Abstract
3,3′-diindolylmethane (DIM) is currently being investigated in many clinical trials including prostate, breast, and cervical cancers and has been shown to possess anticancer effects in several in vivo and in vitro models. Previously, DIM has been reported to possess cancer chemopreventive effects in prostate carcinogenesis in TRAMP mice; however, the in vivo mechanism is unclear. The present study aims to investigate the in vitro and in vivo epigenetics modulation of DIM in TRAMP-C1 cells and in TRAMP mouse model. In vitro study utilizing TRAMP-C1 cells showed that DIM suppressed DNMT expression and reversed CpG methylation status of Nrf2 resulting in enhanced expression of Nrf2 and Nrf2-target gene NQO1. In vivo study, TRAMP mice fed with DIM-supplemented diet showed much lower incidence of tumorigenesis and metastasis than the untreated control group similar to what was reported previously. DIM increased apoptosis, decreased cell proliferation and enhanced Nrf2 and Nrf2-target gene NQO1 expression in prostate tissues. Importantly, immunohistochemical analysis showed that DIM reduced the global CpG 5-methylcytosine methylation. Focusing on one of the early cancer chemopreventive target gene Nrf2, bisulfite genomic sequencing showed that DIM decreased the methylation status of the first five CpGs of the Nrf2 promoter region, corroborating with the results of in vitro TRAMP-C1 cells. In summary, our current study shows that DIM is a potent cancer chemopreventive agent for prostate cancer and epigenetic modifications of the CpG including Nrf2 could be a potential mechanism by which DIM exerts its chemopreventive effects.
Electronic supplementary material
The online version of this article (doi:10.1208/s12248-013-9493-3) contains supplementary material, which is available to authorized users.
KEY WORDS: 3,3′-diindolylmethane (DIM); epigenetic; methylation; Nrf2; prostate cancer
INTRODUCTION
DNA hyper- or hypomethylation of promoter regions of tumor-associated genes plays critical roles in cancer development and progression (1). For example, the hypermethylation status in the promoter regions of tumor-suppressing genes has been demonstrated in various cancers recently, such as estrogen receptor α (ESR1), glutathione S-transferase π1 (GSTP1), retinoid acid receptor β2 (RARβ2), and death-associated protein kinase 1 (DAPK1) in breast cancer; ESR1 and RARβ2 in colorectal cancer; GSTP1 and RARβ2 in prostate cancers; and DAPK1 in lung cancers (2,3).
It has been reported that prostate cancers might involve a cascade of epigenetic and genetic changes associated with chronic inflammatory status and long-term oxidative stress which would drive tissue tumorigenesis in epidemiological, experimental, preclinical, and clinical studies (4–6). Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) plays a critical role in the antioxidative stress response including the coordinated regulation of many detoxifying/antioxidant enzymes (7,8). Recently, a progressive loss of Nrf2 expression as well as Nrf2 downstream target genes has been demonstrated in clinical prostate cancer patients (6). We have also reported that there is a progressive loss of Nrf2 expression and Nrf2 downstream target antioxidative genes, such as NQO1, HO-1, GSTm1, GCLC, and UGT1a1, during prostate cancer progresses in transgenic adenocarcinoma of mouse prostate (TRAMP) mice (5,9,10). Hypermethylated CpGs in the promoter region of Nrf2 gene was further found in TRAMP prostate tumors and TRAMP-C1 cells (11). The enhanced Nrf2 CpG methylation during TRAMP prostate tumor progresses was associated with the attenuated expressions of Nrf2 and its target genes (12). However, dietary feeding of γ-tocopherol rich of tocopherols mixture can inhibit prostate cancer progression in TRAMP mice since it increases the expression of Nrf2 in prostate tumors via the CpG demethylation of Nrf2 promoter epigenetically (12).
Accumulating evidence suggests that epigenetic changes arise earlier than genetic defects, during prostatic carcinogenesis, linking the appearance of epigenetic alterations in some way to disease etiology (3). Polyphenols and isothiocyanates from green tea and cruciferous vegetables as nontoxic dietary phytochemicals have been reported to suppress initiation and development of cancer by epigenetic modifications both in vivo and in vitro (13). Recently, our laboratory showed that curcumin, a potent anticancer agent against many cancers such as prostate cancer, suppresses DNA methyltransferases (DNMT) expression and modifies the histone deacetylases (HDAC) activity in human prostate LNCaP cells, which might contribute to DNA demethylation in promoter region of Nrf2 gene in TRAMP-C1 cells as well as Neurog1 gene in LNCaP cells (14,15). 3,3′-diindolylmethane (DIM), a compound derived from indole-3-carbinol (I3C), is found abundantly in cruciferous vegetables and its multi-target anticancer and cancer chemopreventive effects have been demonstrated in many in vivo and in vitro cancer models (16–19). Currently, there are around ten clinical trials being conducted in early stage of prostate, breast, and cervical cancer patients for this compound (www.clinicaltrial.gov). However, the efficacy and mechanisms of epigenetic modifications of DIM for preventing prostate tumorigenesis in TRAMP mice has not been investigated.
In our current study, we examined the epigenetic modifications of the promoter of Nrf2, a critical transcription factor in mediating cellular defense against oxidative stress and inflammation (8,20), in vitro in TRAMP-C1 cells. Following that, we further investigated the inhibitory efficacy of prostate tumorigenesis and epigenetic modifications of DIM in vivo in TRAMP mouse model. The results show that DIM effectively inhibits the progression and development of prostate cancer in TRAMP mice and DIM demethylates and re-expresses Nrf2 and Nrf2-target genes in vivo and in vitro. This is the first report showing the epigenetic demethylation ability of DIM both in vivo and in vitro, which could be translated to human studies.
MATERIALS AND METHODS
Reagents and Cell Culture
The DIM used in the study contains approximately 98% of DIM purchased from Sigma-Aldrich (St. Louis, MO). TRAMP-C1 cells (provided by Dr. Barbara Foster, Department of Pharmacology and Therapeutics, Roswell Park Cancer Institute, Buffalo, NY), originally derived from TRAMP prostate tumor (21), were maintained in a humidified incubator with 5% CO2 at 37°C, cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen Corp., Carlsbad, CA) supplemented with 10% (v/v) fetal bovine serum (FBS; Invitrogen Corp., Grand Island, NY), penicillin 100 U/ml, and streptomycin 100 μg/ml (Invitrogen Corp., Grand Island, NY). Cells were seeded in 10-cm plates for 24 h and then treated with different concentrations of DIM or 0.1% DMSO (control) in DMEM containing 1% FBS for 5 days and changed every 2 days (14).
Quantitative Real-Time PCR Assays
Total RNA were extracted from TRAMP-C1 cells which were treated with or without DIM and were reversed transcribed to cDNA. DNMT1, DNMT3a, DNMT3b, NQO1, HO-1, GSTm1, UGT1a1, and control β-actin as the primers of quantitative real-time PCR assays (qPCR) are listed in Supplementary Table I (Integrated DNA Technologies, Coralville, IA). The qPCR reactions were carried out with 1 μl reversed RNA product (cDNA), 50 nM of each primer, and Power SYBR Green master mix (Applied Biosystems, Foster City, CA) in 10 μl reactions. An ABI Prism 7900HT sequence detection system was used for performing the reactions; the amplification was verified by first-derivative melting curve (dissociation curve) analysis using the ABI software (SDS 2.3, Applied Biosystems, Foster City, CA), specifically. A ΔΔCt method (RQ manager, Applied Biosystems, Foster City, CA) as we have performed previously was used to calculate relative quantification of gene expression profile (22). The results are presented as mean ± SD including three independent experiments performed showing similar results.
