Crystals soaked with RGD peptides reveal six intermediate conformational states between the closed and higher affinity, fully open state of the integrin αIIbβ3 headpiece.
Abstract
Carefully soaking crystals with Arg-Gly-Asp (RGD) peptides, we captured eight distinct RGD-bound conformations of the αIIbβ3 integrin headpiece. Starting from the closed βI domain conformation, we saw six intermediate βI conformations and finally the fully open βI with the hybrid domain swung out in the crystal lattice. The β1-α1 backbone that hydrogen bonds to the Asp side chain of RGD was the first element to move followed by adjacent to metal ion-dependent adhesion site Ca2+, α1 helix, α1’ helix, β6-α7 loop, α7 helix, and hybrid domain. We define in atomic detail how conformational change was transmitted over long distances in integrins, 40 Å from the ligand binding site to the opposite end of the βI domain and 80 Å to the far end of the hybrid domain. During these movements, RGD slid in its binding groove toward αIIb, and its Arg side chain became ordered. RGD concentration requirements in soaking suggested a >200-fold higher affinity after opening. The thermodynamic cycle shows how higher affinity pays the energetic cost of opening.
Introduction
Among cell surface receptors, integrins undergo the most complex and longest-range conformational changes currently known. These changes function to transmit bidirectional signals over long distances between the ligand-binding integrin headpiece and the actin cytoskeleton. Integrins thus are able to communicate binding to the extracellular matrix or ligands on other cells to the actin cytoskeleton, to discriminate against soluble ligands, and to bind only with high affinity to cell surface or matrix-bound ligands (Luo et al., 2007; Springer and Dustin, 2012).
Integrins contain α and β subunits. The α subunit β-propeller and thigh domains and the β subunit βI, hybrid, PSI (plexin, semaphorin, and integrin), and I-EGF-1 domains form the ligand-binding headpiece, i.e., the head and the upper legs (Fig. 1). The α subunit calf-1 and calf-2 and the β subunit I-EGF-2 to I-EGF-4 and β tail domains form the lower legs (Fig. 1; Xiong et al., 2001; Zhu et al., 2008).
Figure 1.
Integrin domain organization and conformational states. Two lower β leg conformations (one with a dashed line) are shown for the extended states because the lower β leg is highly flexible (Takagi et al., 2002), and these states can exist with the α and β TM domains either associated or separated (Zhu et al., 2007b). However, signal transmission through the membrane, both in the inside-out and outside-in directions, requires TM and cytoplasmic domain separation (Kim et al., 2003; Luo et al., 2004a; Zhu et al., 2007b).
Two distinct types of global conformational changes occur in integrin extracellular domains. Extension at the knees releases integrins from a compact bent conformation (Fig. 1, A, B, D, and E). In integrin headpiece opening, the hybrid domain swings out, the βI domain changes from closed to open conformation, and affinity for ligand increases (Fig. 1, B, C, E, and F; Takagi et al., 2002; Luo et al., 2007; Springer and Dustin, 2012).
βI domains, present in all integrin β subunits, transmit conformational change from their interface with the swinging hybrid domain to a ligand binding site at an interface with the α subunit β-propeller domain (Fig. 1, A–F; Xiao et al., 2004). The βI domain divides the hybrid domain into N- and C-terminal sequence segments. Activation at the metal ion-dependent adhesion site (MIDAS) in the βI domain ligand binding site is communicated to the opposite end of the βI domain by α7 helix pistoning at the C-terminal connection to the hybrid domain (Fig. 1, B, C, E, and F). Pivoting about the N-terminal connection causes the hybrid domain to swing away from the α subunit thigh domain, with an increase in separation at the α and β knees of 70 Å (Takagi et al., 2002; Xiao et al., 2004). Swing out is readily visualized in solution by small angle x-ray scattering for both headpiece fragment (Mould et al., 2003b) and intact, detergent-soluble integrin (Eng et al., 2011) and at typical negative-stain EM or tomography resolution of ∼25 Å for intact integrins or their ectodomain or headpiece fragments (Takagi et al., 2002, 2003; Iwasaki et al., 2005; Eng et al., 2011; Shi et al., 2011; Wang et al., 2012; Yu et al., 2012).
The evidence for integrin headpiece opening and its association with the high affinity state of integrins is extensive. RGD opens the headpiece of αvβ3 ectodomain (Takagi et al., 2002). RGD and fibronectin open the α5β1 headpiece (Mould et al., 2003b; Takagi et al., 2003), and an allosteric, inhibitory Fab lowers affinity for fibronectin by stabilizing the closed headpiece (Luo et al., 2004b; Nagae et al., 2012). The αIIbβ3 headpiece cocrystallizes with ligands and pseudoligands with an open headpiece (Xiao et al., 2004) and in their absence with a closed headpiece (Zhu et al., 2010). RGD mimetics increase the Stokes radius of the αIIbβ3 headpiece by the amount predicted from the hydrodynamic radii of the crystal structures of the closed and open headpieces (Zhu et al., 2010). Headpiece opening induced by ligand mimetics has been demonstrated with detergent-soluble intact αIIbβ3 using EM (Eng et al., 2011), small angle x-ray scattering (Eng et al., 2011), and electron tomography (Iwasaki et al., 2005). Antibodies that stabilize the closed and open headpieces of β2 integrins show that the open conformation is required for cell adhesion and high affinity (Chen et al., 2010; Schürpf and Springer, 2011).
The equilibria between integrin extension and headpiece opening are linked because the hybrid domain, as part of the upper β leg, participates in interfaces with the lower legs that stabilize the bent conformation (Fig. 1). Thus, induction by ligand of headpiece opening in intact integrins or ectodomain fragments is always accompanied by integrin extension. Furthermore, because the lower α and β legs are very close in the ectodomain, and the α and β subunit transmembrane (TM) domains associate with one another, the equilibria governing integrin TM domain association, extension, and headpiece opening are all linked (Takagi et al., 2002).
The complexities of linked equilibria, the lack of a clear order to the steps in integrin activation (Takagi et al., 2002), and the complexity of the structure of the integrin ectodomain itself have all contributed to the difficulty of comprehending the structural basis of integrin function in the cell biology community. Currently, the concept that the open integrin headpiece corresponds to the conformation with high affinity for ligand is well accepted for β1, β2, β6, and β7 integrins, as referenced in the preceding paragraphs. However, this concept remains controversial in the β3 integrin field (Adair et al., 2005; Xiong et al., 2009) and has resulted in a lively dialog in the literature on αIIbβ3 and αvβ3 integrins. One idea, based on the observation that more metal ion sites in the βI domain are occupied in αvβ3 crystals after soaking with ligand than in absence of ligand, is that metal ion binding could somehow regulate ligand binding (Xiong et al., 2002). However, protonation of metal-coordinating Asp and Glu residues at the low pH at which αvβ3 crystallizes is probably responsible for variable absence of bound metal (Dong et al., 2012), and unliganded, closed αIIbβ3, which crystallizes at higher pH, has all metal sites bound (Zhu et al., 2008, 2010). Arguments for regulation by a deadbolt and then an interface between the α and β knees have been raised (Gupta et al., 2007; Xiong et al., 2009) and contradicted (Zhu et al., 2007a; Smagghe et al., 2010; Xie et al., 2010; Dong et al., 2012). In contrast, mutational studies that shorten the βI α7 helix (Yang et al., 2004) or introduce disulfide bonds (Luo et al., 2004c; Kamata et al., 2010), N-glycan wedges (Luo et al., 2003, 2004b), or point mutations (Mould et al., 2003a; Barton et al., 2004; Luo et al., 2009) consistently show that the closed and open headpiece conformations of intact β3 integrins on cell surfaces have low and high affinity for ligand, respectively.
Soaking RGD peptides into closed, bent αVβ3 ectodomain and α5β1 closed headpiece crystals has in each case revealed only a single intermediate and a conformation nearer to closed than open (Xiong et al., 2002; Nagae et al., 2012). Although the βI domain adopted an intermediate conformation in these studies, little α1 helix movement occurred, the α7 helix did not piston, and the hybrid domain did not swing out; thus, the headpiece remained in an overall closed conformation. If ligands added to crystals were able to drive conformational change all the way to the open headpiece, this would provide incontrovertible thermodynamic evidence that RGD peptides bind with higher affinity to the open than closed headpiece conformation.
With purified integrins, conformational change is most readily studied by adding an excess of ligand; however, because of the principle of reversibility of chemical reactions, the same pathway will mediate change in both the outside-in and inside-out directions. Because the difference is so large between the closed and open headpiece, structures trapped at various positions along the pathway between these states would provide important information about how such a large conformational change is accomplished. In this study, we soaked RGD peptide into closed αIIbβ3 headpiece crystals under different conditions. We captured RGD peptide bound to the closed headpiece, six intermediate states between closed and open, complete conversion of the βI domain to the open state, and swing out of the hybrid domain in the crystal lattice.
Results
Soaking RGD peptide into closed headpiece crystals of αIIbβ3
In the closed αIIbβ3 headpiece crystal form studied here, two independent molecules are present in the asymmetric unit. Each molecule contains the αIIb subunit β-propeller domain bound to 10E5 Fab and the β3 subunit PSI, hybrid, βI, and I-EGF-1 domains (Zhu et al., 2010). Crystals were soaked with or without GRGDSP peptide in buffer containing either 5 mM Mg2+/1 mM Ca2+ (Mg/Ca) or 2 mM Mn2+/0.1 mM Ca2+ (Mn/Ca; Table 1). Crystals were stable after soaking for up to 24 h in 10 mM GRGDSP and Mg/Ca; furthermore, diffraction revealed bound peptide. However, soaking with 0.34 mM peptide for 72 h did not show bound ligand. In contrast, strong electron density for ligand was observed after soaking a preformed open αIIbβ3 headpiece crystal with 0.05 mM peptide (Springer et al., 2008).
