Abstract
The first mouse mutation associated with a heritable defect in embryonic mammary gland development was Extratoes. It represents a functional null-mutation of the gene encoding Gli3, which is best known as a transcription factor mediating canonical Hedgehog (Hh) signaling. Here we review the roles of Hh and Gli proteins in murine embryonic mammary development. We propose that an off-state for Hh signaling, mediated by Gli3-repressor, is determinant for induction of a mammary instead of hair follicle fate in the trunk surface ectoderm.
Keywords: Mouse, Embryo, Prenatal, Mammary gland, Breast, Hedgehog, Gli3, Transcriptional repressor
Introduction
Mouse embryos form five pairs of mammary rudiments (MRs). Their formation is preceded by the formation of three Wnt10b-expressing mammary streaks in the axilla, inguen, and in between forelimb and hindlimb respectively around embryonic day (E) 10.5. These streaks fuse into one continuous mammary line (ML) on each flank by E11.5, on which MRs develop in asynchronous fashion and not in numerical sequence, with MR1 in the axilla; MR2, MR3 and MR4 on the flank; and MR5 in the inguen [1].
The Hedgehog (Hh) pathway was among the first signaling cascades tested for a role in murine mammogenesis, but is not required for it [2, 3]. Nonetheless, Extratoes (Xt) mice, which harbor a spontaneous, functional null-mutation for the transcription factor Gli3, exhibit multiple defects in prenatal mammary development [4–7].
Given the role of Gli3 in Hh signaling, we provide a brief overview of Hh signaling and Gli transcription factors, followed by their functions in mammogenesis in mouse embryos.
Signal Transduction via Hedgehog and Gli Proteins
Mammals possess three orthologues of the Drosophila segment polarity gene Hedgehog; Sonic hedgehog (Shh), Indian hedgehog (Ihh), and Desert hedgehog (Dhh) [8, 9]. While embryonic expression and function of Dhh is restricted to spermatogenesis and Schwann cells, Shh and Ihh are widely expressed and encode secreted morphogens with well-known roles for Shh in neural tube, limb and somite development and for Ihh in chondrogenesis [10, 11].
In canonical Hh signaling, Hh molecules bind their twelve-transmembrane receptor Patched1 (Ptch1) or Ptch2. Ptch then derepresses the seven-transmembrane protein Smoothened (Smo), which becomes activated. Through intracellular signaling complexes -whose components have not been studied in embryonic mammary development and are thus not described here- Smo activation converges on the Gli family of Krüppel-type zinc-finger transcription factors, comprising Gli1, Gli2 and Gli3. Gli1 is expressed when Hh signaling is active, and encodes the transcriptional activator Gli1A. Gli2 and Gli3 can be expressed in the absence of Hh signaling, in which case their protein products are cleaved to render transcriptional repressors (Gli2R, Gli3R). Gli2 requires less active Hh signaling than Gli3 to remain uncleaved and function as a transcriptional activator (Gli2A, Gli3A). Among Gli transcriptional targets are Gli1 and Ptch1, which provide regulatory feedback loops for Hh signaling [12, 13].
Recently, deviations from this canonical paradigm have been discovered [14]. They need further exploration, but certainly point to three additional, “non-canonical” signaling scenarios: 1) Signaling involving Hh pathway components independently of Gli-mediated transcription, such as Shh-mediated ERK activation in mammary epithelial cells without Smo-activation [15]; 2) Direct interaction of Hh signalling components with other molecular pathways; potentially relevant for mammogenesis is the role of Wnt signaling in expression and function of Gli proteins [16], and 3) “Non-contiguous” or “atypical” interaction of core Hh pathway components with each other.