Western Blot
Dorsolateral prostate tissues collected from control (AIN-76A diet) and experiment (DIM-supplemented diet) groups were pooled and homogenized in the RIPA buffer (Cell Signaling Technology, Inc., Danvers, MA), or TRAMP-C1 cells with or without DIM treatment were collected and homogenized in the RIPA buffer with 10 μg/ml protease inhibitor cocktail (EMD Chemicals, Philadelphia, PA) for protein extraction. Twenty-microgram protein was loaded onto 4–18% SDS-PAGE gel (Bio-Rad Laboratories, Hercules, CA). After separation by SDS-PAGE, the protein was transferred onto nitrocellulose membrane (Millipore Corp., Billerica, MA). Membranes were blocked in the 5% bovine serum albumin (Fisher Scientific, Fair Law, NJ)/tris-buffered saline solution with Tween 20 followed by being probed using the different mono- or polyclonal primary antibodies overnight at 4°C. After incubation with secondary antibodies for 1 h, respectively, the immune-reactive blots were developed by adding SuperSignal West Femto mix (1:1 mix of stable peroxide buffer and luminol/enhancer solution; Thermo Scientific, Rockford, IL) to detect immune-reactive blots. The blots were visualized and semiquantified by BioRad ChemiDoc XRS system (Hercules, CA). Beta-actin, Nrf2, NQO1 (Santa Cruz Biotechnology, Santa Cruz, CA), HDAC1, HDAC2, HDAC3, HDAC4 (Cell Signaling Technology Inc., Danvers, MA), and HDAC8 (Proteintech Group Inc., Chicago, IL) and DNMT1, DNMT3a, and DNMT3b (Imgenex, San Diego, CA) were applied as the primary antibodies. Goat polyclonal IgG for β-actin and NQO1, rabbit polyclonal IgG for Nrf2, HDAC1, HDAC2, HDAC3, HDAC4, and HDAC8 and mouse polyclonal IgG for DNMT1, DNMT3a, and DNMT3b (Santa Cruz Biotechnology, Santa Cruz, CA) were utilized as the secondary antibodies.
DNA Extraction and Bisulfite Genomic Sequencing
Genomic DNA was isolated from the control or DIM-treated TRAMP-C1 cells and TRAMP dorsolateral prostate tissues collected from control or DIM-treated groups pooled and homogenized using the DNeasy tissue kit (Qiagen, Valencia, CA). The genomic DNAs were extracted and subjected to bisulfite conversion carried out using 750 ng of genomic DNA and applying to EZ DNA Methylation Gold Kits (Zymo Research Corp., Orange, CA) following the manufacturer’s instructions. The converted DNA was amplified by PCR-utilizing Platinum PCR SuperMix (Invitrogen, Grand Island, NY) with a set of specific primers, forward: 5′-AGT TAT GAA GTA GTA GTA AAA A-3′ and reverse: 5′-AAT ATA ATC TCA TAA AAC CCC AC-3′, amplifying the first five CpGs located between −1,266 and −1,086 of the promoter region of Nrf2 with the translation initiation site defined as +1 (11,14). Gel extraction using Qiaquick™ gel extraction kit (Qiagen, Valencia, CA) were used to purify the PCR products, then cloned into pCR4 TOPO vector using a TOPO™ TA Cloning kit (Invitrogen, Grand Island, NY). Plasmids DNA from at least 20 colonies of each treatment were prepared using QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) and sequenced (Genwiz, Piscataway, NJ) as we have previously reported (11,14).
Methylation DNA Immunoprecipitation Analysis
Genomic DNA (8 μg each) extracted from control and DIM-treated TRAMP-C1 cells were used for the methylation DNA immunoprecipitation (MeDIP) analysis (23), analogous to other chromatin immunoprecipitation (ChIP) analysis, modified from previous reports (24,25) and as we have published recently (11,14). Inputs from each sample contained around one tenth of the amount of fragmented DNAs; the remaining DNA were applied for denaturing for 10 min, and immunoprecipitation (IP) in 1× IP buffer (10 mM sodium phosphate at pH 7.0, 140 mM NaCl, and 0.25% Triton X-100) using antimethylcytosine antibody (antimecyt; purchased from Anaspec, Fremont, CA) and anti-c-Myc as a negative control antibody (Santa Cruz, Santa Cruz, CA) for 2 h at 4°C, respectively. Precipitated DNA was purified using miniprep kit from Qiagen (Valencia, CA). The inputs and precipitated DNA were used as templates for PCR amplification of Nrf2 promoter region position from −1,190 to −1,092 covering the first five CpGs as described previously (11,14). A forward primer 5′-GAG GTC ACC ACA ACA CGA AC-3′ and a reverse primer 5′-ATC TCA TAA GGC CCC ACC TC-3′ were used to amplify the Nrf2 fragment. PCR was performed using Platinum PCR SuperMix (Invitrogen, Grand Island, NY) The PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide (EB) staining using a Gel Documentation 2000 system (Bio-Rad Laboratories, Hercules, CA).
Animals
Housing and care of the animals was performed in accordance with the guidelines established by the University’s Animal Research Committee consistent with the NIH Guidelines for the Care and Use of Laboratory Animals. The animals were housed in a temperature-controlled room (68–72°F) with a 12-h light–dark cycle, at a relative humidity of 45% to 55% throughout the studies. Female hemizygous C57BL/TGN TRAMP mice, line PB Tag 8247NG, and male C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). TRAMP mice were bred on the same genetic background C57BL/6 and maintained in the Laboratory Animal Service facility at Rutgers University. Transgenic males (TRAMP × C57BL/6) F1 or (TRAMP × C57BL/6) F2, were used for the studies. Identity of transgenic mice was established by PCR-based DNA genotyping using the primers suggested by The Jackson Laboratory (Supplementary Table II).
Diet and Animal Study Design
DIM was obtained from Sigma-Aldrich (St. Louis, MO). One percent DIM added in the AIN-76A-based diets were prepared by Research Diets Inc. (New Brunswick, NJ) and stored at −20°C. The dose was chosen based in part on studies reported previously (26,27). In the present study, around 30 mg of DIM would be estimated to be administered to each TRAMP mouse per day when consuming AIN-76 diet containing 1% DIM (1% DIM diet). The control TRAMP males (n = 19) received AIN-76A diets without DIM throughout the experiment while the treated TRAMP males received 1% DIM diet starting from 8 weeks of age (n = 9) as groups 1 (D-G1) and 2 (D-G2) starting from 12 weeks of age (n = 8) (Fig. 1a). All groups of TRAMP mice were receiving fresh AIN-76A diets with or without DIM supplement twice weekly. Control and experiment TRAMP males were weighed every week, and the overall health condition of the animals was monitored on a regular basis. All mice were killed at the age of 24 weeks by carbon dioxide euthanasia, and the genitourinary apparatus (GU) consisting of the bladder, prostate, and seminal vesicles were collected for further analyses (Fig. 1b) (10).