Table 1.
Relation of crystal soaking conditions to conformational state
| GRGDSP | Metal ions | Soaking time | Resolution | Chains A + B (molecule 1) | Chains C + D (molecule 2) | PDB accession no. | |
| Mn or Mg | Ca | ||||||
| mM | mM | mM | h | Å | |||
| 0 | Mn, 2 | 0.1 | 4 | 2.45 | State 1 | State 1 | 3ZDX |
| 10 | Mg, 5 | 1.0 | 24 | 2.45 | State 2a | State 1a | 3ZDY |
| 1 | Mn, 2 | 0.1 | 4 | 2.75 | State 7a | State 3a | 3ZDZ |
| 3 | Mn, 2 | 0.1 | 4 | 2.95 | State 7 | State 4a | 3ZE0 |
| 5 | Mn, 2 | 0.1 | 4 | 3.00 | State 7 | State 5a | 3ZE1 |
| 10 | Mn, 2 | 0.1 | 4 | 2.35 | State 6a | State 8a | 3ZE2 |
PDB, Protein Data Bank.
Representative state shown in the main text figures.
Soaking in 10 mM GRGDSP and Mn/Ca for 4 h resulted in no visible cracks in some crystals and microscopically visible cracks in others. Some of these crystals yielded excellent diffraction (Table 1). All crystals had cracks after 6 h, and some began to dissolve. At 12 h, all crystals dissolved. Soaking for 0.5, 1, 1.5, and 2 h failed to yield similarly good diffraction, perhaps because conformational state was not homogenous in the crystal. Therefore, in Mn/Ca, we chose 4 h as an optimum time point and varied the concentration of RGD peptide from 1 to 10 mM (Table 1). Diffraction data from a large number of soaked crystals were examined. A single round of refinement was sufficient to show whether electron density at the ligand binding site corresponding to bound peptide was present. The best diffracting crystals from six soaking conditions were fully refined from 2.35- to 3.00-Å resolution with Rfree ∼0.22 (Table 2).
Table 2.
Statistics of x-ray diffraction data and structure refinement
| Parameters | Ligand | |||||
| 0 mM GRGDSP (Mn/Ca), 4 h | 10 mM GRGDSP (Mg/Ca), 24 h | 1 mM GRGDSP (Mn/Ca), 4 h | 3 mM GRGDSP (Mn/Ca), 4 h | 5 mM GRGDSP (Mn/Ca), 4 h | 10 mM GRGDSP (Mn/Ca), 4 h | |
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 | P21212 |
| Unit cell a, b, c (Å) | 259.7, 145.0, 104.8 | 257.8, 145.3, 106.1 | 259.6, 144.5, 104.8 | 259.6, 144.7, 104.6 | 259.0, 144.6, 104.7 | 233.2, 143.6, 104.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.97934 | 0.97934 | 1.03320 | 1.03320 | 1.03320 | 0.97934 |
| Resolution (Å) | 50–2.45/2.58–2.45a | 50–2.45/2.51–2.45a | 50–2.75/2.82–2.75a | 50–2.95/3.03–2.95a | 50–3.00/3.08–3.00a | 50–2.35/2.41–2.35a |
| Number of reflections (total/unique) | 1,096,275/145,557 | 975,256/146,427 | 676,162/102,899 | 399,094/83,398 | 371,307/78,003 | 1,080,830/146,644 |
| Completeness (%) | 99.9/99.9a | 99.9/99.5a | 99.8/100.0a | 99.7/99.6a | 98.4/96.4a | 100.0/99.8a |
| I/σ(I) | 10.2/2.8a | 7.0/1.6a | 10.1/1.8a | 9.5/1.7a | 9.5/1.6a | 11.0/1.9a |
| Rmerge (%)b | 15.0/81.1a | 19.8/139.1a | 11.8/110.6a | 11.0/112.4a | 10.6/117.8a | 13.9/93.7a |
| Rwork/Rfreec | 0.163/0.197 | 0.182/0.216 | 0.184/0.221 | 0.185/0.243 | 0.175/0.233 | 0.175/0.204 |
| RMSD: bond (Å) | 0.007 | 0.005 | 0.005 | 0.005 | 0.005 | 0.006 |
| Angle (°) | 0.658 | 0.570 | 0.591 | 0.566 | 0.560 | 0.676 |
| Ramachandran plot (%)d | 96.4/3.3/0.3 | 96.2/3.6/0.2 | 95.9/3.6/0.5 | 95.6/4.1/0.3 | 95.1/4.6/0.3 | 94.5/5.3/0.2 |
| Molecules/asymmetric unit | 2 | 2 | 2 | 2 | 2 | 2 |
| Residues, αIIb/β3 | 1–457 (453)/3–466 (471)e | 1–454 (453)/3–466 (471)e | 1–455 (453)/1–466 (471)e | 1–455 (453)/1 (3)–466 (471)e | 1–455 (453)/3–466 (471)e | 1–454 (453)/3 (59)–471 (433)e |
| Numbers of amino acid/carbohydrate/water | 2,703/15/1,324 | 2,711/14/887 | 2,712/15/540 | 2,714/14/327 | 2,713/14/303 | 2,615/13/1,318 |
| Conformational states (molecule 1/molecule 2)f | State 1/State 1 | State 2/State 1 | State 7/State 3 | State 7/State 4 | State 7/State 5 | State 6/State 8 |
| PDB accession no. | 3ZDX | 3ZDY | 3ZDZ | 3ZE0 | 3ZE1 | 3ZE2 |
Mg/Ca, 5 mM Mg2+/1 mM Ca2+; Mn/Ca, 2 mM Mn2+/0.1 mM Ca2+; RMSD, Root-mean-square deviation; PDB, Protein Data Bank.
Numbers correspond to the last resolution shell.
Rmerge = ∑h∑i|Ii(h) − < I(h) > |/∑h∑i|Ii(h)|, in which Ii(h) and < I(h) > are the ith and mean measurement of the intensity of reflection h.
Rwork = ∑h||Fobs (h)| − |Fcalc (h)||/∑h|Fobs (h)|, in which Fobs (h) and Fcalc (h) are the observed and calculated structure factor amplitudes, respectively. No I/σ(I) cutoff was applied. Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 0.6% subset of data excluded from refinement.
Residues in favorable, allowed, and outlier regions of the Ramachandran plot are as reported by MolProbity.
Numbers in parenthesis correspond to chains C and D.
Molecule 1 = chains A and B; and molecule 2 = chains C and D.
All crystals were isomorphous with starting crystals, except for crystals soaked with 10 mM GRGDSP in Mn/Ca. These crystals shrank 26 Å along the a axis (Table 2), correlating with swing out of the hybrid domain in one molecule in the asymmetric unit (see Hybrid domain swing out in the crystal lattice in Results). An almost identical change in unit cell dimension occurred in a crystal soaked with 10 mM of a one-residue-longer GRGDSPK peptide in Mn/Ca, and it also showed hybrid domain swing out; however, because resolution was lower at 2.75 Å and the C-terminal Lys residue of the GRGDSPK peptide was not resolved, this crystal was not fully refined.
Molecules 1 and 2 in the asymmetric unit in each of six crystals result in 12 examples of the αIIbβ3 headpiece bound to GRGDSP peptide (Table 1). Comparisons among these, with a previously published native closed-headpiece structure (Zhu et al., 2012) and with an open-headpiece structure that was cocrystallized with a ligand mimetic (Springer et al., 2008), define eight conformational states. States 1 and 8 correspond to closed and open, respectively, and states 2–7 correspond to intermediate conformations (Table 1).
Overall, there was a close correlation between soaking condition and conformational state (Table 1). The shift in conformation in Mn/Ca was clearly dependent on RGD because no conformational change was observed in Mn/Ca alone (Table 1; Fig. S1, A and B; and Fig. S2, A and B). In Mn/Ca, the conformation of molecule 2 progressively shifted more toward open between 1, 3, 5, and 10 mM RGD (Table 1). In contrast, the conformation of molecule 1 shifted immediately to state 7 at 1 mM RGD in Mn/Ca and continued to be in state 7 in 3 and 5 mM RGD until it shifted to state 6 in 10 mM RGD, coincident with the change in lattice dimensions and the swing out of the hybrid domain in molecule 2.
Insights into ligand binding
RGD binds at the interface between the αIIb β-propeller and β3 βI domains (Fig. 2). The Arg and Asp side chains of RGD extend linearly in opposite directions toward αIIb and β3, respectively (Fig. 2, B–J). A binding pocket is formed by aliphatic and aromatic side chains, water-mediated interaction with the Arg backbone carbonyl oxygen, and specific interactions with the Arg and Asp side chains. The Arg’s positively charged guanidino moiety forms a salt bridge and, in states 4–8, also forms hydrogen bonds to the side chain of αIIb residue Asp-224 (Fig. 2). One Asp carboxyl oxygen coordinates to the MIDAS metal ion, and depending on the conformation, the two Asp carboxyl oxygens hydrogen bond to one to three backbone nitrogens of βI domain residues Tyr-122 and Ser-123 in the β1-α1 loop and Arg-214 (Fig. 2, B–J).
