, 358,655 of which involved active compounds.
Assay Frequency
We also determined the distribution of active compounds over screening assays. Figure 1a shows how many compounds found to be active in at least one assay were tested in how many different assays. Compounds were tested in different assays with high frequency. The distribution in Fig. 1a reveals that more than 77% of all active compounds were tested in more than 50 different assays. Thus, there was broad assay coverage of active compounds, which provided a sound basis for promiscuity analysis.
Fig. 1.
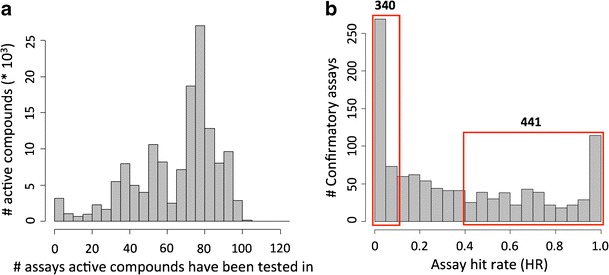
Active compounds from confirmatory assays. a For compounds active in one or more confirmatory assays, the distribution of the total number of assays these compounds have been tested in is shown. For example, more than 25,000 compounds have been tested in 75–80 assays. b For assays with confirmed active compounds, the distribution of hit rates is shown. Subsets of 340 and 441 assays yielded hit rates of less than 10% or more than 40%, respectively
Assay Hit Rates
For the 1,085 qualifying assays, hit rates were calculated by dividing the number of compounds with reported activity by the total number of compounds that were tested. The distribution shown in Fig. 1b reflects a broad range of hit rates. A total of 340 assays yielded very low hit rates of less than 10%, whereas 441 assays produced hit rates higher than 40%.
Assay Relationships
We further analyzed relationships among confirmatory assays. Assay pairs were generated that shared at least five tested compounds. A total of 100,043 assay pairs involving 1,021 assays were identified. On average, each assay formed relationships (i.e., shared at least five compounds) with other 98 assays. These findings indicated that a large number of compounds were tested in many different assays. Furthermore, assay pairs were determined that shared at least five active compounds. Under this condition, 14,753 assay pairs were detected that involved a total of 775 assays. On average, an assay was found to share at least five active compounds with 19 other assays.
Activity Assessment
Figure 2 reports the distribution of compounds over assays in which they were active. Nearly 47% of all active compounds were only active in a single assay, and an additional 21% of the compounds were active in two assays. Overall, compound activity in up to 48 assays was observed, but there was a steady decline in compound numbers with increasing numbers of assays with reported activity. Only a total of 322 of nearly half a million tested compounds were confirmed to be active in more than 20 assays. Figure 3 reports the distribution of active compounds over targets against which they were active. Again, almost 50% of all compounds were only active against a single target and an additional 21% active against two targets. With increasing target numbers, compound numbers were further reduced compared to Fig. 2 because multiple assays were frequently available for individual targets. Compound activity against up to 40 targets was observed, but only a total of 160 compounds were confirmed to be active against more than 20 targets and only seven compounds against more than 30 targets (Fig. 3).
Fig. 2.
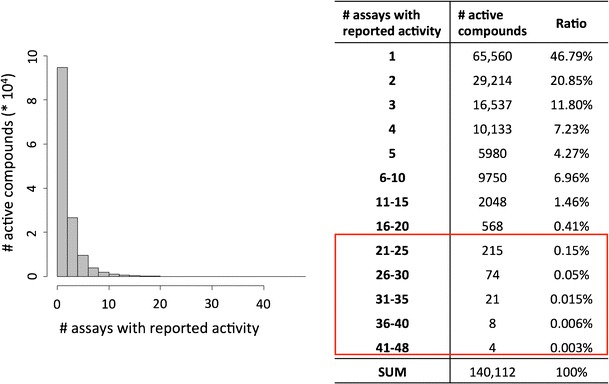
Distribution of compounds and assays with reported activity. For all active compounds, the number of assays in which they were active is reported in a histogram and table format. For example, nearly 100,000 compounds were active in one or two assays. On the right, the distribution of compounds with activity in more than 20 assays is highlighted using a red frame
Fig. 3.