Fig. 1.
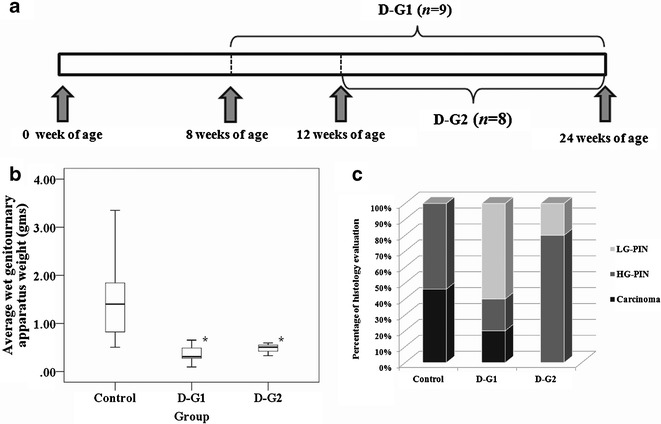
DIM-supplemented diet in TRAMP mice. a Time line: D-G1 =8 weeks of age TRAMP males were put on AIN-76A diet supplemented with 1% DIM and were killed at 24 weeks of age, D-G2 12 weeks old TRAMP males were put on AIN-76A with 1% DIM and were killed at 24 weeks of age. b Effects of DIM on the GU weights of animals treated from 8 and 12 weeks old. *p < 0.05, significantly different from the control based on Mann–Whitney U test. c Histological evaluation of the incidence of PIN and carcinoma. Control group, carcinoma/HG-PIN = 46/54%; D-G1 group, carcinoma/HG-PIN/LG-PIN = 20:20:60%; D-G2 group, HG-PIN/LG-PIN = 80:20%. (Please refer to Supplementary Table III for further detail)
Histopathology
The dorsolateral prostates (n = 5 from each group) were excised and fixed in 10% formaldehyde for 24 h and then transferred to 70% ethanol for 24 h. After dehydration processing and embedding with paraffin, tissue sections (4 μm) were cut from paraffin-embedded prostate tissues and mounted on slides. The sections were stained with hematoxylin and eosin to examine any neoplastic changes and then a histopathologist in a blinded fashion was evaluated to classify prostatic intraepithelial neoplasia (PIN) lesion, as what we have reported previously (5,9,10,28). Lesions were classified as PIN I, II, III, and IV as described by Park et al., as well as what we have reported previously (5,9,28,29). For ease of classification, PIN I and II were combined as low-grade PIN (LG-PIN) while PIN III and IV were combined as high-grade PIN (HG-PIN) as we have performed previously (5,9,28,29).
Immunohistochemistry Staining Assay
Sections (4 μm) were cut from the paraffin-embedded prostate tissues and mounted on glass slides. The slides (n = 5 from each animal and five animals from each group for each biomarker staining) were deparaffinized in xylene and rehydrated. After antigen retrieval was performed by applying proteinase K digestion directly on the slides for 15 min, endogenous peroxidase was blocked by incubating in 3% H2O2 for 5 min, and ApopTag Plus Peroxidase In Situ Apoptosis Detection Kit (Millipore, Temecula, CA) was used to detect apoptotic cells. For detection of proliferative cells, monoclonal mouse antiproliferating cell nuclear antigen (PCNA) antibody (Clone PC10, 1:50, Dako North America, Carpinteria, CA) was used (9). Anti-5-methylcytosine (5-MC) mouse monoclone antibody (Clone 162 33 D3, 1:50, EMD Chemicals, Philadelphia, PA) was used to detect genome-wide methylated DNA. Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, CA) was used to detect apoptotic, proliferative, or methylated cells by applying enzyme-conjugated avidin/biotin complex, peroxidase substrate, and 3,3′-diaminobenzidine to develop color for visualization (9,10). The staining was performed following the manufacturer’s protocols.
Assessment of IHC Staining
Aperio ScanScope® GL system was applied for quantitation of immunohistochemistry (IHC) staining according to the manufacture’s protocol (Aperio Technologies Inc., Vista, CA). The instrument is a single-slide scanning, digital pathological analysis system for IHC of tissue and tumor samples to analyze the IHC-stained slides for the various markers. Prostate tumor samples obtained from untreated control versus the DIM-supplemented TRAMP mice were unbiased quantified and quantitated to analyze the IHC staining biomarkers using the Aperio ImageScopoe software (v 10.1.3.2028).
Statistical Analysis
The means and the standard deviation of the results were presented as means ± SD in the figures. Data were analyzed statistically using SPSS 17 (SPSS Inc., Chicago, IL). A nonparametric Mann–Whitney U test (30) was employed for in vivo animal study. Data distribution was presented by box plots. The upper edge of the box indicates the 75th percentile of the dataset, and the lower edge indicates the 25th percentile. The line in the box indicates the median value of the data. The error bars represents the 95% confidence intervals. The Student’s t test was used to determine the statistical differences for the in vitro study.
RESULTS
DIM Activated Nrf2 and Induced Nrf2-Target Genes Expression in TRAMP-C1 Cells
DIM enhanced the mRNA expression of Nrf2 and Nrf2-targert genes NQO1 and GSTm1 in TRAMP-C1 cells (Fig. 2a), which was originally derived from TRAMP prostate tumor (11). In agreement with the results of mRNA expressions, Nrf2 and Nrf2-mediate protein, NQO1, protein levels were significantly induced in TRAMP-C1 cells treated with DIM (Fig. 2b). These results suggest that DIM may be able to modify the epigenetic status of CpG methylation of Nrf2 and restore Nrf2 and Nrf2-target genes mRNA and protein expressions in TRAMP-C1 cells in vitro.
Fig. 2.