Figure 2.
The RGD-binding pocket. (A–J) The αIIbβ3 headpiece states are native closed (A; Zhu et al., 2012); the states indicated in Table 1 (B–I), and native open (J; Springer et al., 2008) are shown. Residues that contribute to the RGD-binding pocket are shown both as sticks and transparent surfaces in light blue (αIIb) and wheat (β3). Metal ions are shown as yellow (SyMBS and ADMIDAS) or cyan (MIDAS) spheres. Waters are smaller red spheres. GRGDSP peptides are shown in stick with green carbons. Oxygens and nitrogens are red and blue, respectively. Composite omit simulated-annealing electron density is in black mesh contoured at 3 σ for SyMBS and MIDAS metal ions, 1 σ (except 0.5 σ in E and F) for ADMIDAS metal ion, and 0.5 σ for waters and GRGDSP peptide. Hydrogen bonds and metal ion coordination bonds are dashed.
Electron density for the ligand in general correlated with the ligand concentration used in soaking and the amount of conformational change (Fig. 3). The real space correlation coefficient (RSCC) of electron density for Arg, Gly, and Asp residues in RGD is an estimate of occupancy and order of the ligand at the ligand binding site. The overall trend in increase of RSCC for each of Arg, Gly, and Asp in molecule 2 after soaking for 4 h in Mn/Ca with 1, 3, 5, and 10 mM RGD (states 3, 4, 5, and 8) shows that occupancy by RGD increases over this concentration range (Fig. 3). For comparison, we show RSCC for open αIIbβ3 headpiece crystals formed with cacodylate ion bound to the βI MIDAS; the cacodylate was replaced by soaking with 0.05 mM RGD peptide for 96 h in Mg/Ca (Fig. 3, state 8). The RSCC values for Asp and Gly of the latter are similar to those for molecule 1 in state 6 and molecule 2 in state 8, which suggests that saturation with RGD is nearly complete after soaking with 10 mM RGD for 4 h in Mn/Ca (Fig. 3).
Figure 3.
Occupation of the ligand binding site by RGD. As an estimate of binding of each residue of RGD, and their order, we measured the real space cross-correlation between composite omit simulated-annealing electron density and the molecular model of bound RGD, as explained in the Materials and methods.
Among the residues of the ligand, the order of RSCC is Arg < Gly < Asp for all molecules (Fig. 3). This is consistent with Asp as the primary driver of RGD binding and disorder or multiple conformations of the Arg side chain. Two alternative Arg side chain conformations were evident in molecules 1 and 2 after soaking with 10 mM peptide for 24 h in Mg/Ca (Fig. 2, B and C). Electron density for Arg was markedly weaker in molecule 2, and a single side chain conformation was modeled, after soaking with 1, 3, and 5 mM peptide for 4 h in Mn/Ca (Fig. 2, D–F). Molecule 1 has greater accessibility of its ligand binding pocket in the crystal lattice than molecule 2, and more completely bound ligand when soaking was limited to 4 h (Fig. 3).
Strong electron densities were present for the metals at the three βI domain metal ion binding sites. When Mn2+ was present, it largely replaced the Mg2+ at the MIDAS and the Ca2+ at the synergistic metal binding site (SyMBS) and adjacent to MIDAS (ADMIDAS); electron density was fit best when metals at all three sites were modeled as Mn2+ (Fig. 2, D–I; and Fig. S1). In crystals soaked with Mn/Ca alone, the RGD-binding pockets of both molecule 1 and 2 were occupied with solvent molecules, which often occupied the same positions as polar atoms of the ligand (Fig. S1, A and B).
Remarkably, the structures reveal movements in position of the bound RGD. One of the most important movements during opening is the strengthening of hydrogen bonds between the RGD Asp side chain and the βI β1-α1 loop backbone as the distance between these elements decreases (Fig. 2 and Fig. 4). However, both elements also move together toward the α subunit, with the Asp side chain and β1-α1 loop moving 1.3 and 2.2 Å, respectively (Fig. 4). Thus, during the opening process, the entire RGD backbone slides in its groove closer to Asp-224 in the β-propeller domain (Fig. 2). The Arg side chain forms strong hydrogen bonds to αIIb Asp-224 only in the final stages of RGD backbone sliding. In states 1–3, the distances are too great for hydrogen bonds, and intervening water molecules are explicitly visible in states 1 and 2 in which RGD density is strong (Fig. 2, B and C). Arg density is weak in states 3–5 (Fig. 2, D–F), and two Arg conformations are present in state 6 (Fig. 2 G). It is only by state 7 that two strong hydrogen bonds develop between the RGD Arg and αIIb Asp-224 side chains (Fig. 2 H and Fig. S1, C and D).
Figure 4.
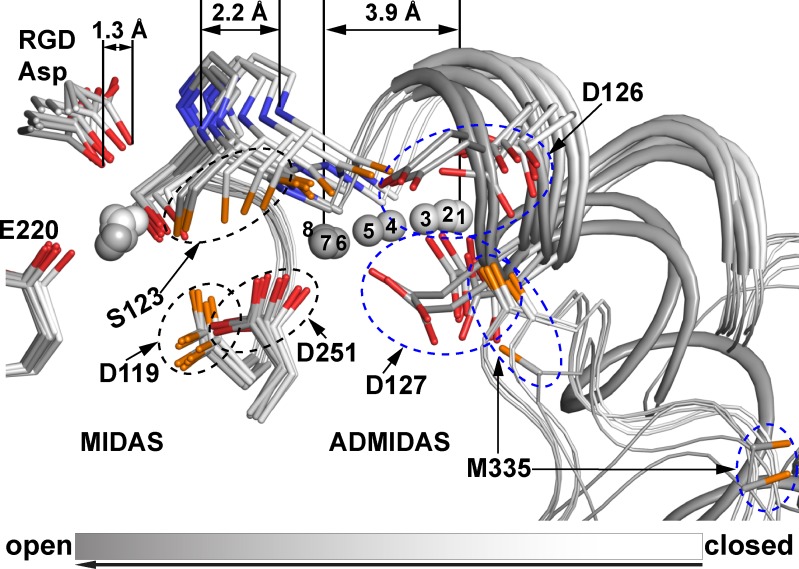
Conformational transition from closed to open around the MIDAS and ADMIDAS. States 1–8 are shown superimposed and shaded on their carbons and metal ions over a grayscale from closed state 1 (white) to open state 8 (dark gray). Key side chain, RGD Asp, and β1-α1 loop backbone atoms are shown in sticks. The remaining backbone is shown as a wormlike trace, with the α1 and merged α1/α1′ helices thicker. Distances show overall movements. MIDAS and ADMIDAS metal ions are spheres with states numbered for the ADMIDAS. Some side chains and the Met-335 carbonyl group are circled, and their oxygens are shown in orange or red to tell them apart. Nitrogens are shown in blue.
The pathway of RGD-induced headpiece opening
Our eight states reveal a detailed pathway for headpiece opening (Fig. 5 and Fig. 6). Movements occur only in the β subunit. From state 1 to 8, the Ser-123 backbone in the β1-α1 loop moves 2.2 Å, the ADMIDAS metal ion moves 3.9 Å, and the carbonyl oxygen of Met-335 moves 9.0 Å (Fig. 4). Fig. 5 shows a view of each state (Fig. 5, S1–S8) that includes the ligand Asp and the moving portions of the βI domain. Fig. 6 shows comparisons between each consecutive state, with more detail in the most relevant region. In each figure, structures from crystals formed in the absence of ligand (closed; Protein Data Bank accession no. 3T3P; Zhu et al., 2012) and the presence of ligand (open; Protein Data Bank accession no. 2VDR; Springer et al., 2008) are included for comparison. The key transitions in each step are described as follows.
Figure 5.
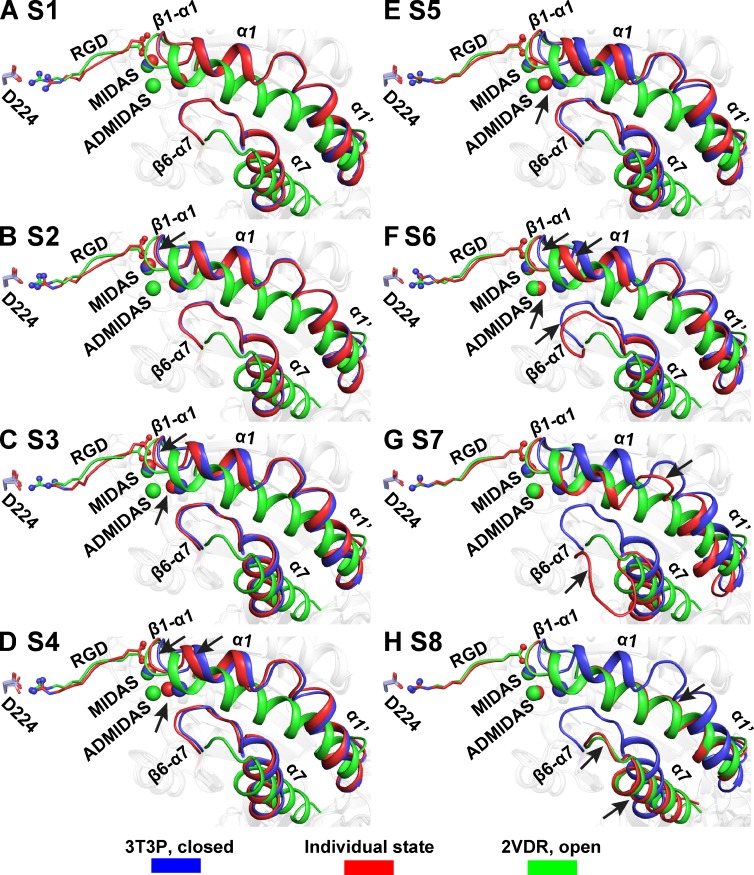
Overview of the moving portions of the βI domain. (A–H) βI domain regions that undergo the largest movements are shown in cartoon. Asp-224 of the αIIb subunit and RGD are shown in stick. MIDAS and ADMIDAS metal ions are shown as spheres. States 1–8, S1–S8.