Distribution of compounds and targets with reported activity. For all active compounds, the number of targets they were active against is reported in a histogram and table format. For example, nearly 100,000 compounds were active against one or two targets. On the right, the distribution of compounds with activity against more than 20 targets is highlighted using a red frame
Highly Promiscuous Compounds
As shown in Figs. 2 and 3, highly promiscuous compounds were only detected with low frequency. There were only a total of 2,037 compounds with activity against more than 10 targets (Fig. 3). Representative examples are provided in Fig. 4. For these structurally diverse compounds, assays in which they were tested and found to be active were compared to evaluated and confirmed targets. In each case, the number of assays exceeded the number of targets by ∼20 or more and the compounds were found to be active against approximately half of the targets they were tested against. These compounds represented the most promiscuous compounds detected in PubChem confirmatory assays. Because the compounds displayed dose–response behavior for all activities, the probability of artifacts was low.
Fig. 4.
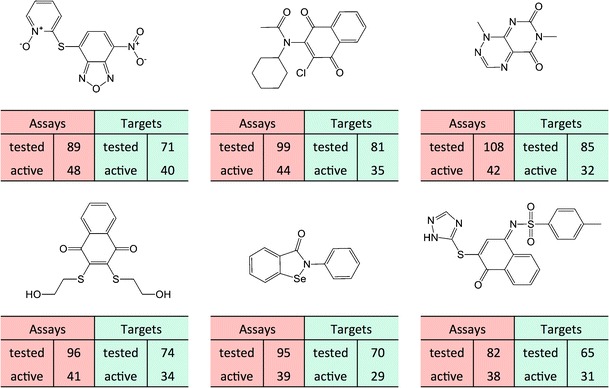
Promiscuous compounds. Six exemplary highly promiscuous compounds are shown. For each compound, the numbers of assays in which it was tested and in which it displayed activity are reported. Furthermore, the numbers of targets against which the compound was tested and against which it was active are given
In addition, the probability that a highly promiscuous compound was reported to be active in a given assay was assessed by API calculations. The distribution of API values is reported in Fig. 5a. The majority of the highly promiscuous compounds yielded an API between 0.15 and 0.25, indicating that these compounds were confirmed as active in 15% to 25% of all assays that they have been tested in. Furthermore, the correlation between API and the number of targets with reported compound activity is shown in Fig. 5b. Only a weak correlation was observed (with a correlation coefficient of 0.62). In addition, we also analyzed the relationship between API and the average hit rate of all assays in which a compound was tested. The results are reported in Fig. 5c. Nearly all promiscuous compounds originated from assays with an average hit rate of less than 20%.
Fig. 5.

Highly promiscuous compounds. a For a total of 2,037 highly promiscuous compounds with activity against more than 10 targets, the distribution of their API values is shown. In addition, the correlations between API and the number of targets with reported activity and the average hit rate of assays that each compound was tested in are reported using scatter plots in b and c, respectively. In these scatter plots, each data point represents a highly promiscuous compound. Correlation coefficients are also reported
Promiscuous Targets
The 2,037 most promiscuous compounds were found to be active against a total of 365 targets. For these targets, the distribution of promiscuous compounds is reported in Fig. 6. The number of promiscuous compounds active against a target ranged from 1 to 1,947. Among these, 133 targets had less than 20 promiscuous compounds and 70 targets more than 1,000 promiscuous compounds. The most promiscuous targets are listed in Table III. These targets belonged to a variety of different families.
Fig. 6.
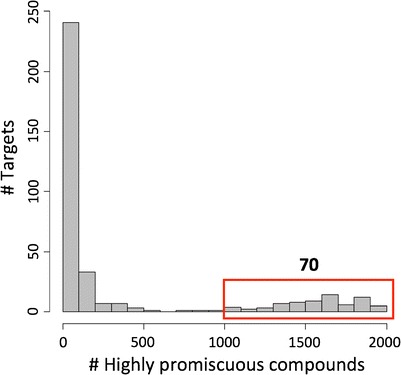
Target sets containing highly promiscuous compounds. For targets of highly promiscuous compounds, the distribution of the number of promiscuous compounds is shown. A total of 70 targets had more than 1,000 promiscuous compounds
Table III.