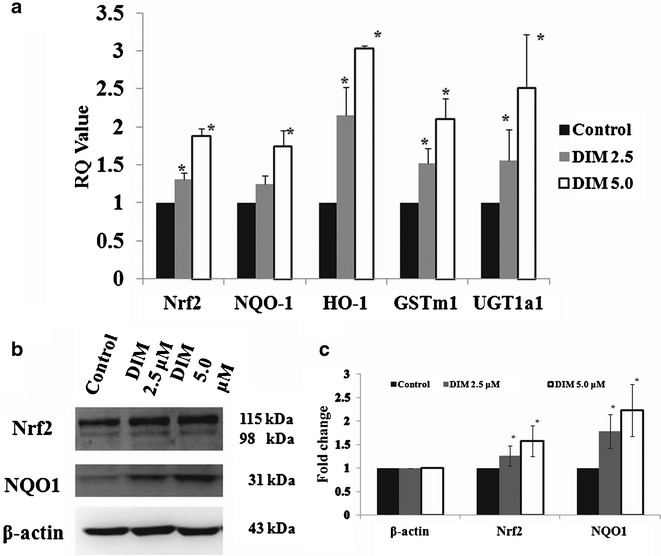
DIM restored expressions of Nrf2 and Nrf2-target genes on TRAMP-C1 cells treated for 5 days. a The mRNA expression levels were significantly enhanced in DIM-treated TRAMP-C1 cells. b Western blots of Nrf2 and NQO1 expression in TRAMP-C1 cells; c the bands (western blots) were visualized and quantified using Gel Documentation 2000 system (Bio-Rad, Hercules, CA). Bars represent mean fold change ± SD (normalized with β-actin and compared with control value). *p < 0.05, significantly different from the control by Student’s t test
DIM Decreased the Hypermethylation of First Five CpGs in the Promoter Region of Nrf2 in TRAMP-C1 Cells
To further understand the epigenetic modifications, bisulfite genomic sequencing (BGS) was performed to test if DIM could reverse the methylation status of the first five CpGs in the promoter region of Nrf2 genes. In agreement with our previous study (11), the first five CpGs were hypermethylated in TRAMP-C1 cells (96.8% methylated; Fig. 3a). Treatment of cells with 2.5 or 5 μM of DIM for 5 days significantly decreased the methylation status of these five CpGs on the Nrf2 promoter region in a dose-dependent manner (73.7% and 55.8% methylation, respectively, Fisher’s exact test, p < 0.001; Fig. 3a).
Fig. 3.
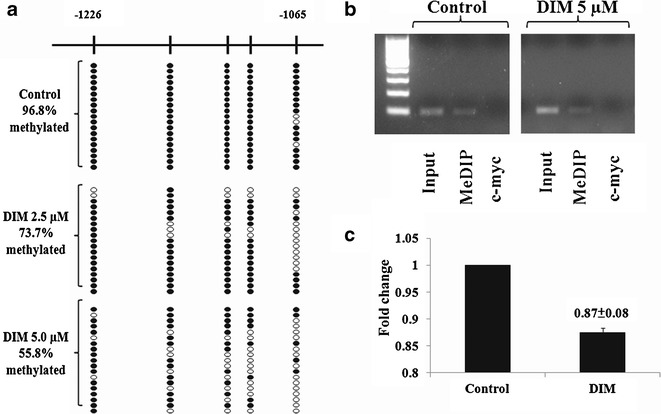
Demethylation effects of DIM treated on TRAMP-C1 cells for 5 days. a The methylation patterns of the first five CpGs of promoter Nrf2 gene in TRAMP-C1 cells was performed using BGS as described in the “Materials and methods.” Filled circles indicate methylated CpGs, and open circles indicate unmethylated CpGs. The five CpGs were hypermethylated in TRAMP-C1 cells which were untreated control (96.8% methylated). For cells treated with either 2.5 or 5 μM of DIM for 5 days, the methylation status of these five CpGs was reversed significantly (73.7% and 55.8% methylation, respectively, Fisher’s exact test, p < 0.001). Methylation status of TRAMP-C1 cells treated with DIM at 5 μM was significantly lower than DIM at 2.5 μM (Fisher’s exact test, p = 0.015). b Methylation DNA immunoprecipation (MeDIP) analysis was performed as described in the “Materials and methods.” Semiquantative PCR was performed to compare the immunoprecipatated DNA with their inputs and negative control (c-myc); c the bands (MeDIP) were visualized and quantified using Gel Documentation 2000 system (Bio-Rad, Hercules, CA). Bars represent mean fold change ± SD (normalized with inputs and compared with control value)
MeDIP/ChIP analysis has been commonly used to enrich the methylated CpG DNA in an unbiased manner (24). DNMT add a methyl group to the five-carbon atom of the DNA-based cytosine (C) to generate methylcytosine (31). Antimecyt antibody which binds specifically to the methylated cytosine (MC) was used to immunoprecipitate (IP) the genomic DNA harvested from control and DIM-treated TRAMP-C1 cells. The IP DNA was purified and used for PCR to amplify the Nrf2 promoter region containing the first five CpGs. MeDIP/ChIP results showed that DIM reduced the methylated DNA bound by antimecyt antibody to the first five CpGs of Nrf2 gene promoter (Fig. 3b, c).
DIM Suppressed DNMT Expression in TRAMP-C1 Cells
The in vitro results from TRAMP-C1 cells above clearly show that DIM treatment of TRAMP-C1 cells reduced the CpG methylation status of the Nrf2 gene promoter region. To elucidate the potential molecular epigenetic mechanism by which DIM exerts its DNA hypomethylation effect, the effect of DIM on DNMT and HDAC mRNA and protein expression was examined. Figure 4a shows the effect of DIM on the mRNA expression of DNMT1, DNMT3a, and DNMT3b quantitated by qPCR in TRAMP-C1 cells. DIM significantly suppressed the mRNA expression of DNMT1 at both 5 μM and 10 μM concentrations (p < 0.05) whereas DNMT3a was suppressed by DIM at 5 μM concentration more significantly (p < 0.05). Corroborating with the mRNA expression of DNMT1, 3a and 3b (Fig. 4a), western blotting showed that DIM suppressed the protein levels of DNMT1, and DNMT3b in TRAMP-C1 cells as well (Fig. 4b). Furthermore, western blotting also showed that DIM could suppress the protein expression of HDAC2 and HDAC3 in TRAMP-C1 cells (Fig. 4b).
Fig. 4.
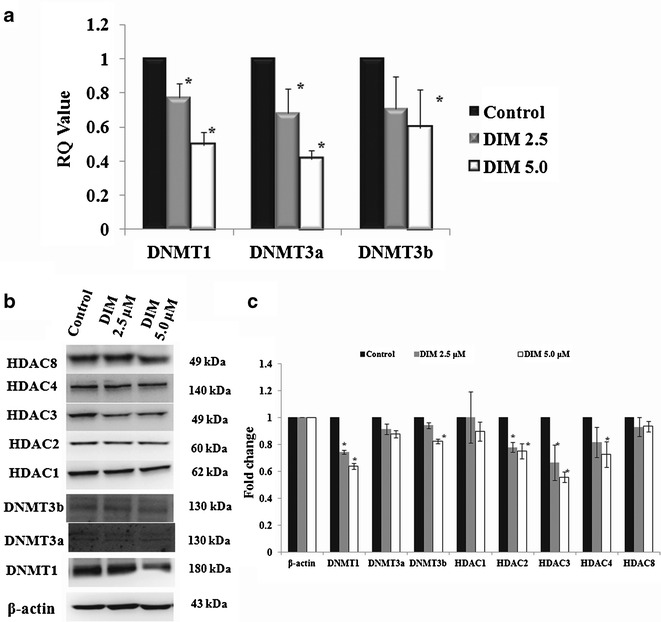
DIM suppressed DNMT and HDAC in TRAMP-C1 cells. a The mRNA expression levels of DNMT1, DNMT3a, and DNMT3b were significantly suppressed by DIM in TRAMP-C1 cells at the concentrations of 5 and 10 μM (p < 0.05). DNMT3a was also significantly suppressed by 5 μM DIM treatments (p < 0.05). DNMT3b was also suppressed by DIM, but there is no significant difference. b Western blots of the DNMT and HDAC protein levels in TRAMP-C1 cells treated with DIM; c the bands (western blots) were visualized and quantified using Gel Documentation 2000 system (Bio-Rad, Hercules, CA). Bars represent mean fold change ± SD (normalized with β-actin and compared with control value). *p < 0.05, significantly different from the control by Student’s t test
Effects of DIM-Supplemented Diet and Overall Health of TRAMP Mice
The overall health of all the mice was monitored during the study period and found to be in good health. All mice were weighed and checked weekly during the course of this study. No significant change in the body weights of all the mice was found throughout the study period. In addition, the liver, kidney, and spleen of the DIM treatment and control groups were collected and weighed after the animals were sacrificed and there was no significant change on the weights of these organs.