Figure 6.
Detailed comparisons between nearest-neighbor states. (A) The region around the ligand Asp, β1-α1 loop, α1-helix, and β6-α7 loop where movement is greatest between states 1 and 6 (S1–S6). (B) The region around the α1 helix, α1′ helix, β6-α7 loop, and α7 helix where movement is greatest between states 6 and 8. Each panel compares two nearest-neighbor states. For economy, and to compare the two rotamers of W129 in states 1–5, image 1 in B compares states 4 and 6. The carbons and metal ions of each state are in the same colors as the names of each state or the reference structures 3T3P (closed) and 2VDR (open). For clarity, water molecules as spheres and metal coordination bonds as red dashes are shown only for the second named structure in each image. Nitrogens and oxygens are shown in blue and red, respectively.
The structure in state 1 of molecule 2 in 10 mM RGD in Mg/Ca (Fig. 2 B) is remarkable because despite strong density for bound RGD, there is no significant difference from the closed conformation in absence of RGD (Fig. 5 A). One RGD Asp carboxyl oxygen directly coordinates the MIDAS Mg2+ (Fig. 2 B) by replacing a water molecule (Fig. 2 A), whereas the other Asp carboxyl oxygen hydrogen bonds to the backbone nitrogen of Tyr-122 (Fig. 2 B and Fig. 6 A, image 1). However, the Asp side chain orients differently than in all other states (Fig. 6 A).
In state 2, the RGD Asp side chain rotates to the same orientation as in states 3–8 (Fig. 6 A, image 2). Furthermore, the βI domain β1-α1 loop moves 0.5 Å toward the Asp of RGD.
In state 3, the β1-α1 loop moves still closer to the Asp of RGD (Fig. 6 A, image 3). The closer approach of the β1-α1 backbone nitrogens, together with the rotation of the Asp side chain in preceding state 2, enables formation of a second backbone hydrogen bond to the RGD Asp (Fig. 6 A, image 3). These two hydrogen bonds, between each of the two Asp oxygens and the Tyr-122 and Ser-123 backbone NH groups, remain intact in all subsequent states and grow in strength with closer approach of the β1-α1 backbone to the Asp side chain carboxyl oxygens. The approach of the Ser-123 backbone to the Asp side chain is accompanied by approach of the Ser-123 side chain oxygen to the MIDAS metal ion. These two atoms form a direct coordination for the first time in state 3 (Fig. 6 A, image 3), which is maintained in all subsequent states. The Ser-123 side chain takes the position of a water, which in states 1 and 2 intervenes between the MIDAS metal ion and the Ser-123 side chain (Fig. 6 A, images 1 and 2). To follow the movement of the β1-α1 backbone, the ADMIDAS metal ion shifts 0.6 Å to maintain coordination to the Ser-123 carbonyl oxygen (Fig. 6 A, image 3). Furthermore, the β6-α7 loop moves with the ADMIDAS metal to maintain close coordination with the Met-335 carbonyl oxygen (Fig. 6 A, image 3).
In state 4, the β1-α1 loop and the ADMIDAS metal ion continue to approach the RGD Asp and MIDAS metal ion (Fig. 6 A, image 4). The RGD Asp side chain also moves, allowing RGD backbone sliding toward αIIb. The α1 helix is divided by a nonhelical segment into α1 and α1′ helices (Fig. 5). In state 4, the α1 portion begins to follow the movements of the β1-α1 loop and the ADMIDAS metal ion (Fig. 5 D and Fig. 6 A, image 4). The ADMIDAS metal ion continues to follow the movement of Ser-123 carbonyl oxygen and shifts an additional 1.0 Å, and ADMIDAS-coordinating residues in the α1 helix, Asp-126 and Asp-127, begin to follow the movement of the ADMIDAS metal ion (Fig. 6 A, image 4).
State 5 marks further movements of the ADMIDAS metal ion and a change in its coordination status. This metal ion now loses its coordination to the β6-α7 loop backbone; however, the β6-α7 loop maintains the position adopted since state 3 (Fig. 6 A, image 5). Furthermore, the ADMIDAS gains a direct coordination to the side chain of Asp-251 (Fig. 6 A, image 5). This direct coordination remains all the way to open state 8 and replaces an indirect coordination through a water molecule to Asp-251 in states 1–4 (Fig. 6 A).
State 6 marks the completion of the movements of the β1-α1 loop and ADMIDAS metal ion, which bring them into positions that are essentially the same as in states 7 and 8 (Fig. 5, F–H; and Fig. 6 A, image 6). The α1 helix moves in position more between states 5 and 6 than between previous states (Fig. 5, A–F), accompanied by changes in positions of ADMIDAS-coordinating α1 helix side chains Asp-126 and Asp-127 (Fig. 6 A, image 6). However, the ADMIDAS metal ion moves further than these side chains, and direct ADMIDAS metal ion coordinations are lost to Asp-126 and Asp-127. In their place, two water molecules appear that mediate indirect ADMIDAS metal ion coordinations to Asp-126 and Asp-127 (Fig. 6 A, image 6). The β6-α7 loop reverses its movement toward the ADMIDAS metal ion seen in state 3 and moves away from the metal ion, making way for the α1 helix to move toward the ADMIDAS (Fig. 6 A, image 6).
The three examples of state 7 each show a single RGD Arg side chain conformation (Fig. 2 H and Fig. S1, C and D) and essentially identical βI domain conformations (Fig. 5 G and Fig. S2, C and D). In state 7, the epicenter of conformational change shifts to the α1 helix and the β6-α7 loop (Fig. 6 B, image 2). The α1 helix with its Asp-126 and Asp-127 residues moves almost as a rigid body toward the ADMIDAS metal ion, enabling direct coordinations of each Asp side chain to reform with the ADMIDAS metal ion. Additionally, Trp-129 in the α1 helix changes rotamer and invades the space of the β6-α7 loop even more than the α1 helix backbone (Fig. 6 B, image 2). These invasions completely displace the β6-α7 loop, which flips far away to make way for the α1 helix (Fig. 6 B, image 2).
In state 8, the action shifts to the α1′ and α7 helices. Between states 7 and 8, the α1′ helix moves as a rigid body toward the α1 helix, with large movements of its buried, hydrophobic Leu-134 and Leu-138 side chains (Fig. 6 B, image 3). The loop between the α1 and α1′ helices becomes helical, and the α1 and α1′ helices with a bend between their axes merge into a single α1 helix with a single helical axis. Trp-129, near the merger, changes rotamer for the final time (Fig. 6 B, image 3). To make way for the merger of the α1 and α1′ helices, the β6-α7 loop reshapes, and the α7 helix pistons along its helical axis toward the hybrid domain (Fig. 6 B, image 3).
Thus, as in a series of falling dominoes, successive movements of the β1-α1 loop, the ADMIDAS metal ion, the α1 helix, and the α1′ helix propagate to the β6-α7 loop and α7 helix. Remarkably, the conformation of the βI domain in state 8, achieved by soaking closed headpiece crystals with 10 mM RGD peptide for 4 h in Mn/Ca, is indistinguishable from the fully open state seen after cocrystallization with a ligand mimetic in Mg/Ca (Fig. 6 B, image 4). Even details of loop positions and rotamers of key side chains are identical. Opening is complete.
Hybrid domain swing out in the crystal lattice
βI domain α7 helix pistoning is accommodated by swing out of the hybrid domain. Fig. 7 (A–C) shows the environments in the crystal lattice, with one integrin molecule shown as a Cα trace with each domain in a different color. Fab fragments and neighboring integrin molecules in the crystal lattice are shown as white, semitransparent, solvent-accessible surfaces. The native crystal lattice has room for the hybrid domain of molecule 2 to move to the right in the orientation shown in Fig. 7 A. In contrast, similar movement of molecule 1 is prevented by its lattice (Fig. 7 B). Indeed, soaking with 10 mM RGD in Mn/Ca for 4 h induced swing out of the hybrid domain of molecule 2 (Fig. 7 C) but not molecule 1 (Fig. 7 B). The change in position of the hybrid domain after swing out is emphasized by placing the label “hybrid” in identical positions in Fig. 7 (A–D). Although the dominant position of the hybrid domain in the lattice was clearly evident from simulated-annealing composite omit map (mesh; Fig. 7 C), this density was weaker than for the hybrid domain of molecule 1, showing some variation in the position in the crystal lattice of the swung out hybrid domain in molecule 2. Density for the PSI and I-EGF-1 domains was insufficient to include them in the molecular model. Enough space is present in the lattice to place the PSI and I-EGF-1 domains based on the position of hybrid domain, as shown by models of the PSI and I-EGF-1 in Fig. 7 C. Further, outward swing of the hybrid, PSI, and I-EGF-1 domains appears to be constrained by the crystal lattice (Fig. 7 C).
