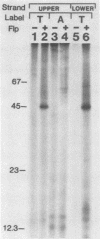
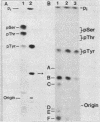
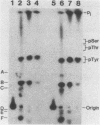
Abstract
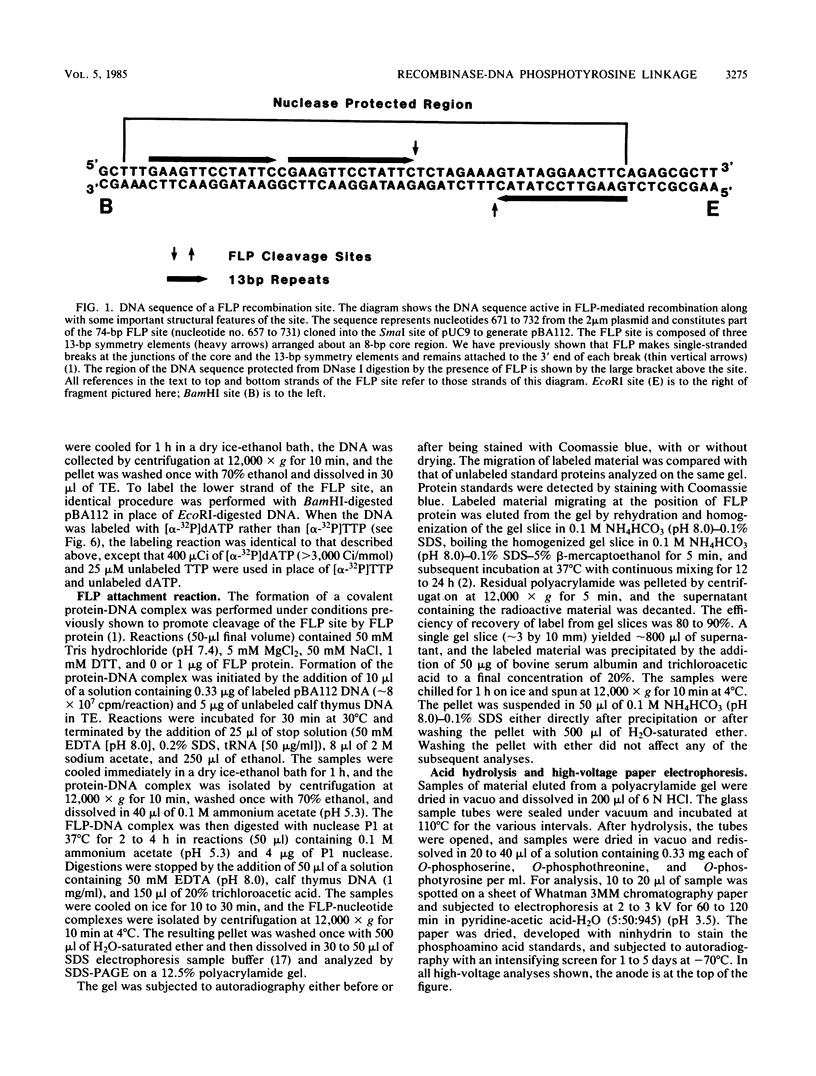
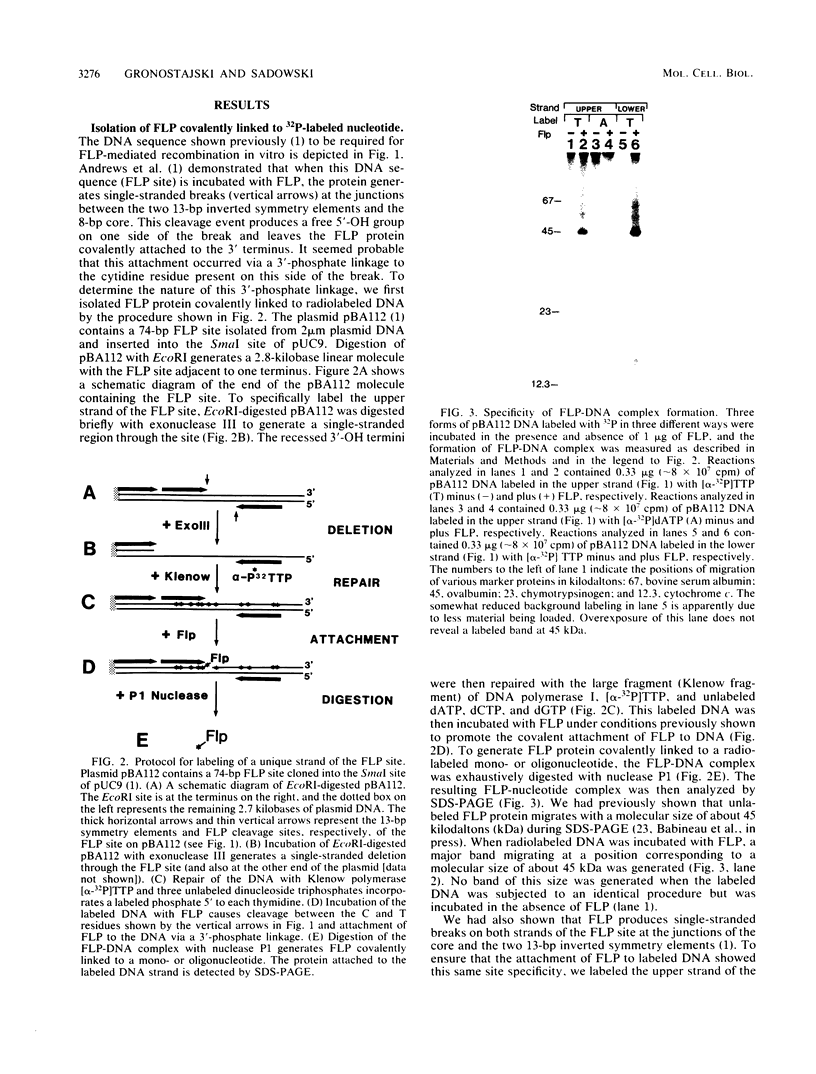
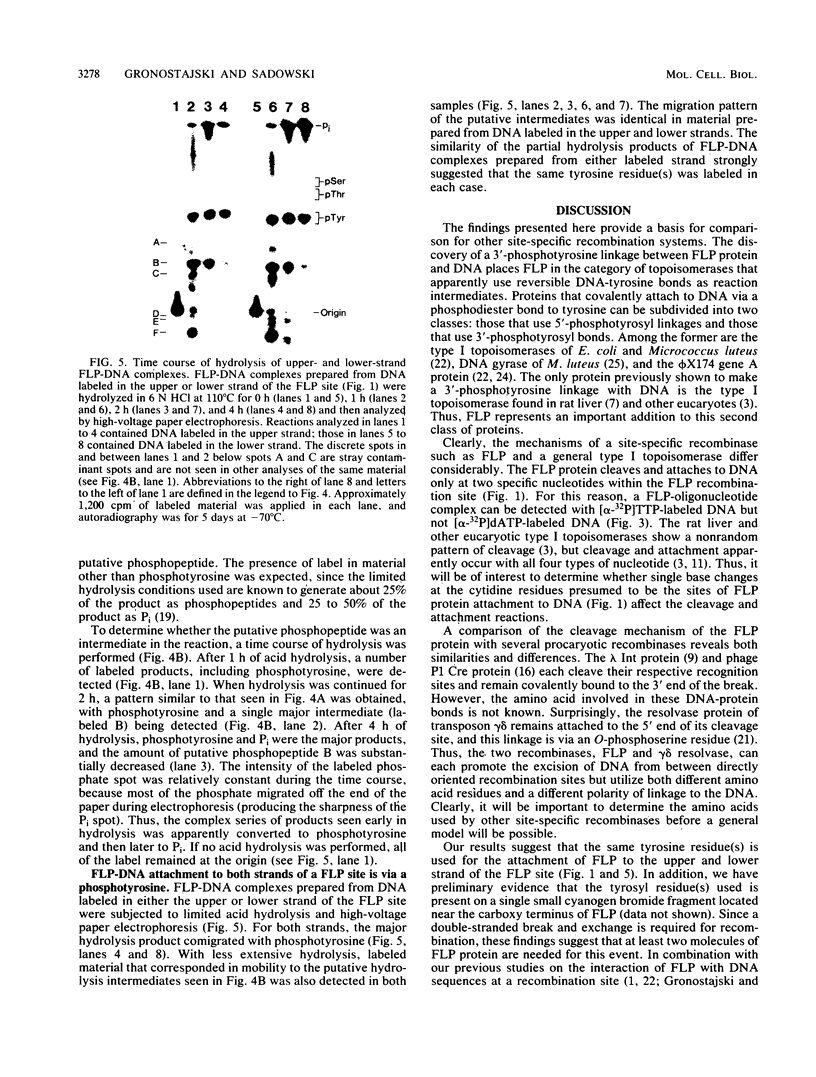
The FLP recombinase, encoded by the 2 micron plasmid of Saccharomyces cerevisiae, promotes efficient recombination in vivo and in vitro between its specific target sites (FLP sites). It was previously determined that FLP interacts with DNA sequences within its target site (B. J. Andrews, G. A. Proteau, L. G. Beatty, and P. D. Sadowski. Cell 40:795-803, 1985), generates a single-stranded break on both DNA strands within the FLP site, and remains covalently attached to the 3' end of each break. We now show that the FLP protein is bound to the 3' side of each break by an O-phosphotyrosyl residue and that it appears that the same tyrosyl residue(s) is used to attach to either DNA strand within the FLP site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Beemon K., Hunter T. Characterization of Rous sarcoma virus src gene products synthesized in vitro. J Virol. 1978 Nov;28(2):551–566. doi: 10.1128/jvi.28.2.551-566.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard O., Hozumi N., Tonegawa S. Sequences of mouse immunoglobulin light chain genes before and after somatic changes. Cell. 1978 Dec;15(4):1133–1144. doi: 10.1016/0092-8674(78)90041-7. [DOI] [PubMed] [Google Scholar]
- Bitte L., Kabat D. Isotopic labeling and analysis of phosphoproteins from mammalian ribosomes. Methods Enzymol. 1974;30:563–590. doi: 10.1016/0076-6879(74)30056-0. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Champoux J. J. DNA is linked to the rat liver DNA nicking-closing enzyme by a phosphodiester bond to tyrosine. J Biol Chem. 1981 May 25;256(10):4805–4809. [PubMed] [Google Scholar]