DIM-Supplemented Diet Inhibited TRAMP Prostate Tumorigenesis
DIM treated groups (D-G1 and D-G2, p < 0.05) showed statistically significant decrease in the wet GUT weight as compared with the untreated control group (Fig. 1b). Seven control untreated mice were found to have hyperplasia and lesions of the prostate tissues and or the seminal vesicles and six mice were found to have primary palpable prostate tumors (Supplementary Table III). DIM decreased the incidence of palpable tumor and metastasis (Supplementary Table III). Six untreated control mice had primary palpable tumors and five were associated with distinct lymph nodes metastases with no lung or liver metastasis (Supplementary Table III). The remaining mice in the control group were found to have either HG-PIN or carcinoma by histological analysis (Fig. 1c). DIM significantly reduced the incidence of palpable tumor in D-G2 (p < 0.05) and lymph nodes metastasis in both D-G1 and D-G2 (p < 0.05) (Supplementary Table III). In D-G1, although one mouse had a palpable tumor that was confirmed histologically as carcinoma, overall the mice treated with DIM starting from 8-week of age (D-G1) showed 60% incidence of LG-PIN and 20% of HG-PIN (Fig. 1c). In contrast, mice treated with DIM starting from 12 weeks of age (D-G2) had 80% HG-PIN and 20% LG-PIN and no malignant carcinoma (Fig. 1c). Compared with the control group, DIM-treated mice had an overall lower tumor incidence and PIN lesions (Fig. 1c), suggesting that DIM suppresses prostate cancer tumor formation and progression. DIM had no suppression effect of SV-40 transgene expression (data not shown).
DIM Inhibited Cell Proliferation and Enhanced Apoptosis in TRAMP Prostate Tissues
PCNA is an auxiliary protein for DNA polymerase known to be cell cycle regulated (9). TRAMP males treated with DIM for 16 (D-G1) or 12 (D-G2) weeks resulted in significantly lower levels of PCNA staining (p = 0.042, 0.030, respectively) measured by IHC analysis (Supplementary Fig. 1). The percent of apoptotic cells in the dorsolateral prostates of the TRAMP males fed with DIM-supplemented diet in D-G1 and D-G2 was significantly higher than the control group (p < 0.001) (Supplementary Fig. 2). Moreover, the percent of apoptotic cells in D-G1 was significantly higher than in D-G2 (p = 0.029) (Supplementary Fig. 2). These results indicate that the tumor inhibitory effect of DIM is correlated in part with increased apoptosis and suppressed proliferation of prostatic epithelial tumor cells.
DIM Induced Protein Expression of Antioxidative Stress Genes Nrf2 and Nrf2-Target Gene NQO1 in TRAMP Prostate Tissues
Figure 5 shows that DIM induced Nrf2 proteins expression in both D-G1 and D-G2 in the dorsolateral prostate tissues of the TRAMP mice. Moreover, NQO1, an Nrf2-targeted downstream antioxidant enzyme, was also induced by DIM in both D-G1 and D-G2 (Fig. 5), as observed in the TRAMP-C1 cells above. Nrf2 and NQO1 protein expression in the prostate tumor samples from the control untreated TRAMP mice was undetectable (Fig. 5), consistent with our previous reported findings (5,10,12,32).
Fig. 5.
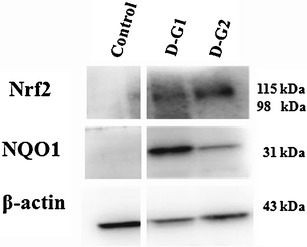
Western blots of biomarkers for Nrf2 and Nrf2-regulated NQO1. Nrf2 and NQO1 were re-activated by DIM in D-G1 and D-G2 significantly different from the control. There was no expression of Nrf2 and NQO1 found in the control animals obviously
DIM Suppressed Global CpG Methylation Staining by 5-MC in TRAMP Prostate Tissues
Aberrant CpG DNA methylation is acquired during carcinogenesis (33). 5-methylcytosine (5-MC) is generated when a methyl group is added to the 5-carbon atom of the DNA-based C. In mammalian cells, 5-MC is found predominantly within CpG dinucleotides (31). Aberrant hypermethylation of CpG islands of many tumor suppressor genes has been linked to the development of cancer (33). Furthermore, 5-MC has been proposed to be a critical clinical biomarker for the diagnosis of cancer and tumors formation (33). Figure 6a shows that DIM significantly decreased 5-MC IHC staining of the prostate tissue in both group D-G1 and D-G2 (p < 0.001, respectively). In addition, the percentage of 5-MC in D-G1 was significantly lower than in D-G2 (p < 0.05).
Fig. 6.
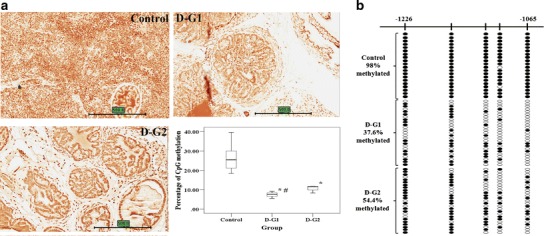
Demethylation effects of DIM-supplemented diet on TRAMP mice. a Immunohistochemical analysis on the methylation marker, 5-methylcytosine. Representative photomicrographs (×40 magnification) of 5-MC-stained (dark orange to red color) TRAMP prostate tissue section and percentage levels of methylation. The scale bar stands for 500 μm. *p < 0.05, significantly different from the control by Mann–Whitney U test. # indicates significant difference between D-G1 and D-G2. b The methylation patterns of the first five CpGs of promoter Nrf2 gene in TRAMP prostate tissues and tumors was performed using BGS as described in the “Materials and methods.” Filled circles indicate methylated CpGs, and open circles indicate unmethylated CpGs. The five CpGs were hypermethylated in the control group (98% methylation) and either D-G1 or D-G2 was found to significantly reduce the methylation of the five CpGs (37.6% and 54.4%, respectively; Fisher’s exact test, p < 0.001). Moreover, compared with the two treated groups, it was found that D-G1 has significantly lower level of methylation than D-G2 (Fisher’s exact test, p = 0.011)
DIM Demethylated the First Five CpGs of the Nrf2 Gene Promoter of TRAMP Prostate Tissues
We have shown that the first five CpGs on the Nrf2 gene promoter region are hypermethylated in TRAMP prostate tumors and in TRAMP-C1 but not in normal prostate tissues, previously (11). In the present study, BGS analysis showed that, in TRAMP prostate tissues or tumors, the first five CpGs were hypermethylated in the control group (control, 98% methylation; Fig. 6b), in agreement with our in vitro and previous studies (11). In contrast to the control group, DIM treatment groups of D-G1/D-G2 had significantly lower methylation of the first five CpGs of Nrf2 (37.6% and 54.4%, respectively, p < 0.001; Fig. 6b), suggesting that DIM inhibits the CpG methylation of the first five CpGs of Nrf2 promoter, which may enhance the transcription of Nrf2 in the TRAMP prostate tissues as we have shown previously (11). Furthermore, comparing the two DIM treated groups, D-G1 showed a significantly lower methylation level of the first five CpGs of Nrf2 than D-G2 (p < 0.05), which substantiates the in vitro results in TRAMP-C1 cells shown above.