Figure 7.
Hybrid domain swing out. (A–D) One integrin molecule is shown as a Cα trace, with different colors for each domain. The hybrid domain (red) and PSI and I-EGF-1 domains (yellow) are shown as thicker traces for emphasis. Other integrin molecules and all Fabs in the crystal lattice are shown as white, semitransparent, solvent accessible surfaces. The label hybrid is placed in identical positions in A–D. (A) Molecule 2 before soaking (3T3P closed structure). (B) Molecule 1 after soaking with 10 mM RGD and Mn/Ca. (C) Molecule 2 after soaking with 10 mM RGD and Mn/Ca. Composite omit simulated-annealing electron density contoured at 0.5 σ around the hybrid domain is shown as purple mesh. PSI and I-EGF-1 domains are missing in density, and superposition on the hybrid domain is used to show their approximate location in the lattice. (D) The native open headpiece (Protein Data Bank [PDB] accession no. 2VDR) superimposed based on the β-propeller and βI domains in C and shown in the same lattice as in C. Severe clashes are evident. (E) Superposition of αIIbβ3 headpieces. Similar regions in gray and colored shape-shifting portions in cartoon; metal ions are shown as spheres, and RGD is shown in stick. Structures are molecule 2 after soaking with 10 mM RGD and Mn/Ca (red), native closed (PDB accession no. 3T3P; blue), and native open (PDB accession no. 2VDR; green).
Fig. 7 E compares the positions of the hybrid domain obtained in previous closed (Fig. 7 E, blue) and open (Fig. 7 E, green) crystal structures with that attained here by soaking (Fig. 7 E, red). Binding of RGD has clearly induced swinging of the hybrid domain toward its position in open crystal structures. However, further movement to the same open position as seen when the headpiece is cocrystallized with ligand results in severe clashes with the lattice. This is illustrated by superposition of the open conformation seen in cocrystallization into the lattice, which shows that much of the ribbon trace of the hybrid domain is obscured by the semitransparent surface of the lattice (Fig. 7 D). Clashes would be even more severe for the other molecule in the lattice (Fig. 7 B) were its hybrid domain to swing out, consistent with cracking and dissolution of crystals soaked for 6 and 12 h.
Discussion
Previous crystal structures of integrin αIIbβ3 defined two conformational states of its headpiece, closed and open. The same closed headpiece conformation was seen in crystals of the complete αIIbβ3 ectodomain without Fab (Zhu et al., 2008) and in crystals of the αIIbβ3 headpiece complex with 10E5 Fab identical to those studied here (Zhu et al., 2010). Similarly, the same open conformation of the αIIbβ3 headpiece was seen with a cacodylate pseudoligand or RGD mimetics, with or without bound 10E5 Fab (Xiao et al., 2004; Springer et al., 2008). Fabs have no carbohydrate and minimal flexibility compared with integrins and can stabilize crystal lattices and improve resolution. 10E5 Fab binds to the β-propeller domain, far from shape-shifting portions of the β subunit, and has no influence on β-propeller or integrin conformation as shown by comparisons among these many views of the αIIbβ3 structure.
Conformational changes have previously been visualized by soaking RGD into crystals containing the αVβ3 ectodomain (Xiong et al., 2002) or α5β1 headpiece (Nagae et al., 2012) with closed conformations. The previous RGD/αVβ3 structure in Mn2+ is closest to our state 5. The RGD/α5β1 structure in Mg/Ca is closest to our states 3 and 4. These structures showed one intermediate trapped at a particular point in the shape-shifting pathway, in contrast to the eight RGD-bound states in a conformational continuum studied here.
Although the term hinge opening was used in molecular dynamics (MD) simulations of αVβ3 (Puklin-Faucher et al., 2006; Puklin-Faucher and Vogel, 2009), the simulations did not result in the open βI domain or open headpiece conformations seen here or previously (Xiao et al., 2004). Compared with a 62° increase in angle between βI and hybrid domains in the open headpiece crystal structure, MD showed a 23° increase starting with a model of Fn3 domain 10 bound to a RGD-liganded state 5–like structure and a 13° increase starting with an unliganded state 1 structure lacking metal ions at the MIDAS and SyMBS. The α1 and α1′ helices merged and approached the α7 helix; however, merger occurred in the absence of α7 helix pistoning and displacement of β6-α7 loop residue Val-340 from its ratchet pocket and occupation of this pocket by Leu-134 in the α1/α1′ helix. Because the simulations began with state 5, they were silent on RGD sliding and β1-α1 loop movement, which are largely complete by states 5–6. Dependence on RGD was not demonstrated by omitting RGD from the state 5–like structure. RGD was not sufficient for α1 and α1′ helix merger because it required a modeled interaction between the body of Fn3 domain 10 and the Trp-129 side chain in the α1 helix itself. Large changes in position and rotamer of Trp-129 between our states 4, 6, 7, and 8 (Fig. 6 B) are incompatible with maintenance of a side chain hydrogen bond to the Fn3 domain 10 in the MD simulations. MD simulations of integrins are challenging because the force fields lack descriptions of the highly directional nature of octahedral metal coordination and hydrogen bonds. MD lost physiological coordinations at the MIDAS and SyMBS (Craig et al., 2004) and an invariant water at the MIDAS (Puklin-Faucher et al., 2006). Additionally, the αVβ3 structures used in simulations lacked cis-Pro at βI domain residues 163 and 169 and contained a sequence-to-structure frameshift at specificity-determining loop residues 168–176 (Dong et al., 2012) that lie near the Fn3 docking site and α1 helix and could have affected MD results.
Surprisingly, βI domain reshaping is related to a change in position of RGD in the ligand-binding pocket. Movement of the β1-α1 loop toward the Asp of RGD permitted the entire RGD moiety to slide in its groove away from the β3 subunit and toward the αIIb subunit. Sliding enabled a water-mediated interaction between the RGD Arg side chain and αIIb Asp-224 to be converted to a much stronger direct, charged interaction through multiple hydrogen bonds. Sliding also correlated with a shift from multiple Arg side chain conformations to a single conformation. Sliding and disorder or multiple conformations of the Arg side chain show that binding of the Asp of RGD to the MIDAS metal ion and the β1-α1 loop backbone is energetically more important than Arg interactions with the αIIb subunit.
Placing our structural snapshots into sequence in a movie provides a plausible shape-shifting pathway for conformational change in integrins (Videos 1, 2, and 3). In contrast to previous soaking studies, our state 1 structure shows that RGD can bind to αIIbβ3 with no appreciable change in structure. The early parts of the pathway from state 1 to 6 are captured in atomic detail as movements of <1 Å, whereas those in states 7 and 8 involve large concerted changes. Ligand-induced integrin shape shifting begins in the β1-α1 loop and then works its way more C-terminally in the α1 and α1′ helices until they finally merge and push the β6-α7 loop and α7 helix out of the way toward the hybrid domain.
Linkage between the MIDAS and ADMIDAS coordination shells grows between states 3 and 6 (Video 2). In state 3, the Ser-123 side chain replaces a water molecule in the inner MIDAS coordination sphere; the Ser-123 backbone oxygen directly coordinates the ADMIDAS through states 1–8. In state 5, the ADMIDAS metal ion directly coordinates the Asp-251 side chain. In state 6, the same Asp-251 oxygen indirectly coordinates the MIDAS through the Ser-123 side chain; the other Asp-251 oxygen indirectly coordinates the MIDAS through a water molecule throughout states 1–8 (Fig. 6). By state 6, interaction between the MIDAS and ADMIDAS coordination shells is strong enough to leave behind the direct ADMIDAS coordinations to the Asp-126 and Asp-127 side chains, which are reformed in state 7.
Saltatory motions between states 1 and 7 thus include numerous steps of removal, addition, and removal yet again of waters that provide indirect MIDAS and ADMIDAS coordinations to side chains. Another movement not predicted by structural interpolation is that of the β6-α7 loop. Its coordinating Met-335 carbonyl oxygen moves toward the fleeing ADMIDAS in state 3 and stays in this new position until state 6, when it moves in the opposite direction.
Presumably, the movement of the β1-α1 loop places strain on the α1 helix, which contains ADMIDAS-coordinating residues Asp-126 and Asp-127. In state 7, there is a large rigid-body movement of the α1 helix, as Asp-126 and Asp-127 catch up with and reform direct coordinations to the ADMIDAS. In turn, the movement of the α1 helix is likely to place strain on the α1′ helix. In final state 8, the α1′ helix catches up with and joins to the α1 helix. Alignment of the α1 and α1′ helices squeezes at their junction the β6-α7 loop. The hydrophobic ratchet pocket occupied by β6-α7 residue Val-340 in states 1–6 is occupied by α1 helix residue Leu-134 in state 8, with Val-340 in an intermediate position in state 7 (Fig. 6). Merging of the α1 and α1′ helices appears to be the final straw that breaks the camel’s back and pushes the α7 helix toward the hybrid domain, causing it to swing out. The side chain of Trp-129 near the end of the α1 helix appears to buffer the large α1 and α1′ conformational movements between states 6 and 8. Between states 1 and 6, the Trp-129 side chain can occupy either of the two rotamers shown in Fig. 6 B (image 1). In state 7, it adopts a very different buried rotamer and acts as a placeholder for the α1′ helix. Then, in state 8, Trp-129 moves outward again and adopts yet another rotamer to make way for the merge of the α1 and α1′ helices and the side chain of Leu-134.