Top-Ranked Promiscuous Targets
| Target name | Number of highly promiscuous compounds |
|---|---|
| Nonstructural protein 1 | 1,947 |
| Thioredoxin reductase | 1,928 |
| Chain A, ampc beta-lactamase in complex with 4-methanesulfonylamino benzoic acid | 1,922 |
| Relaxin/insulin-like family peptide receptor 1 | 1,917 |
| Beta-2 adrenergic receptor | 1,916 |
| Acid alpha-glucosidase preproprotein | 1,896 |
| Ubiquitin carboxyl-terminal hydrolase 2 isoform a | 1,882 |
| Niemann-Pick C1 protein precursor | 1,865 |
| Heat shock 70 kDa protein 5 (glucose-regulated protein, 78 kDa) | 1,864 |
| Ras-related protein rab-9A | 1,864 |
| Isocitrate dehydrogenase 1 | 1,856 |
| T cell receptor | 1,847 |
| Chaperonin-containing T-complex protein 1 beta subunit homolog | 1,847 |
| Eyes absent homolog 2 isoform a | 1,838 |
| Acid sphingomyelinase | 1,815 |
| Serine-protein kinase ATM isoform 1 | 1,810 |
| Glucocerebrosidase | 1,808 |
| Tumor susceptibility gene 101 protein | 1,796 |
| Pyruvate kinase | 1,790 |
| Pyruvate kinase, muscle isoform M2 | 1,762 |
Shown are top 20 targets having the largest number of highly promiscuous compounds with activity against more than 10 targets
Stringent Activity Annotations
Figure 7 reports the numbers of assays for the 439 targets available in PubChem. For 181 targets, only a single assay was available, but for the remaining 258 targets, multiple assays were available, with up to 17 assays per target. Due to the availability of multiple assays for many targets, it was possible to further refine activity annotations by evaluating their consistency in different assays. Therefore, we determined for all relevant compounds if they were consistently active in all assays for a given target they were tested in. For example, if a compound was tested in three assays available for a particular target and found to be active in only one or two of them, the target annotation was discarded. This consistency assessment further increased the stringency of activity annotations. To derive a statistic comparable to the one reported in Fig. 3, active compounds tested in single assays were also considered consistently active. The distribution of active compounds over targets on the basis of stringent/consistent activity annotations is reported in Fig. 8. For ∼8.6% of all active compounds, no consistent activity annotations in multiple assays were identified. On the basis of stringent activity annotations, the degree of target promiscuity was further reduced compared to original annotations, as one would expect. As shown in Fig. 8, there were 1,446 compounds with consistent activity annotations against more than 10 targets (compared to 2,037 on the basis of original annotations) and the maximal number of targets of an active compound was 39.
Fig. 7.
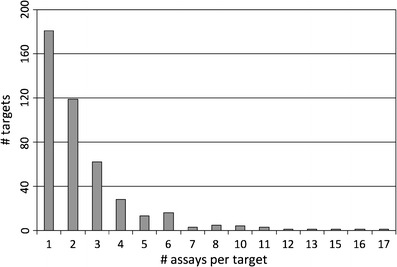
Assays per target. The distribution of assays over targets is shown. For more than 180 targets, a single assay was available. The largest number of assays per target was 17
Fig. 8.
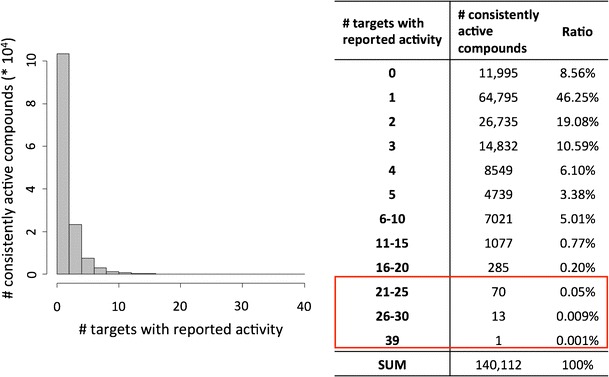
Compounds with consistent assay activity. For all active compounds, the number of targets is reported against which the compounds were consistently active in all assays they were tested in. Approximately 8.6% of all compounds were not consistently active in two or more assays for a given target (i.e., “# targets” is 0)
Comparison with ChEMBL
A comparative promiscuity analysis was also carried out for ChEMBL (12), the major public domain resource of compounds from the medicinal chemistry literature. Because ChEMBL contains a variety of activity measurements, different from PubChem confirmatory bioassays, separate ChEMBL compound subsets were generated for explicit Ki and IC50 measurements. From ChEMBL release 14, 36,542 and 80,522 compounds with Ki and IC50 values were extracted, respectively, following a previously reported protocol focusing on high-confidence data (15). The Ki and IC50 subsets were then subjected to promiscuity analysis in analogy to PubChem compounds. The results are reported in Table IV. Here, 62% and 75% of all compounds from the Ki and IC50 subsets, respectively, were found to be active against a single target. Compound activity against up to 40 (Ki) and 47 (IC50) targets was observed, respectively, but compound numbers rapidly declined for increasing numbers of targets. In the Ki and IC50 subsets, only 192 and 158 compounds were found to be active against more than 10 targets, respectively.