Pleiotropic Functions of Gli3 in Mammary Development
Studies of the role of Hh signaling components in mammogenesis began in the late 1960’s, when D.R. Johnson identified and characterized a mouse strain which he named Extra-toes (Xt), after the prominent polydactyly in all four paws of heterozygotes [4]. In E13 embryos homozygous for this spontaneous, semi-dominant mutation, he also found a reduced number of MRs between the forelimb and hindlimb. Because his wild type control had a supernumerary MR near MR3, he concluded that “the number of nipples visible in Xt/Xt mice at this stage is reduced from four to two pairs, the most anterior and posterior ones persisting” [4]. Note that Johnson was mistaken in three aspects: 1) what he referred to as nipples were in fact mammary buds, which precede nipple formation [17]; 2) had he taken a proper control, he would have concluded that only one MR (MR3) was missing on the flank; 3) had he known that wild-types also form MRs behind the forelimb (MR1) and hindlimb (MR5), he would have noticed an absence of MR5 in Xt/Xt embryos [5–7, 18]. Additional Xt/Xt mammary defects are described below.
At The Jackson Laboratory arose a spontaneous mouse mutant, named Xt J because it phenocopies Johnson’s Xt mutant [19]. Xt J mice carry a 51.5 kb deletion downstream of nucleotide 1670 of Gli3, including the DNA-binding zinc fingers and thus abolishing both Gli3A and Gli3R functions [19–21], without loss of other known functional sequences. These renamed Gli3 Xt-J mice represent a good model for the human Greig cephalopolysyndactyly syndrome (GCPS), which can be a sporadic or inherited trait, caused by heterozygous loss-of-function mutations of the GLI3 gene. The syndrome is characterized by polysyndactyly of hands and feet, hypertelorism, and skull abnormalities including craniosynostosis at variable penetrance [19, 22][OMIM 165240, 175700], the latter seen in mice only when homozygous mutant [23].
While no mammary phenotypes have been reported for GCPS patients and Gli3 Xt-J/+ mouse mutants, Gli3 Xt-J/Xt-J embryos fail to induce MR3 and MR5 [5–7, 18]. Moreover, their MR2 and MR4 are hypoplastic; MR2 protrudes in females [18] (compare Fig. 1t with 1l), resembling MRs of wild-type male embryos [17], and also exhibits impaired branching morphogenesis; while MR1 is hardly affected [18]. The Xt J mutation thus exemplifies that all mammary glands are different and can develop inter-independently [24]. The question then arises via which molecular mechanisms and tissue-interactions Gli3 regulates formation and development of MR2-5.
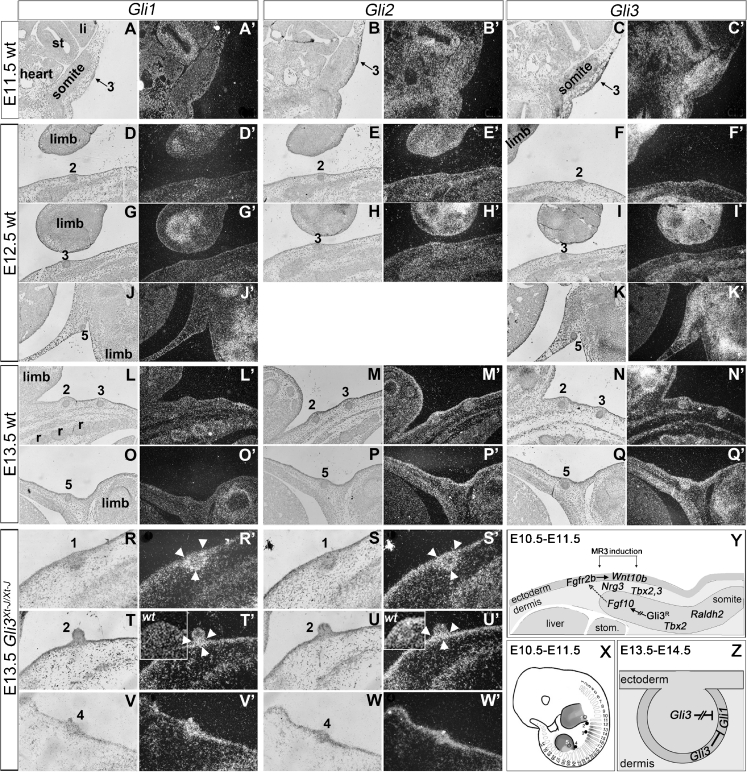
Fig. 1.