These movements illustrate the separability yet interdependence of the MIDAS and ADMIDAS coordination shells, the β1-α1 loop, α1 helix, α1′ helix, and β6-α7 loop. The final movements of the α1′ helix and α7 helix are highly concerted (Video 3). Compared with the earlier movements in states 1–6, those of the α1 helix and β6-α7 loop in state 7 are much larger, whereas the movements in the α1′ helix, β6-α7 loop, and α7 helix in state 8 are cataclysmic. The extraordinary movements that follow state 7 likely correspond to a large decrease in free energy and suggest that state 7 must be close to the transition state for conformational change.
Among the caveats in our study is the difficulty of modeling the segments undergoing conformational change as single conformations. Particularly for the β6-α7 loop in state 7, density is consistent with multiple backbone conformations. Resolution of most of our structures was sufficient to model alternative conformations of the side chains of some residues; however, attempts to include alternative backbone conformations did not improve model fit to density. In general, the quality of the density in our structures is high. It is difficult to explain the gradual shift in positions described here in each structure as an averaging of structural ensembles that change in occupancy and not in position. Further evidence against this idea is the saltatory nature of many of the movements.
Another caveat is that the states seen here are trapped in crystals and may be perturbed by lattice contacts. However, changes occurred in two molecules in different lattice environments. Furthermore, despite presence of states 2, 6, and 7 in molecule 1 and states 1, 3, 4, 5, and 8 in molecule 2, these states fall on a single pathway of headpiece opening. Constraints limiting conformational change differ in crystals and on cell surfaces. Nonetheless, hybrid domain swing out is the largest motion in headpiece opening and thus may be rate limiting for conformational change on cell surfaces as well as in crystals. Furthermore, the different moving elements, i.e., α1 helix, α1′ helix, β6-α7 loop, α7 helix, and hybrid domain are key on cell surfaces as well, as shown by mutational studies in these elements (Luo et al., 2003, 2004b,c, 2009; Mould et al., 2003a; Barton et al., 2004; Yang et al., 2004; Kamata et al., 2010).
It is extremely difficult to obtain information on conformational change on cell surfaces at any similar resolution, and structures determined here will likely offer the best models for understanding integrin allostery on cell surfaces for many years. The principle of the reversibility of chemical reactions implies that inside-out activation of integrins may proceed by a similar pathway as outside-in activation studied here but in the opposite direction.
Two alternative mechanisms for achieving conformational change in proteins, selection by ligand of preexisting conformational states and ligand-induced fit (Henzler-Wildman and Kern, 2007), are linked in a thermodynamic cycle (Fig. 8). In the crystals studied here, no conformational change is required for RGD binding to state 1, and thus, induced fit (Fig. 8, k1 and k2) is the mechanism driving conformational change. However, on cell surfaces, interconversion between conformational states driven by thermal motion (Fig. 8, k3 and k−3) will be far faster than in crystals, and inside-out signals may also increase k3. Therefore, the relative fluxes of integrins to ligand-bound states on cell surfaces through induced fit (k1 and k2), and selection of preexisting states (k3 and k4) will depend on their relative rates, as well as ligand concentration, and remains to be determined.
Figure 8.
The thermodynamic cycle for ligand binding and conformational change in integrins. The ΔG values are for reactions in the direction shown by the arrows. From the unliganded closed state in the bottom left, the induced-fit mechanism proceeds clockwise, and the preexisting conformational change mechanism with selection by ligand proceeds counterclockwise, to the liganded open state in the top right. The cycle for headpiece fragments can be modified for intact integrins on cell surfaces by adding additional conformational states such as those shown in Fig. 1.
RGD must bind with higher affinity to the open than closed headpiece to drive conversion to the higher energy open headpiece conformation. This may be formally demonstrated using the thermodynamic cycle (Fig. 8). The difference in energy between any two states in this cycle is identical, whether conversion occurs by clockwise or counterclockwise routes. Therefore, ΔGopen+RGD = ΔGclosed+RGD + ΔGinduced fit − ΔGconf. Crystal soaking experiments demonstrate that ΔGinduced fit is highly negative. A large number of experiments referenced in the Introduction demonstrate that in absence of ligand, [Closed] >> [Open]; therefore, ΔGconf is highly positive. It follows from the aforementioned equation that ΔGopen+RGD << ΔGclosed+RGD; i.e., that the affinity of the open headpiece is much higher than that of the closed headpiece for RGD.
Conclusions based on soaking experiments in Mn/Ca and the thermodynamic cycle are supported by soaking results in Mg/Ca (Fig. 3). Based on soaking with 10 mM RGD in Mn/Ca for different times, near-equilibrium concentrations of RGD should have been reached inside the crystal lattice after soaking with 10 mM RGD in Mg/Ca for 24 h. Incomplete occupation of the ligand binding site in Mg/Ca as shown by the RSCC values in Fig. 3 suggests an affinity insufficient to saturate binding in 10 mM RGD, consistent with lack of detectable binding after soaking for 72 h with 0.34 mM RGD in Mg/Ca. In contrast, when crystals with the open headpiece were soaked for 96 h with 0.05 mM RGD in Mg/Ca, RGD peptide completely replaced a cacodylate pseudoligand (Fig. 3; Springer et al., 2008). Complete saturation of the open headpiece at 0.05 mM RGD and incomplete saturation of the closed headpiece at 10 mM RGD support the conclusion that the affinity of the open αIIbβ3 headpiece for RGD peptide is ≥200-fold higher than that of the closed headpiece in Mg/Ca.
The ability of the lattice around molecule 2 to hold the integrin headpiece in the closed conformation after binding RGD in Mg/Ca provides a thus far unique estimate of affinity of the closed headpiece for RGD. This affinity is between 0.34 and 10 mM and closer to 10 mM. It is not possible to estimate the affinity of the closed headpiece for RGD from measurements of binding to integrins in solution or on cell surfaces because, as discussed in the previous two paragraphs, the higher affinity for the open headpiece is more than sufficient to pay the energetic penalty of inducing or selecting the open headpiece conformation. Conversion to the open headpiece is consistent with 50% effective concentration estimates in the range of 7 µM to 1 mM RGD in Mg/Ca for binding of antibodies to ligand-induced binding sites or increased protease sensitivity using isolated, intact αIIbβ3 or αIIbβ3 and αVβ3 on cell surfaces (Parise et al., 1987; Frelinger et al., 1988, 1990). 50% inhibitory concentration values for inhibition by RGD of activated platelet binding to ligands are in the range of 0.01–0.2 mM (Plow et al., 1985), suggesting that the headpiece has been opened.
The thermodynamic cycle characterized here for headpiece fragments has wide applicability. In extended integrins (Fig. 1, B, C, E, and F), as in headpiece fragments, none of the headpiece domains are in buried interfaces, and the lower β leg is highly flexible (Fig. 1, dashed lines). Therefore, the relative energies of the four states in the cycle (Fig. 8) are expected to be very similar in headpiece fragments and in extended integrins on cell surfaces. Bent integrins on cell surfaces have extensive interfaces that are exposed upon extension; the hybrid domain is in one of these buried interfaces (Fig. 1, A and D). Also, the α and β subunit C termini are close to one another in the bent conformation, and the TM domains associate (Luo et al., 2007; Springer and Dustin, 2012). Because headpiece opening exposes most of the same interfaces that become exposed upon integrin extension, ΔGinduced fit and ΔGconf in Fig. 8 (which each require headpiece opening) are higher (by similar amounts) for cell surface integrins than for headpiece fragments. If one is willing to accept that ligand-induced binding site epitopes in β3 integrins measure either integrin extension induced by headpiece opening or headpiece opening itself, one could conclude that ΔGinduced fit is negative and ΔGconf is positive (as for integrin headpieces) and deduce that the open headpiece conformation has higher affinity for RGD than the closed headpiece conformation in intact integrins, just as demonstrated here for headpiece fragments.
A remaining question is whether headpiece opening upon ligand binding is general for integrins or is dependent on the integrin or the ligand. MAdCAM-1 binding to the α4β7 headpiece can yield either an intermediate or open state as seen by EM (Yu et al., 2012), which appear to mediate rolling adhesion (intermediate affinity) and firm adhesion (high affinity), respectively (Chen et al., 2004). Thus, there may be differences among integrins. However, headpiece opening has been demonstrated for all integrins yet tested, including those containing the β1, β2, β3, β6, and β7 subunits (Shi et al., 2011; Springer and Dustin, 2012; Yu et al., 2012). An obvious exception would be when a carboxyl group is absent in the ligand because this is the moiety that interacts with the integrin β subunit in which allostery occurs. Thus, antagonists that mimic the Arg moiety of RGD and do not bind the MIDAS, or instead displace Mg2+ from the MIDAS, do not activate opening and stabilize the closed headpiece against opening, respectively (Zhu et al., 2010, 2012).