Table IV.
Target Promiscuity of ChEMBL Compounds
| Degree of target promiscuity | ChEMBL 14 | |||
|---|---|---|---|---|
| K i | IC50 | |||
| Number of compounds | Percentage | Number of compounds | Percentage | |
| 1 | 22,700 | 62.12 | 60,624 | 75.29 |
| 2 | 7323 | 20.04 | 13,097 | 16.27 |
| 3 | 3963 | 10.85 | 3993 | 4.96 |
| 4 | 1634 | 4.47 | 1696 | 2.11 |
| 5 | 476 | 1.30 | 468 | 0.58 |
| 6–10 | 254 | 0.70 | 486 | 0.60 |
| 11–15 | 133 | 0.36 | 84 | 0.10 |
| 16–20 | 33 | 0.09 | 38 | 0.05 |
| 21–25 | 15 | 0.04 | 19 | 0.02 |
| 26–30 | 3 | 0.008 | 6 | 0.007 |
| 31–35 | 5 | 0.014 | 2 | 0.002 |
| 36–40 | 3 | 0.008 | 5 | 0.006 |
| 41–47 | – | – | 4 | 0.005 |
| Sum | 36,542 | 100 | 80,522 | 100 |
For ChEMBL release 14, the number (and percentage) of compounds active against different numbers of targets is reported for the K i and IC50 subsets, respectively
Overall Degree of Compound Promiscuity
We then determined the degree of target promiscuity among confirmed active compounds from PubChem and, for comparison, the Ki and IC50 subsets from ChEMBL. Given the distribution of compounds active against single and multiple targets reported in Fig. 3, the likelihood of an active PubChem compound to be promiscuous (i.e., active against two or more targets) was ∼50.9%. The probability of activity against more than five targets was significantly reduced to ∼7.6%. In the Ki and IC50 subsets of ChEMBL, the corresponding likelihoods were ∼37.9% (1.2%; more than five targets) and ∼24.7% (0.8%), respectively. Thus, as expected, confirmatory screening data yielded a higher probability of promiscuity among active compounds than compound optimization data. Furthermore, although most of the growth in promiscuous compounds observed in ChEMBL overtime was attributed to IC50 data (15), which is also reflected by the presence of 13,842 vs. 19,898 multi-target compounds in the Ki and IC50 subsets in Table IV, the ChEMBL Ki subset yielded a higher promiscuity rate than the IC50 subset.
For stringent activity annotations (Fig. 8), the likelihood for an active PubChem compound to be promiscuous was ∼45.2% (6.0%; more than five targets). On average, an active PubChem compound interacted with ∼2.3 (stringent activity annotations) to ∼2.5 targets (original annotations), whereas an active compound from the Ki and IC50 subsets of ChEMBL interacted on average with ∼1.7 and ∼1.4 targets, respectively.