Expression patterns of Gli1, Gli2 and Gli3, and models of tissue- and molecular interactions involving Gli3 in mammary development. a–q Cross-sections of wild-type embryos and (r–w) Gli3 Xt-J/Xt-J embryos at E11.5-E13.5 hybridized with 35S-labeled RNA-probes for Gli1, Gli2 and Gli3. Magnification of r-w is twice that of a–q. Insets in t’ and u’ present wild-type MR2 from l’ respectively m’ at the same magnification. MRs are indicated by number. Black arrow in a–c indicates emerging MR3 at E11.5. White arrowheads in r’, t’ s’ and u’ point to MM expression. x Cartoon of a lateral view of a mouse embryo. Between E10.5 and E11.5 Gli3 is expressed in the limb buds and all somites (outlined in gray and numbered). Gray shades represent overlapping in expression with Fgf10, the intensity positively correlating with level of Fgf10 expression. Note the proximity of the overlapping somitic expression to MR3 and MR2, and overlapping limb mesenchymal expression to MR1, MR2, MR4 and MR5. MR1 and MR5 are hidden behind the limbs. y Model exclusively for formation of MR3: See main text for explanation. z Model for Gli3-mediated repression of Gli1 in the ME (bud shape) and contiguous MM (darker shade of gray surrounding mammary bud), based on expression data in panels a–w and in [5, 6]. In the absence of Gli3, Gli1 is misexpressed in the MM of MR1, MR2 and MR4, indicating Gli3R normally represses Gli1 expression. Whether this occurs tissue-autonomously (intact block arrow) or via tissue interactions (broken block-arrow) remains unclear. Abbreviations: li: liver, r: rib primordium, st: stomach, wt: wild-type
Prior to and at the time of ML and MR formation, Gli3 is not expressed in the surface ectoderm [5], from which the MRs derive [25], but is expressed in the limb mesenchyme, and in the somites in the region between fore- and hindlimbs [5] (Fig. 1c). Given the earlier implications of somitic involvement in the establishment of the mammary streak along the flank and in MR induction [1, 7], the somitic Gli3 expression was of interest. Indeed, the ventral elongation of the somites including expression of Gli3 in all thoracic and lumbar somites, and Fibroblast growth factor (Fgf)10 in thoracic somites #12 to #18, determines the position of the ML on the dorso-ventral body axis [5]. The absence of MR3 in Gli3 Xt-J/Xt-J embryos [5] is due to reduction of somitic Fgf10 expression, in particular the relatively high Fgf10 expression in somite #15 underlying MR3 [5]. As MR2 is located above somite #12 [5], reduced somitic Fgf10 expression may be involved in impaired development of MR2 as well.
It is most likely that somitic Gli3 indirectly activates somitic Fgf10 independent of Shh signaling ([5] and references therein). Somitic FGF10 may reach and activate the ectodermal FGFR2b via diffusion and/or transportation by dermal precursor cells that delaminate from the somites. This cascade leads to ectodermal cell elongation, Wnt10b marker expression, and canonical Wnt signaling as required for ML and MR formation, but only at the level of MR3 [5], and possibly MR2. These insights finally gave some molecular explanation for the capacity of the dermal mesenchyme to induce MR formation as recognized four decades earlier [25–27].
However, at the level of MR4 and MR5, Fgf10 is not expressed in the somites. Given that null mutants for either Gli3 or Fgf10 fail to induce MR5 and both genes are expressed in the adjacent limb mesenchyme [5] (Fig. 1x), it is tempting to speculate that an epistatic interaction between these molecules may also exist in the limb mesenchyme and be required for induction of MR5. The compromised induction of MR2 and MR4 in the absence of Gli3 can perhaps be attributed to Gli3-independent residual expression of Fgf10 and other genes in the thoracic somites and limb mesenchyme.