- Clark S. P., Yoshikai Y., Taylor S., Siu G., Hood L., Mak T. W. Identification of a diversity segment of human T-cell receptor beta-chain, and comparison with the analogous murine element. 1984 Sep 27-Oct 3Nature. 311(5984):387–389. doi: 10.1038/311387a0. [DOI] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Early P., Huang H., Davis M., Calame K., Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980 Apr;19(4):981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitaki J. M., Smith R. A. Techniques in the detection and characterization of phosphoramidate-containing proteins. Methods Enzymol. 1984;107:23–36. doi: 10.1016/0076-6879(84)07004-x. [DOI] [PubMed] [Google Scholar]
- Gascoigne N. R., Chien Y., Becker D. M., Kavaler J., Davis M. M. Genomic organization and sequence of T-cell receptor beta-chain constant- and joining-region genes. Nature. 1984 Aug 2;310(5976):387–391. doi: 10.1038/310387a0. [DOI] [PubMed] [Google Scholar]
- Guo L. H., Wu R. New rapid methods for DNA sequencing based in exonuclease III digestion followed by repair synthesis. Nucleic Acids Res. 1982 Mar 25;10(6):2065–2084. doi: 10.1093/nar/10.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayday A. C., Saito H., Gillies S. D., Kranz D. M., Tanigawa G., Eisen H. N., Tonegawa S. Structure, organization, and somatic rearrangement of T cell gamma genes. Cell. 1985 Feb;40(2):259–269. doi: 10.1016/0092-8674(85)90140-0. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Abremski K. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol. 1985 Feb 5;181(3):351–362. doi: 10.1016/0022-2836(85)90224-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Martensen T. M. Chemical properties, isolation, and analysis of O-phosphates in proteins. Methods Enzymol. 1984;107:3–23. doi: 10.1016/0076-6879(84)07003-8. [DOI] [PubMed] [Google Scholar]