Is binding to the MIDAS alone sufficient to open the headpiece, or must another moiety such as the Arg of RGD pull the βI domain β1-α1 loop toward the α subunit to open the headpiece? The primary role of the Asp shown here in allostery suggests that Arg might not be required. Some small molecules based on RGD, selected by the pharmaceutical industry for their ability to bind equally well to the low and high affinity states of αIIbβ3, are reported not to induce reactivity with antibodies to ligand-induced binding sites (Aga et al., 2004) and, therefore, appear not to induce headpiece opening. These mimetics have unique chemical features that may set them apart from the Asp and Glu side chains present in physiological integrin ligands.
Further investigation is important of the principles that ligand binding induces integrin headpiece opening and that the open headpiece corresponds to the high affinity state of integrins. The large movement at the integrin knees upon headpiece opening is thought to be important for transmission of allostery through long legs that are flexible except when elongational force is applied in cell adhesion (Zhu et al., 2008; Springer and Dustin, 2012). We have shown here a remarkable pathway for transmission of signals in a cell surface protein with an exceptionally complex and large structure. A movement of only 2 Å at the β1-α1 loop in the ligand binding site is transmitted through an intricate shape-shifting pathway a distance of 40 Å across the βI domain. A 10-Å α7 helix movement like that of a connecting rod in the βI domain causes the hybrid domain to swing out by pivoting at its other connection to the βI domain. The length of the hybrid domain is 40 Å, and the PSI and I-EGF1 domains attached at the end opposite the βI domain make the total length of the upper integrin β leg ∼70 Å. The leverlike swing of the upper leg amplifies the 2-Å movement of the β1-α1 loop to a 75-Å increase in separation at the integrin knees (Fig. 1). The integrin headpiece is a marvelous biological machine that appears designed for the difficult job of transmitting allostery in extracellular environments. If in general the open headpiece is the high affinity state of integrins, induction by inside-out signals of headpiece opening can be a general mechanism for integrin activation on cell surfaces.
Materials and methods
Protein expression, purification, and crystallization
The soluble αIIbβ3 headpiece construct was expressed in CHO-Lec cells and purified using its His6 tag, treated with chymotrypsin and carboxypeptidase to remove the thigh domain as well as the C-terminal acid-base coiled coil and His6 tag, and further purified exactly as described in detail in the supplemental materials in Xiao et al. (2004). Crystallization in complex with 10E5 Fab in the absence of ligand was as previously described (Zhu et al., 2010); purified αIIbβ3–10E5-Fab complex at 10 mg/ml was crystallized at 4°C in hanging drops by adding an equal volume of 11–13% PEG 8000, 0.2 M ammonium sulfate, and 0.1 M Tris-HCl, pH 8.9. Crystals did not always appear under these conditions. In this case, crystal seeding was used. Seeds were obtained by breaking a single crystal with a seed bead (Hampton Research). 0.1-µl diluted crystal seeds were added to 0.9 µl of a hanging drop that had preequilibrated overnight. Crystals were grown at 4°C for >1 mo before soaking.
Crystal soaking and diffraction data collection
Crystals were harvested in cryosolution containing 15% PEG 8000, 0.2 M ammonium sulfate, and 0.1 M Tris-HCl, pH 8.9, with the addition of glycerol as a cryoprotectant in 5% increments up to a 20% final concentration. Harvested crystals were soaked in the same cryosolution plus desired concentrations of GRGDSP peptide and metal ions shown in Table 1 for the indicated times at 4°C before plunge vitrification in liquid nitrogen. Diffraction data were collected at beamline 23-ID (Advanced Photon Source) and processed with MOSFLM (Leslie and Powell, 2007) and SCALA (Evans, 2006) or the XDS package (Kabsch, 2001). Resolution cutoff was chosen to give I/σ(I) > 1.6 in the highest resolution shell.
Structure refinement
For soaked crystals with little difference in the unit cell compared with the nonsoaked crystal, we used the closed-headpiece αIIbβ3–10E5-Fab structure (Protein Data Bank accession no. 3NIG; 2.25 Å) as the starting structure and subjected each domain to rigid body refinement with PHENIX (Adams et al., 2010). Electron densities at the ligand binding pocket corresponding to RGD peptides were readily identified after the first round of rigid body refinement. For crystals soaked with 10 mM RGD peptide in Mn2+, the unit cell shrank in a dimension, and structures were solved by molecular replacement with PHASER (McCoy et al., 2007) using the αIIb β-propeller and β3 βI domains as search models. Structures of soaked ligands were built into their electron densities with COOT (Emsley and Cowtan, 2004). The complete structures were then finalized by several rounds of rebuilding with COOT and refinement with PHENIX (Adams et al., 2010). High resolution αIIbβ3 structures (3T3P, 2.2 Å; or 3NIG, 2.25 Å) were used as reference structures when rebuilding the lower resolution soaked structures with COOT. Metal ion coordination restraints were used for MIDAS and SyMBS, but not ADMIDAS metals, during refinement with PHENIX. Because the two molecules of the asymmetric unit were trapped in different conformations after crystal soaking, noncrystallographic symmetry restraints were not used during structure refinement. The structures were validated with the MolProbity server (Davis et al., 2007).
Simulated-annealing composite omit maps and RSCCs
Both simulated-annealing composite omit maps and RSCCs with simulated-annealing composite omit density were calculated by PHENIX (Adams et al., 2010). When dual conformations were present, the structures were refined with either conformation or a third intermediate conformation as a single conformation, and the highest RSCC values were plotted. Although RSCC values provide a good estimate of occupancy, they also are dependent on the resolution and quality of the diffraction data and refinement.
Superpositions and figures
All superpositions were on Cα atoms of the αIIb β-propeller domain and nonmoving portions of the β3 βI domain. Panels in multipanel figures (Fig. 2, Fig. 4, Fig. 6 [A and B], and Fig. 7 [A–D]) are shown in identical orientations and are aligned vertically and horizontally on the page. Figures were prepared with PyMOL (Schrödinger, LLC). All main text figures use the states indicated in Table 1.
Online supplemental material
Fig. S1 shows electron density map and the RGD-binding pocket for examples of states 1 and 7 not depicted in Fig. 2. Fig. S2 is an overview of the moving portions of the βI domain for examples of states 1 and 7 not depicted in Fig. 5. Video 1 shows an overview of headpiece opening. Video 2 shows a detailed view of the ligand and metals. Video 3 focuses on the α1 and α1′ helices, β6-α7 loop, and α7 helix. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201212037/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Barry Coller for providing the purified mAb 10E5.
This work was supported by National Institutes of Health grant HL-103526. Jieqing Zhu is a recipient of a Scientist Development Grant from the American Heart Association and an ASH Scholar Award for junior faculty from the American Society of Hematology.
Footnotes
Abbreviations used in this paper:
- ADMIDAS
- adjacent to MIDAS
- MD
- molecular dynamics
- MIDAS
- metal ion-dependent adhesion site
- RSCC
- real space correlation coefficient
- SyMBS
- synergistic metal binding site
- TM
- transmembrane
References
- Adair B.D., Xiong J.P., Maddock C., Goodman S.L., Arnaout M.A., Yeager M. 2005. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J. Cell Biol. 168:1109–1118 10.1083/jcb.200410068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aga Y., Baba K., Tam S., Nakanishi T., Yoneda K., Kita J., Ueno H. 2004. UR-3216: a new generation oral platelet GPIIb/IIIa antagonist. Curr. Pharm. Des. 10:1597–1601 10.2174/1381612043384592 [DOI] [PubMed] [Google Scholar]
- Barton S.J., Travis M.A., Askari J.A., Buckley P.A., Craig S.E., Humphries M.J., Mould A.P. 2004. Novel activating and inactivating mutations in the integrin β1 subunit A domain. Biochem. J. 380:401–407 10.1042/BJ20031973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.F., Takagi J., Xie C., Xiao T., Luo B.-H., Springer T.A. 2004. The relative influence of metal ion binding sites in the I-like domain and the interface with the hybrid domain on rolling and firm adhesion by integrin α4β7. J. Biol. Chem. 279:55556–55561 10.1074/jbc.M407773200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xie C., Nishida N., Li Z., Walz T., Springer T.A. 2010. Requirement of open headpiece conformation for activation of leukocyte integrin αXβ2. Proc. Natl. Acad. Sci. USA. 107:14727–14732 10.1073/pnas.1008663107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig D., Gao M., Schulten K., Vogel V. 2004. Structural insights into how the MIDAS ion stabilizes integrin binding to an RGD peptide under force. Structure. 12:2049–2058 10.1016/j.str.2004.09.009 [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., III, Snoeyink J., Richardson J.S., Richardson D.C. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35(Suppl. 2):W375–W383 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Mi L.-Z., Zhu J., Wang W., Hu P., Luo B.H., Springer T.A. 2012. αVβ3 integrin crystal structures and their functional implications. Biochemistry. 51:8814–8828 10.1021/bi300734n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Eng E.T., Smagghe B.J., Walz T., Springer T.A. 2011. Intact αIIbβ3 integrin is extended after activation as measured by solution X-ray scattering and electron microscopy. J. Biol. Chem. 286:35218–35226 10.1074/jbc.M111.275107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82 10.1107/S0907444905036693 [DOI] [PubMed] [Google Scholar]