DISCUSSION
DIM is a metabolically active product of I3C found abundantly in the dietary cruciferous vegetables. It has been shown in many in vitro and in vivo cancer models that it is a potent anticancer agent, and currently, there are more than ten clinical trials in human prostate, breast, and cervical cancers. DIM appears to elicit multiple targets anticancer effects through many potential mechanisms including anti-inflammation, antioxidative stress effect, cell-cycle regulation, and apoptosis depending on the context of tissues and cell types (10,34–38). Several naturally occurring anticancer phytochemicals have been shown to possess the ability to prevent cancer via epigenetic modifications (14,15,39–41). A previous study showed that DIM possesses the ability to inhibit HDAC activity which correlated with increased expression of p21in both human PC-3 and LNCaP cells (19). However, no study has been reported so far on the potential epigenetic modifications, and the anticancer efficacy of DIM in a transgenic prostate cancer mouse model such as TRAMP mice remains unknown.
Nrf2 is a redox sensitive transcriptional factor and plays a very important role in the cellular defense against oxidative and inflammatory stresses (8). Our previous studies show that, as prostatic tumor progresses in TRAMP mice, there is a decrease in the expression of Nrf2 and Nrf2 downstream target antioxidative stress genes (5,32). When TRAMP mice were supplemented with (γ)-tocopherol-enriched mixed tocopherol or I3C in the diets, Nrf2 and Nrf2-mediated antioxidative stress phase II detoxifying/antioxidant enzymes were induced (5,10,32). I3C and DIM have been shown to be potent inducers of Nrf2-mediated antioxidant enzymes in TRAMP-C1 cells and human hepatoma HepG2C8 cells (10,42). In the current study, DIM enhances the expression of Nrf2 and Nrf2-target gene NQO1 proteins in TRAMP prostate tissues in vivo as compared with the controls which was undetectable (Fig. 5).
To better understand the underlying epigenetic mechanisms of DIM, in vitro study utilizing the TRAMP-C1 cell line was conducted. In vitro BGS of TRAMP-C1 cells also showed that DIM reduction of the methylation status of the first five CpGs on the Nrf2 promoter region (Fig. 2a) which was consistent with the results obtained from the in vivo BGS of TRAMP prostate tissues and tumors (Fig. 6b). In agreement with the BGS results, MeDIP/ChIP assay also showed that DIM reversed the CpG methylation on the Nrf2 gene promoter region in TRAMP-C1 cells (Fig. 2b, c). The demethylation of Nrf2 gene was found to be associated with the enhanced mRNA expression of Nrf2 and Nrf2-target genes such as NQO1, GSTm1 (Fig. 3a), as well as increased protein levels of Nrf2 and NQO1 (Fig. 3b) in TRAMP-C1 cells. Interestingly, in TRAMP prostate tissues, both D-G1 and D-G2 showed significantly lower percentage of genome-wide 5-MC IHC staining significantly in the IHC analysis (Fig. 6a), suggesting that DIM may impact on the global CpG methylation epigenomic profiles. From the BGS results, DIM substantially reduced the methylation status of the first five CpGs of the Nrf2 promoter region (Fig. 6b). The results of 5-MC (Fig. 6a) and BGS (Fig. 6b) suggests that DIM possesses the ability to reduce the methylation status of the first five CpGs on the Nrf2 gene promoter region may lead to increase or re-expression of Nrf2 and Nrf2-target gene NQO1 proteins in vivo as shown in Fig. 5. Furthermore, the demethylation effects of DIM correlated with DIM’s ability to suppress the expression of DNMT and HDAC in TRAMP-C1 cells (Fig. 4). DNMT1 is a key maintenance DNMT, with the most abundance in mammalian cells comparing with others (43). In human cancer cells, DNMT1 has been shown to be responsible for both the de novo and the maintenance of promoter CpG islands methylation of tumor suppressor genes (44–46). Coincidentally, our previous studies with other DNA demethylating agents such as curcumin and 5-aza+TSA also show decreased CpG methylation of the first five CpGs of the promoter of Nrf2 gene and enhanced the expression of Nrf2 and Nrf2-target genes in TRAMP-C1 cells (11,14), which suggest this may be a common epigenetic modification mechanism.
Our present study shows that TRAMP mice fed with DIM-supplemented diet had lower percentage of palpable tumor and incidence of lymph node metastasis as compared with the control diet (5.8% versus 31.6% for palpable tumor in treated versus control and 0% versus 26.3% for lymph node metastasis in treated versus control) (Supplementary Table III). The GU weights were significantly lower in DIM treated groups as compared with the control (Fig. 1b). To investigate the in vivo anticancer and chemopreventive effect of DIM in different stages of prostate tumorigenesis, DIM was supplemented in the diet to TRAMP mice starting at 8 weeks of age (D-G1), when the LG-PIN starts to form, and 12 weeks of age (D-G2) when some of the LG-PINs progress into HG-PINs (29,47). In addition, administration of DIM at the early stage of tumorigenesis starting at 8 weeks old when low-grade PIN lesions started to form achieve better anticancer chemopreventive effect than when given later at 12 weeks old when some of the LG-PINs have progressed to HG-PINs (Fig. 1c). Immunohistochemistry (IHC) analysis of the prostatic sections shows that DIM-fed mice had lower percentage of PCNA (cell proliferation marker) (Supplementary Fig. 1) and higher percentage of apoptotic cells (Supplementary Fig. 2), in agreement with previous reports (35,38). These results show that DIM is an effective prostate cancer chemopreventive agent that blocks the development and progression of prostate cancer in TRAMP mice correlated in part with increased apoptosis and suppressed proliferation of prostatic epithelial tumor cells.