The cellular functions of Gli3 during early mammary growth and morphogenesis are fourfold, and the same in all MRs [18]: downregulation of 1) cell proliferation and 2) cell apoptosis in epithelial cells of the ML and MRs; 3) mediation of ectodermal cell recruitment into MRs, which contributes to MR growth until at least E14.5; 4) hypertrophy of the basal/peripheral cells of the ME, which provides a large proportion of the growth between E12.5 and E13.5 [18]. While similar regulatory functions for Gli3 have been identified in other cell types (references in [18]), it remains to be investigated how Gli3 mediates so many distinct effects in the same cell population; and whether these epithelial effects rely on Gli3 expression prior to mammary induction, on cell-autonomous Gli3 function in the ME, or via tissue-interactions with and Gli3 expression in the MM (Fig. 1z).
Mammogenesis Does not Require Gli1 and Gli2, but Only Gli3R
At E11.5, the somites also express Gli2, but not Gli1 (Fig. 1a, b) [6]. Expression of Gli genes in the surface ectoderm or mammary tissues is arguable at E12.5, except for notable Gli3 expression in MR5 (Fig. 1d–k) as confirmed by RNA-profiling [18]. By E13.5, Gli3 and Gli2 are convincingly expressed in overlap in the ectoderm, mammary epithelium (ME) and mammary mesenchyme (MM) (Fig. 1m,n,p,q), the latter confirmed for Gli3 by RNA-profiling [18] and immunohistochemistry [6]. Thus, the initial growth defects of Gli3 Xt-J/Xt-J MR2 and MR4 at E11.5 could subsequently be compounded by the lack of Gli3 in any of the tissue compartments.
Although Gli2 and Gli3 overlap in expression, homozygous Gli2-null mutants have no mammary induction defect [6, 28]. Neither have Gli1-null, Gli2 lkni/+ or Gli2 lkn/lkni mutants which express Gli1A from the endogenous Gli2 promoter [6]. However, removing one Gli3 allele in Gli2 lkni/lkni embryos abolishes induction of MR3 and MR5 as in Gli3 Xt-J/Xt-J embryos. Moreover, Gli1 is misexpressed in the MM of MR2 in E13.5 Gli3 Xt-J/Xt-J embryos (Fig. 1t). The high Gli1 expression in MR1 and MR4 at E13.5 (Fig. 1r, v) may also reflect misexpression in the MM, similar to such misexpression reported at E14.5 [6]. Together, these data indicate that Gli3 acts as Gli3R during MR formation [6] (Fig. 1y, z), and is the only Gli family member with functional relevance for embryonic mammary development, by repressing Gli1 transcription, and antagonizing Gli2A function [6]. Moreover, these data indicate that canonical Hh signaling needs to be in an off-state to allow mammogenesis [6], at least at the positions of MR3 and MR5.
Mammogenesis Requires Absence of Hh Signaling, Likely to Prevent Acquisition of a Hair Follicle Fate
The above-described repressor function of Gli3 and absence of Shh, Ihh, Gli1 and Ptc1 expression in the surface ectoderm or dermal mesenchyme at the time of ML and MR induction [6] (our unpublished data and Fig. 1a), explain why Shh −/− and Ihh −/− mutant mice have no mammary induction defect [2, 3]. All together, these data indicate Hh signaling is not required for induction of any of the five MRs. While normally, Shh, Ihh and Ptc1 are expressed in the ME from E12.5 onward and in postnatal stages [3], the normal outgrowth of transplanted E12.5 Shh −/− and Ihh −/− ME in cleared wild-type mammary fat pads or under the kidney capsule, indicates that epithelial Hh signaling or mesenchymal Shh is not either required during later embryonic and postnatal stages [2].
Interestingly, activating components of the Hh signaling cascade become expressed in the surface ectoderm and ME only after the formation of MR, but prior to hair follicle formation [3]. Moreover, Shh is required for hair follicle development [2, 3]. We therefore propose that the off-state of Hh signaling is a discriminatory factor for the choice of a skin appendage between mammary and either hair or non-mammary fate, and that therefore MRs must develop prior to hair follicle formation, i.e. prior to activation of Hh signaling in the trunk ectoderm [29, 30]. Such a decisive role for Hh signaling appendage fate requires further investigation, but seems to be confirmed by the glandular appearance of hair follicles when Hh signaling in the skin is ablated in K14Cre;Smo fl/fl embryos [31].