- Meyer-Leon L., Senecoff J. F., Bruckner R. C., Cox M. M. Site-specific genetic recombination promoted by the FLP protein of the yeast 2-micron plasmid in vitro. Cold Spring Harb Symp Quant Biol. 1984;49:797–804. doi: 10.1101/sqb.1984.049.01.090. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Brinkman A., van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. R., Moser C. D. Resolvase-mediated recombination intermediates contain a serine residue covalently linked to DNA. Cold Spring Harb Symp Quant Biol. 1984;49:245–249. doi: 10.1101/sqb.1984.049.01.028. [DOI] [PubMed] [Google Scholar]
- Roth M. J., Brown D. R., Hurwitz J. Analysis of bacteriophage phi X174 gene A protein-mediated termination and reinitiation of phi X DNA synthesis. II. Structural characterization of the covalent phi X A protein-DNA complex. J Biol Chem. 1984 Aug 25;259(16):10556–10568. [PubMed] [Google Scholar]
- Sadowski P. D., Lee D. D., Andrews B. J., Babineau D., Beatty L., Morse M. J., Proteau G., Vetter D. In vitro systems for genetic recombination of the DNAs of bacteriophage T7 and yeast 2-micron circle. Cold Spring Harb Symp Quant Biol. 1984;49:789–796. doi: 10.1101/sqb.1984.049.01.089. [DOI] [PubMed] [Google Scholar]
- Sanhueza S., Eisenberg S. Bacteriophage phi X174 A protein cleaves single-stranded DNA and binds to it covalently through a tyrosyl-dAMP phosphodiester bond. J Virol. 1985 Feb;53(2):695–697. doi: 10.1128/jvi.53.2.695-697.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Y. C., Kirkegaard K., Wang J. C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980 Jun 25;255(12):5560–5565. [PubMed] [Google Scholar]
- Vetter D., Andrews B. J., Roberts-Beatty L., Sadowski P. D. Site-specific recombination of yeast 2-micron DNA in vitro. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7284–7288. doi: 10.1073/pnas.80.23.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]