DIM is a multi-targeting anticancer agent as reported previously (10,34–38). Among the molecular targets, DIM has been shown to strongly activate Nrf2 and induces Nrf2-mediated downstream target genes (42). Furthermore, the ability of DIM in restoring the expression of Nrf2 and its downstream genes via epigenetic mechanism may play an important role in preventing the development and progression of prostate tumor in TRAMP mice in vivo. In conclusion, our results suggest that DIM as a cancer epigenetic-modifying agent may contribute to the future clinical development of DIM as a cancer epigenetic-modifying cancer chemopreventive and therapeutic drug.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Immunohistochemical analysis of the effects of cell proliferation, PCNA. Representative photomicrographs (×40 magnification) of PCNA-stained (dark brown color) TRAMP prostate tissue section and percentage levels of cell proliferation. The scale bar stands for 500 μm. p < 0.05, significantly different from the control and was based on Mann–Whitney U test. (JPEG 43 kb)
Immunohistochemical analysis of the effects of apoptosis, TUNEL. Representative photomicrographs (×40 magnification) of TUNEL stained (dark brown color) TRAMP prostate tissue section and percentage levels of apoptosis. The scale bar stands for 500 μm. *p < 0.05, significantly different from the control by Mann–Whitney U test. Number sign indicates significantly different between D-G1 and D-G2 (p = 0.043). (JPEG 46 kb)
Murine primers for qPCR (DOC 32 kb)
Confirmation of genotype of the TRAMP mice (DOC 28 kb)
DIM inhibit palpable tumor and metastasis in TRAMP males. aNumbers represent the presence of palpable tumor showed at the end of the experiment at 24 weeks of age. Fisher’s exact test was used to compare the incidence of palpable tumor between the control and the DIM-treated mice killed at 24 weeks of age. *p values < 0.05 were considered as significant. bNumbers represent the presence of lymph nodes metastasis showed at the end of experiment when the mice were killed. Fisher’s exact test was used to compare the incidence of lymph node metastasis between the control and the DIM treated mice sacrificed at 24 weeks of age. #p values < 0.05 were considered as significant. (DOC 27 kb)
TRAMP mice body weights, wet GU weights, and normalized wet GU weights by body weights measured weekly. (PDF 12 kb)
ACKNOWLEDGMENTS
We thank Dr. Chungxiou Wang as a histopathologist for her assistance in histology evaluation. We also thank Dr. Barbara Foster, Rowell Park Cancer Institute, Buffalo, NY, who generously provided the TRAMP-C1 cells. We thank all members in Dr. Kong’s group for their generous help in discussion and preparation of this manuscript.
Conflict of Interest Statement
None declared
Funding
This work was supported by Institutional Funds to Dr. Ah-Ng Tony Kong.
REFERENCES
- 1.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J, Plass C, Gerhauser C. Cancer chemoprevention by targeting the epigenome. Curr Drug Targets. 2011;12(13):1925–1956. doi: 10.2174/138945011798184155. [DOI] [PubMed] [Google Scholar]
- 3.Nelson WG, De Marzo AM, Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology. 2009;150(9):3991–4002. doi: 10.1210/en.2009-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pathak SK, Sharma RA, Steward WP, Mellon JK, Griffiths TR, Gescher AJ. Oxidative stress and cyclooxygenase activity in prostate carcinogenesis: targets for chemopreventive strategies. Eur J Cancer. 2005;41(1):61–70. doi: 10.1016/j.ejca.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Barve A, Khor TO, Nair S, Reuhl K, Suh N, Reddy B, et al. Gamma-tocopherol-enriched mixed tocopherol diet inhibits prostate carcinogenesis in TRAMP mice. Int J Cancer. 2009;124(7):1693–1699. doi: 10.1002/ijc.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frohlich DA, McCabe MT, Arnold RS, Day ML. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27(31):4353–4362. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- 7.Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13(11):1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 8.Hu R, Saw CL, Yu R, Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal. 2010;13(11):1679–1698. doi: 10.1089/ars.2010.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barve A, Khor TO, Hao X, Keum YS, Yang CS, Reddy B, et al. Murine prostate cancer inhibition by dietary phytochemicals–curcumin and phenyethylisothiocyanate. Pharm Res. 2008;25(9):2181–2189. doi: 10.1007/s11095-008-9574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu TY, Saw CL, Khor TO, Pung D, Boyanapalli SS, Kong AN. In vivo pharmacodynamics of indole-3-carbinol in the inhibition of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: involvement of Nrf2 and cell cycle/apoptosis signaling pathways. Mol Carcinog. 2012;51(10):761–70. [DOI] [PubMed]
- 11.Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One. 2010;5(1):e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Khor TO, Shu L, Saw CL, Wu TY, Suh N, et al. A gamma-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. J Nutr. 2012;142(5):818–23. doi: 10.3945/jn.111.153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010;17(20):2141–2151. doi: 10.2174/092986710791299966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khor TO, Huang Y, Wu TY, Shu L, Lee J, Kong AN. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol. 2011;82(9):1073–1078. doi: 10.1016/j.bcp.2011.07.065. [DOI] [PubMed] [Google Scholar]
- 15.Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, et al. Epigenetic CpG Demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011;13(4):606–14. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng JR, Tsai CH, Kulp SK, Chen CS. Indole-3-carbinol as a chemopreventive and anti-cancer agent. Cancer Lett. 2008;262(2):153–163. doi: 10.1016/j.canlet.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar FH, Li Y. Indole-3-carbinol and prostate cancer. J Nutr. 2004;134(12 Suppl):3493S–3498S. doi: 10.1093/jn/134.12.3493S. [DOI] [PubMed] [Google Scholar]
- 18.Banerjee S, Kong D, Wang Z, Bao B, Hillman GG, Sarkar FH. Attenuation of multi-targeted proliferation-linked signaling by 3,3′-diindolylmethane (DIM): from bench to clinic. Mutat Res. 2011;728(1–2):47–66. doi: 10.1016/j.mrrev.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaver LM, Yu TW, Sokolowski EI, Williams DE, Dashwood RH, Ho E. 3,3′-diindolylmethane, but not indole-3-carbinol, inhibits histone deacetylase activity in prostate cancer cells. Toxicol Appl Pharmacol. 2012;263(3):345–351. doi: 10.1016/j.taap.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. When NRF2 talks, who’s listening? Antioxid Redox Signal. 2010;13(11):1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster BA, Gingrich JR, Kwon ED, Madias C, Greenberg NM. Characterization of prostatic epithelial cell lines derived from transgenic adenocarcinoma of the mouse prostate (TRAMP) model. Cancer Res. 1997;57(16):3325–3330. [PubMed] [Google Scholar]
- 22.Barve A, Khor TO, Nair S, Lin W, Yu S, Jain MR, et al. Pharmacogenomic profile of soy isoflavone concentrate in the prostate of Nrf2 deficient and wild-type mice. J Pharm Sci. 2008;97(10):4528–4545. doi: 10.1002/jps.21311. [DOI] [PubMed] [Google Scholar]