- Frelinger A.L., III, Lam S.C.T., Plow E.F., Smith M.A., Loftus J.C., Ginsberg M.H. 1988. Occupancy of an adhesive glycoprotein receptor modulates expression of an antigenic site involved in cell adhesion. J. Biol. Chem. 263:12397–12402 [PubMed] [Google Scholar]
- Frelinger A.L., III, Cohen I., Plow E.F., Smith M.A., Roberts J., Lam S.C.T., Ginsberg M.H. 1990. Selective inhibition of integrin function by antibodies specific for ligand-occupied receptor conformers. J. Biol. Chem. 265:6346–6352 [PubMed] [Google Scholar]
- Gupta V., Gylling A., Alonso J.L., Sugimori T., Ianakiev P., Xiong J.P., Arnaout M.A. 2007. The beta-tail domain (betaTD) regulates physiologic ligand binding to integrin CD11b/CD18. Blood. 109:3513–3520 10.1182/blood-2005-11-056689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler-Wildman K., Kern D. 2007. Dynamic personalities of proteins. Nature. 450:964–972 10.1038/nature06522 [DOI] [PubMed] [Google Scholar]
- Iwasaki K., Mitsuoka K., Fujiyoshi Y., Fujisawa Y., Kikuchi M., Sekiguchi K., Yamada T. 2005. Electron tomography reveals diverse conformations of integrin αIIbβ3 in the active state. J. Struct. Biol. 150:259–267 10.1016/j.jsb.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2001. Chapter 25.2.9: XDS. International Tables for Crystallography, Volume F: Crystallography of Biological Macromolecules. Rossmann M.G., Arnold E.V., Kluwer Academic Publishers, Dordrecht, Netherlands: 730–734 [Google Scholar]
- Kamata T., Handa M., Ito S., Sato Y., Ohtani T., Kawai Y., Ikeda Y., Aiso S. 2010. Structural requirements for activation in αIIbβ3 integrin. J. Biol. Chem. 285:38428–38437 10.1074/jbc.M110.139667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Carman C.V., Springer T.A. 2003. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 301:1720–1725 10.1126/science.1084174 [DOI] [PubMed] [Google Scholar]
- Leslie A.G.W., Powell H.R. 2007. Processing Diffraction Data With MOSFLM. In Evolving Methods for Macromolecular Crystallography. Vol. 245 Read R.J., Sussman J.L., Springer; 41–51 [Google Scholar]
- Luo B.-H., Springer T.A., Takagi J. 2003. Stabilizing the open conformation of the integrin headpiece with a glycan wedge increases affinity for ligand. Proc. Natl. Acad. Sci. USA. 100:2403–2408 10.1073/pnas.0438060100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.-H., Springer T.A., Takagi J. 2004a. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2:e153 10.1371/journal.pbio.0020153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.-H., Strokovich K., Walz T., Springer T.A., Takagi J. 2004b. Allosteric β1 integrin antibodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J. Biol. Chem. 279:27466–27471 10.1074/jbc.M404354200 [DOI] [PubMed] [Google Scholar]
- Luo B.-H., Takagi J., Springer T.A. 2004c. Locking the β3 integrin I-like domain into high and low affinity conformations with disulfides. J. Biol. Chem. 279:10215–10221 10.1074/jbc.M312732200 [DOI] [PubMed] [Google Scholar]
- Luo B.-H., Carman C.V., Springer T.A. 2007. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25:619–647 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.H., Karanicolas J., Harmacek L.D., Baker D., Springer T.A. 2009. Rationally designed integrin β3 mutants stabilized in the high affinity conformation. J. Biol. Chem. 284:3917–3924 10.1074/jbc.M806312200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. 2007. Phaser crystallographic software. J. Appl. Cryst. 40:658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould A.P., Barton S.J., Askari J.A., McEwan P.A., Buckley P.A., Craig S.E., Humphries M.J. 2003a. Conformational changes in the integrin β A domain provide a mechanism for signal transduction via hybrid domain movement. J. Biol. Chem. 278:17028–17035 10.1074/jbc.M213139200 [DOI] [PubMed] [Google Scholar]
- Mould A.P., Symonds E.J., Buckley P.A., Grossmann J.G., McEwan P.A., Barton S.J., Askari J.A., Craig S.E., Bella J., Humphries M.J. 2003b. Structure of an integrin-ligand complex deduced from solution x-ray scattering and site-directed mutagenesis. J. Biol. Chem. 278:39993–39999 10.1074/jbc.M304627200 [DOI] [PubMed] [Google Scholar]
- Nagae M., Re S., Mihara E., Nogi T., Sugita Y., Takagi J. 2012. Crystal structure of α5β1 integrin ectodomain: Atomic details of the fibronectin receptor. J. Cell Biol. 197:131–140 10.1083/jcb.201111077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parise L.V., Helgerson S.L., Steiner B., Nannizzi L., Phillips D.R. 1987. Synthetic peptides derived from fibrinogen and fibronectin change the conformation of purified platelet glycoprotein IIb-IIIa. J. Biol. Chem. 262:12597–12602 [PubMed] [Google Scholar]
- Plow E.F., Pierschbacher M.D., Ruoslahti E., Marguerie G.A., Ginsberg M.H. 1985. The effect of Arg-Gly-Asp-containing peptides on fibrinogen and von Willebrand factor binding to platelets. Proc. Natl. Acad. Sci. USA. 82:8057–8061 10.1073/pnas.82.23.8057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklin-Faucher E., Vogel V. 2009. Integrin activation dynamics between the RGD-binding site and the headpiece hinge. J. Biol. Chem. 284:36557–36568 10.1074/jbc.M109.041194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklin-Faucher E., Gao M., Schulten K., Vogel V. 2006. How the headpiece hinge angle is opened: New insights into the dynamics of integrin activation. J. Cell Biol. 175:349–360 10.1083/jcb.200602071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürpf T., Springer T.A. 2011. Regulation of integrin affinity on cell surfaces. EMBO J. 30:4712–4727 10.1038/emboj.2011.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Zhu J., Wang R., Chen X., Mi L., Walz T., Springer T.A. 2011. Latent TGF-β structure and activation. Nature. 474:343–349 10.1038/nature10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagghe B.J., Huang P.S., Ban Y.-E., Baker D., Springer T.A. 2010. Modulation of integrin activation by an entropic spring in the β-knee. J. Biol. Chem. 285:32954–32966 10.1074/jbc.M110.145177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer T.A., Dustin M.L. 2012. Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24:107–115 10.1016/j.ceb.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer T.A., Zhu J., Xiao T. 2008. Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin αIIbβ3. J. Cell Biol. 182:791–800 10.1083/jcb.200801146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J., Petre B.M., Walz T., Springer T.A. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 110:599–11 10.1016/S0092-8674(02)00935-2 [DOI] [PubMed] [Google Scholar]
- Takagi J., Strokovich K., Springer T.A., Walz T. 2003. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 22:4607–4615 10.1093/emboj/cdg445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhu J., Dong X., Shi M., Lu C., Springer T.A. 2012. GARP regulates the bioavailability and activation of TGFβ. Mol. Biol. Cell. 23:1129–1139 10.1091/mbc.E11-12-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T., Takagi J., Coller B.S., Wang J.H., Springer T.A. 2004. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 432:59–67 10.1038/nature02976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Zhu J., Chen X., Mi L., Nishida N., Springer T.A. 2010. Structure of an integrin with an αI domain, complement receptor type 4. EMBO J. 29:666–679 10.1038/emboj.2009.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.-P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D.L., Joachimiak A., Goodman S.L., Arnaout M.A. 2001. Crystal structure of the extracellular segment of integrin αVβ3. Science. 294:339–345 10.1126/science.1064535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J.-P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S.L., Arnaout M.A. 2002. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 296:151–155 10.1126/science.1069040 [DOI] [PubMed] [Google Scholar]
- Xiong J.-P., Mahalingham B., Alonso J.L., Borrelli L.A., Rui X., Anand S., Hyman B.T., Rysiok T., Müller-Pompalla D., Goodman S.L., Arnaout M.A. 2009. Crystal structure of the complete integrin αVβ3 ectodomain plus an α/β transmembrane fragment. J. Cell Biol. 186:589–600 10.1083/jcb.200905085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Shimaoka M., Chen J.F., Springer T.A. 2004. Activation of integrin β-subunit I-like domains by one-turn C-terminal α-helix deletions. Proc. Natl. Acad. Sci. USA. 101:2333–2338 10.1073/pnas.0307291101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Zhu J., Mi L.Z., Walz T., Sun H., Chen J.-F., Springer T.A. 2012. Structural specializations of α4β7, an integrin that mediates rolling adhesion. J. Cell Biol. 196:131–146 10.1083/jcb.201110023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Boylan B., Luo B.-H., Newman P.J., Springer T.A. 2007a. Tests of the extension and deadbolt models of integrin activation. J. Biol. Chem. 282:11914–11920 10.1074/jbc.M700249200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Carman C.V., Kim M., Shimaoka M., Springer T.A., Luo B.-H. 2007b. Requirement of α and β subunit transmembrane helix separation for integrin outside-in signaling. Blood. 110:2475–2483 10.1182/blood-2007-03-080077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., Zhang C., Nishida N., Springer T.A. 2008. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 32:849–861 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Zhu J., Negri A., Provasi D., Filizola M., Coller B.S., Springer T.A. 2010. Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood. 116:5050–5059 10.1182/blood-2010-04-281154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Choi W.-S., McCoy J.G., Negri A., Zhu J., Naini S., Li J., Shen M., Huang W., Bougie D., et al. 2012. Structure-guided design of a high-affinity platelet integrin αIIbβ3 receptor antagonist that disrupts Mg2+ binding to the MIDAS. Sci. Transl. Med. 4:125ra32 10.1126/scitranslmed.3003576 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.