Conclusion and Perspectives
Gli3 function is critical for embryonic development of MR2-5; in particular GliR-mediated repression of canonical Hh signaling is critical for induction of MR3 and MR5. Its variable requirement for the five MRs in mouse gives perhaps some insight in mechanisms underlying the variation in number of mammary glands among mammalian species. For example, MR3 requires somitic Gli3 as the earliest known effector in a cascade involving FGF10/FGFR2b and subsequent Wnt signaling (Fig. 1y), but other tissue-interactions and perhaps molecular mechanisms act downstream of Gli3 in early development of other MRs, and remain to be elucidated. Examples of additional interesting avenues for investigation pertain to possible interactions with other important mesenchymal factors in MR3 formation (e.g. Nrg3, Tbx3, or Raldh2 that mediates retinoic acid signaling) [32–34] (Fig. 1y); whether the sustained protrusion of MR2 in Gli3 Xt-J/Xt-J embryos bears any mechanistic similarity to the protrusion of MRs in wild-type males; and whether Gli3 is involved in non-canonical Hh signaling in mammogenesis.
Given the many molecular similarities between embryonic mammary development and mammary/breast tumors [35, 36], it is of interest whether GLI3 plays a role in breast cancer. It is noteworthy that GLI(1) was discovered as an oncogene, and like GLI2 and SHH, is upregulated in various cancers [13]. HH signaling is considered a therapeutic target for breast cancer, as GLI(1) upregulation predicts a poor prognosis for estrogen receptor negative or triple negative breast cancers [37–39]. Yet, GLI3 is seldom reported in association with cancer [13]. In absolute number and relative Gli1, only few somatic mutations in Gli3 have been found in breast cancer samples [COSMIC database, Sanger institute]. It would be of interest to determine whether these Gli3 mutations relate to Gli1 upregulation, and whether they have any diagnostic or prognostic value.
Acknowledgments
The authors thank A*STAR (Agency for Science, Technology and Research) and its Institute of Molecular and Cell Biology for funding and research facilities, and Bradley Spencer-Dene for generating in situ hybridization data.
Abbreviations
- Dhh
Desert Hedgehog
- E
Embryonic day
- FGF
Fibroblast Growth Factor
- FGFR2b
FGF Receptor 2-IIIb
- Gli
Glioma-associated oncogene
- Gli1A,Gli2A, Gli3A
Gli1, Gli2 or Gli3 transcriptional activators
- Gli2R,Gli3R
Gli2 or Gli3 transcriptional repressors
- Hh
Hedgehog
- Ihh
Indian Hedgehog
- ME
Mammary epithelium
- ML
Mammary line
- MM
Mammary mesenchyme
- MR
Mammary rudiment (pair)
- OMIM
Online Mendelian Inheritance in Man (database)
- Ptch
Patched
- Shh
Sonic Hedgehog
- Smo
Smoothened
- UTR
Untranslated region
- Xt-J, XtJ
Extratoes mutation (mouse) identified at The Jackson Laboratory
References
- 1.Veltmaat JM, Van Veelen W, Thiery JP, Bellusci S. Identification of the mammary line in mouse by Wnt10b expression. Dev Dyn Off Publ Am Assoc Anatomists. 2004;229(2):349–356. doi: 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]
- 2.Gallego MI, Beachy PA, Hennighausen L, Robinson GW. Differential requirements for shh in mammary tissue and hair follicle morphogenesis. Dev Biol. 2002;249(1):131–139. doi: 10.1006/dbio.2002.0761. [DOI] [PubMed] [Google Scholar]
- 3.Michno K, Boras-Granic K, Mill P, Hui CC, Hamel PA. Shh expression is required for embryonic hair follicle but not mammary gland development. Dev Biol. 2003;264(1):153–165. doi: 10.1016/S0012-1606(03)00401-9. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DR. Extra-toes: a new mutant gene causing multiple abnormalities in the mouse. J Embryol Exp Morphol. 1967;17(3):543–581. [PubMed] [Google Scholar]