- 23.Thu KL, Pikor LA, Kennett JY, Alvarez CE, Lam WL. Methylation analysis by DNA immunoprecipitation. J Cell Physiol. 2010;222(3):522–531. doi: 10.1002/jcp.22009. [DOI] [PubMed] [Google Scholar]
- 24.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 25.Cheung HH, Lee TL, Davis AJ, Taft DH, Rennert OM, Chan WY. Genome-wide DNA methylation profiling reveals novel epigenetically regulated genes and non-coding RNAs in human testicular cancer. Br J Cancer. 2010;102(2):419–427. doi: 10.1038/sj.bjc.6605505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderton MJ, Manson MM, Verschoyle R, Gescher A, Steward WP, Williams ML, et al. Physiological modeling of formulated and crystalline 3,3′-diindolylmethane pharmacokinetics following oral administration in mice. Drug Metab Dispos. 2004;32(6):632–638. doi: 10.1124/dmd.32.6.632. [DOI] [PubMed] [Google Scholar]
- 27.Reed GA, Sunega JM, Sullivan DK, Gray JC, Mayo MS, Crowell JA, et al. Single-dose pharmacokinetics and tolerability of absorption-enhanced 3,3′-diindolylmethane in healthy subjects. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2619–2624. doi: 10.1158/1055-9965.EPI-08-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khor TO, Yu S, Barve A, Hao X, Hong JL, Lin W, et al. Dietary feeding of dibenzoylmethane inhibits prostate cancer in transgenic adenocarcinoma of the mouse prostate model. Cancer Res. 2009;69(17):7096–7102. doi: 10.1158/0008-5472.CAN-09-0597. [DOI] [PubMed] [Google Scholar]
- 29.Park JH, Walls JE, Galvez JJ, Kim M, Abate-Shen C, Shen MM, et al. Prostatic intraepithelial neoplasia in genetically engineered mice. Am J Pathol. 2002;161(2):727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saw CL, Olivo M, Chin WW, Soo KC, Heng PW. Transport of hypericin across chick chorioallantoic membrane and photodynamic therapy vasculature assessment. Biol Pharm Bull. 2005;28(6):1054–1060. doi: 10.1248/bpb.28.1054. [DOI] [PubMed] [Google Scholar]
- 31.Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286(21):18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barve A, Khor TO, Reuhl K, Reddy B, Newmark H, Kong AN. Mixed tocotrienols inhibit prostate carcinogenesis in TRAMP mice. Nutr Cancer. 2010;62(6):789–794. doi: 10.1080/01635581003605896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taberlay PC, Jones PA. DNA methylation and cancer. Prog Drug Res. 2011;67:1–23. doi: 10.1007/978-3-7643-8989-5_1. [DOI] [PubMed] [Google Scholar]
- 34.Kim EJ, Shin M, Park H, Hong JE, Shin HK, Kim J, et al. Oral administration of 3,3′-diindolylmethane inhibits lung metastasis of 4T1 murine mammary carcinoma cells in BALB/c mice. J Nutr. 2009;139(12):2373–2379. doi: 10.3945/jn.109.111864. [DOI] [PubMed] [Google Scholar]
- 35.Cho HJ, Park SY, Kim EJ, Kim JK, Park JH. 3,3′-diindolylmethane inhibits prostate cancer development in the transgenic adenocarcinoma mouse prostate model. Mol Carcinog. 2011;50(2):100–112. doi: 10.1002/mc.20698. [DOI] [PubMed] [Google Scholar]
- 36.Fan S, Meng Q, Saha T, Sarkar FH, Rosen EM. Low concentrations of diindolylmethane, a metabolite of indole-3-carbinol, protect against oxidative stress in a BRCA1-dependent manner. Cancer Res. 2009;69(15):6083–6091. doi: 10.1158/0008-5472.CAN-08-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YH, Kwon HS, Kim DH, Shin EK, Kang YH, Park JH, et al. 3,3′-diindolylmethane attenuates colonic inflammation and tumorigenesis in mice. Inflamm Bowel Dis. 2009;15(8):1164–1173. doi: 10.1002/ibd.20917. [DOI] [PubMed] [Google Scholar]
- 38.Chinnakannu K, Chen D, Li Y, Wang Z, Dou QP, Reddy GP, et al. Cell cycle-dependent effects of 3,3′-diindolylmethane on proliferation and apoptosis of prostate cancer cells. J Cell Physiol. 2009;219(1):94–99. doi: 10.1002/jcp.21650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137(1 Suppl):223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 40.Degner SC, Papoutsis AJ, Selmin O, Romagnolo DF. Targeting of aryl hydrocarbon receptor-mediated activation of cyclooxygenase-2 expression by the indole-3-carbinol metabolite 3,3′-diindolylmethane in breast cancer cells. J Nutr. 2009;139(1):26–32. doi: 10.3945/jn.108.099259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Li X, Guo B. Chemopreventive agent 3,3′-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer Res. 2010;70(2):646–654. doi: 10.1158/0008-5472.CAN-09-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saw CL, Cintron M, Wu TY, Guo Y, Huang Y, Jeong WS, et al. Pharmacodynamics of dietary phytochemical indoles I3C and DIM: induction of Nrf2-mediated phase II drug metabolizing and antioxidant genes and synergism with isothiocyanates. Biopharm Drug Dispos. 2011;32(5):289–300. doi: 10.1002/bdd.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kho MR, Baker DJ, Laayoun A, Smith SS. Stalling of human DNA (cytosine-5) methyltransferase at single-strand conformers from a site of dynamic mutation. J Mol Biol. 1998;275(1):67–79. doi: 10.1006/jmbi.1997.1430. [DOI] [PubMed] [Google Scholar]
- 44.Smith SS, Kaplan BE, Sowers LC, Newman EM. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jair KW, Bachman KE, Suzuki H, Ting AH, Rhee I, Yen RW, et al. De novo CpG island methylation in human cancer cells. Cancer Res. 2006;66(2):682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- 46.Shukla V, Coumoul X, Lahusen T, Wang RH, Xu X, Vassilopoulos A, et al. BRCA1 affects global DNA methylation through regulation of DNMT1. Cell Res. 2010;20(11):1201–1215. doi: 10.1038/cr.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gingrich JR, Barrios RJ, Morton RA, Boyce BF, DeMayo FJ, Finegold MJ, et al. Metastatic prostate cancer in a transgenic mouse. Cancer Res. 1996;56(18):4096–4102. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunohistochemical analysis of the effects of cell proliferation, PCNA. Representative photomicrographs (×40 magnification) of PCNA-stained (dark brown color) TRAMP prostate tissue section and percentage levels of cell proliferation. The scale bar stands for 500 μm. p < 0.05, significantly different from the control and was based on Mann–Whitney U test. (JPEG 43 kb)
Immunohistochemical analysis of the effects of apoptosis, TUNEL. Representative photomicrographs (×40 magnification) of TUNEL stained (dark brown color) TRAMP prostate tissue section and percentage levels of apoptosis. The scale bar stands for 500 μm. *p < 0.05, significantly different from the control by Mann–Whitney U test. Number sign indicates significantly different between D-G1 and D-G2 (p = 0.043). (JPEG 46 kb)
Murine primers for qPCR (DOC 32 kb)
Confirmation of genotype of the TRAMP mice (DOC 28 kb)
DIM inhibit palpable tumor and metastasis in TRAMP males. aNumbers represent the presence of palpable tumor showed at the end of the experiment at 24 weeks of age. Fisher’s exact test was used to compare the incidence of palpable tumor between the control and the DIM-treated mice killed at 24 weeks of age. *p values < 0.05 were considered as significant. bNumbers represent the presence of lymph nodes metastasis showed at the end of experiment when the mice were killed. Fisher’s exact test was used to compare the incidence of lymph node metastasis between the control and the DIM treated mice sacrificed at 24 weeks of age. #p values < 0.05 were considered as significant. (DOC 27 kb)
TRAMP mice body weights, wet GU weights, and normalized wet GU weights by body weights measured weekly. (PDF 12 kb)