Concluding Remarks
Herein, we have presented a systematic analysis of PubChem confirmatory bioassays to determine the degree of promiscuity among active compounds and compared the results to ChEMBL compounds. Given the large number of assay results available for confirmed active PubChem compounds and their screening hit character (i.e., taking weakly potent compounds into account), the analysis was designed to yield an upper-level assessment of compound promiscuity. Moreover, it is important to emphasize that most active compounds in confirmatory assays were tested in more than 50 different assays. Therefore, the analysis focused on compounds having the largest degree of apparent promiscuity in original assays, which further supports the upper-level assessment of promiscuity presented herein. Activity annotations of confirmed active compounds from more than 1,000 biological screens against more than 400 targets were analyzed. Because the majority of PubChem compounds have been tested in many different assays, the current analysis was based on a more complete data array than previous investigations. Hence, our analysis of PubChem confirmatory bioassays and its comparison with ChEMBL likely provides the most comprehensive assessment of compound promiscuity that is currently available. For active compounds from PubChem, we observed a ∼50% probability to be active against two or more targets, which was significantly reduced to ∼7.6% for activity against more than five targets. On average, an active PubChem compound interacted with ∼2.5 targets, with only minor differences between original and stringent activity annotations. For ChEMBL, the corresponding IC50 data subset yielded on average a ratio of only ∼1.4 targets per active compound. Thus, although the degree of promiscuity was higher for PubChem than for ChEMBL compounds, as expected, it was low in both cases and considerably lower than previous estimates made for drugs that included predicted drug–target interactions or operated on the basis of ligand–target interaction networks. Recent investigations of compound promiscuity and drug polypharmacology referred to in the introductory sections of our study are methodologically distinct and not directly comparable in their details. However, our large-scale and data-driven analysis taking both compounds from screening campaigns and medicinal chemistry projects into account clearly indicates that the degree of target promiscuity among active compounds is lower than often discussed for drugs. These findings are relevant from several viewpoints. The results imply that many compounds act selectively on a primary target. Thus, in medicinal chemistry, compound optimization can first and foremost concentrate on primary target of interest, without taking promiscuity as a potential caveat early on into account. Moreover, for drug discovery, there is good news. While polypharmacology has substantially expanded drug development opportunities, for example, in oncology, many compounds with a high degree of selectivity are available. Given the overall very limited degree of compound promiscuity detected in an analysis focusing on upper-level promiscuity, the likelihood of developing compounds with sufficient target selectivity for therapeutic areas such as infectious diseases or for the treatment of chronic disease states should continue to be high. In summary, a more conservative view of compound promiscuity should be promoted on the basis of currently available compound activity data.
REFERENCES
- 1.Paolini GV, Shapland RHB, van Hoorn WP, Mason JS, Hopkins AL. Global mapping of pharmacological space. Nat Biotechnol. 2006;24(7):805–15. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 2.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25(2):197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–90. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 4.Mestres J, Gregori-Puigjane E, Valverde S, Sole RV. Data completeness—the achilles heel of drug–target networks. Nat Biotechnol. 2008;26(9):983–4. doi: 10.1038/nbt0908-983. [DOI] [PubMed] [Google Scholar]
- 5.Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10(2):130–7. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merino A, Bronowska AK, Jackson DB, Cahill DJ. Drug profiling: knowing where it hits. Drug Discov Today. 2010;15(17–18):749–56. doi: 10.1016/j.drudis.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Xie L, Xie L, Kinnings SL, Bourne PE. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu Rev Pharmacol Toxicol. 2012;52:361–79. doi: 10.1146/annurev-pharmtox-010611-134630. [DOI] [PubMed] [Google Scholar]
- 8.Jalencas X, Mestres J. On the origins of drug polypharmacology. Med Chem Commun. 2013;4(1):80–7. doi: 10.1039/c2md20242e. [DOI] [Google Scholar]
- 9.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321(5886):263–6. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 10.Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, Lavan P, Weber E, Doak AK, Côté S, Shoichet BK, Urban L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486(7403):361–7. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH. PubChem’s BioAssay database. Nucleic Acids Res. 2012;40(Database issue):D400–12. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(D1):D1100–7. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–41. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Bajorath J. Many structurally related drugs bind different targets whereas distinct drugs display significant target overlap. RSC Adv. 2012;2(8):3481–9. doi: 10.1039/c2ra01345b. [DOI] [Google Scholar]
- 15.Hu Y, Bajorath J. Growth of ligand-target interaction data in ChEMBL is associated with increasing and activity measurement-dependent compound promiscuity. J Chem Inf Model. 2012;52(10):2550–8. doi: 10.1021/ci3003304. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y, Bajorath J. How promiscuous are pharmaceutically relevant compounds? a data-driven assessment. AAPS J. 2013;15(1):104–11. doi: 10.1208/s12248-012-9421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J Med Chem. 1996;39(15):2887–93. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]