- 5.Veltmaat JM, Relaix F, Le LT, Kratochwil K, Sala FG, van Veelen W, et al. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development. 2006;133(12):2325–2335. doi: 10.1242/dev.02394. [DOI] [PubMed] [Google Scholar]
- 6.Hatsell SJ, Cowin P. Gli3-mediated repression of Hedgehog targets is required for normal mammary development. Development. 2006;133(18):3661–3670. doi: 10.1242/dev.02542. [DOI] [PubMed] [Google Scholar]
- 7.Mailleux AA, Spencer-Dene B, Dillon C, Ndiaye D, Savona-Baron C, Itoh N, et al. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129(1):53–60. doi: 10.1242/dev.129.1.53. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S, Balczarek KA, Lai ZC. Evolution of the hedgehog gene family. Genetics. 1996;142(3):965–972. doi: 10.1093/genetics/142.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 10.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/S0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 11.Resende TP, Ferreira M, Teillet MA, Tavares AT, Andrade RP, Palmeirim I. Sonic hedgehog in temporal control of somite formation. Proc Natl Acad Sci U S A. 2010;107(29):12907–12912. doi: 10.1073/pnas.1000979107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz i Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90(2):193–196. doi: 10.1016/S0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 13.Stecca B, Ruiz IAA. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2(2):84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins D. Hedgehog signalling: emerging evidence for non-canonical pathways. Cell Signal. 2009;21(7):1023–1034. doi: 10.1016/j.cellsig.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 15.Chang H, Li Q, Moraes RC, Lewis MT, Hamel PA. Activation of Erk by sonic hedgehog independent of canonical hedgehog signalling. Int J Biochem Cell Biol. 2010;42(9):1462–1471. doi: 10.1016/j.biocel.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borycki A, Brown AM, Emerson CP., Jr Shh and Wnt signaling pathways converge to control Gli gene activation in avian somites. Development. 2000;127(10):2075–2087. doi: 10.1242/dev.127.10.2075. [DOI] [PubMed] [Google Scholar]
- 17.Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S. Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation. 2003;71(1):1–17. doi: 10.1046/j.1432-0436.2003.700601.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee MY, Racine V, Jagadpramana P, Sun L, Yu W, Du T, et al. Ectodermal influx and cell hypertrophy provide early growth for all murine mammary rudiments, and are differentially regulated among them by Gli3. PLoS One. 2011;6(10):e26242. doi: 10.1371/journal.pone.0026242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet. 1993;3(3):241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 20.Buscher D, Grotewold L, Ruther U. The XtJ allele generates a Gli3 fusion transcript. Mamm Genome: Off J Int Mamm Genome Soc. 1998;9(8):676–678. doi: 10.1007/s003359900845. [DOI] [PubMed] [Google Scholar]
- 21.Maynard TM, Jain MD, Balmer CW, LaMantia AS. High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm Genome: Off J Int Mamm Genome Soc. 2002;13(1):58–61. doi: 10.1007/s00335-001-2115-X. [DOI] [PubMed] [Google Scholar]
- 22.Vortkamp A, Franz T, Gessler M, Grzeschik KH. Deletion of GLI3 supports the homology of the human Greig cephalopolysyndactyly syndrome (GCPS) and the mouse mutant extra toes (Xt) Mamm Genome: Off J Int Mamm Genome Soc. 1992;3(8):461–463. doi: 10.1007/BF00356157. [DOI] [PubMed] [Google Scholar]
- 23.Rice DP, Connor EC, Veltmaat JM, Lana-Elola E, Veistinen L, Tanimoto Y et al. Gli3Xt-J/Xt-J mice exhibit lambdoid suture craniosynostosis which results from altered osteoprogenitor proliferation and differentiation. Hum Mol Genet. 2010;19(17):3457–67. [DOI] [PMC free article] [PubMed]
- 24.Veltmaat JM, Ramsdell A. F., Sterneck E. Positional variations in mammary gland development and cancer. J Mammary Gland Biol Neoplasia. 2013 (this issue). doi:10.1007/s10911-013-9287-3. [DOI] [PMC free article] [PubMed]
- 25.Cunha GR, Young P, Christov K, Guzman R, Nandi S, Talamantes F, et al. Mammary phenotypic expression induced in epidermal cells by embryonic mammary mesenchyme. Acta Anat (Basel) 1995;152(3):195–204. doi: 10.1159/000147698. [DOI] [PubMed] [Google Scholar]
- 26.Propper A, Gomot L. Tissue interactions during organogenesis of the mammary gland in the rabbit embryo. C R Acad Sci Hebd Seances Acad Sci D. 1967;264(22):2573–2575. [PubMed] [Google Scholar]
- 27.Kratochwil K. Organ specificity in mesenchymal induction demonstrated in the embryonic development of the mammary gland of the mouse. Dev Biol. 1969;20(1):46–71. doi: 10.1016/0012-1606(69)90004-9. [DOI] [PubMed] [Google Scholar]
- 28.Lewis MT, Ross S, Strickland PA, Sugnet CW, Jimenez E, Hui C, et al. The Gli2 transcription factor is required for normal mouse mammary gland development. Dev Biol. 2001;238(1):133–144. doi: 10.1006/dbio.2001.0410. [DOI] [PubMed] [Google Scholar]
- 29.Lewis MT, Veltmaat JM. Next stop, the twilight zone: hedgehog network regulation of mammary gland development. J Mammary Gland Biol Neoplasia. 2004;9(2):165–181. doi: 10.1023/B:JOMG.0000037160.24731.35. [DOI] [PubMed] [Google Scholar]
- 30.Widelitz RB, Veltmaat JM, Mayer JA, Foley J, Chuong CM. Mammary glands and feathers: comparing two skin appendages which help define novel classes during vertebrate evolution. Semin Cell Dev Biol. 2007;18(2):255–266. doi: 10.1016/j.semcdb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gritli-Linde A, Hallberg K, Harfe BD, Reyahi A, Kannius-Janson M, Nilsson J, et al. Abnormal hair development and apparent follicular transformation to mammary gland in the absence of hedgehog signaling. Dev Cell. 2007;12(1):99–112. doi: 10.1016/j.devcel.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard B, Panchal H, McCarthy A, Ashworth A. Identification of the scaramanga gene implicates Neuregulin3 in mammary gland specification. Genes Dev. 2005;19(17):2078–2090. doi: 10.1101/gad.338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho KW, Kim JY, Song SJ, Farrell E, Eblaghie MC, Kim HJ, et al. Molecular interactions between Tbx3 and Bmp4 and a model for dorsoventral positioning of mammary gland development. Proc Natl Acad Sci U S A. 2006;103(45):16788–16793. doi: 10.1073/pnas.0604645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho KW, Kwon HJ, Shin JO, Lee JM, Cho SW, Tickle C, et al. Retinoic acid signaling and the initiation of mammary gland development. Dev Biol. 2012;365(1):259–266. doi: 10.1016/j.ydbio.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 35.Howard B, Ashworth A. Signalling pathways implicated in early mammary gland morphogenesis and breast cancer. PLoS Genet. 2006;2(8):e112. doi: 10.1371/journal.pgen.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zvelebil M, Oliemuller E, Gao Q, Wansbury O, Mackay A, Kendrick H, et al. Embryonic mammary signature subsets are activated in Brca1−/− and basal-like breast cancers. Breast Cancer Res: BCR. 2013;15(2):R25. doi: 10.1186/bcr3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64(17):6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 38.ten Haaf A, Bektas N, von Serenyi S, Losen I, Arweiler EC, Hartmann A, et al. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer. 2009;9:298. doi: 10.1186/1471-2407-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L, Kwon YJ, Frolova N, Steg AD, Yuan K, Johnson MR, et al. Gli1 promotes cell survival and is predictive of a poor outcome in ERalpha-negative breast cancer. Breast Cancer Res Treat. 2010;123(1):59–71. doi: 10.1007/s10549-009-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]