Abstract
Most mammals develop their mammary glands in pairs of which the two counterparts are symmetrically displaced away from the ventral midline. Based on this symmetry and the same functional outcome as a milk-producing organ, the mammary glands are easily presumed to be mere copies of one another. Based on our analysis of published data with inclusion of new results related to mammary development and pathology in mice, we argue that this presumption is incorrect: Between and within pairs, mammary glands differ from one another, and tumor incidence and biology depend on the position along the anterior-posterior and the left-right axis as well. This insight has implications for experimental designs with mouse models and for data extrapolation between mammary glands within and between species. We suggest that improved documentation of location-specific mammary gland features will lead to more insights into the molecular mechanisms of mammary gland development and cancer biology in both mice and humans.
Keywords: Embryonic development, Body axis, Anterior-posterior, Asymmetry, Mammary gland, Breast cancer
Introduction
Within the animal Class of Mammals the number of mammary glands (MGs) varies considerably, from two in most primates to 25 in some opossums [1]. Mammary glands emerge along bilateral mammary lines, that are in some species distinct as ridges in the surface ectoderm, one on each flank of the embryo [2]. In mouse embryos, the mammary lines do not form ridges, yet become histologically and molecularly distinct between embryonic day (E) 10.5 and E11.5 (Fig. 1), and yield five mammary rudiments (MRs) on each side of the embryo by E12 [3]. A few species have a double row of mammary glands on each body half [1].
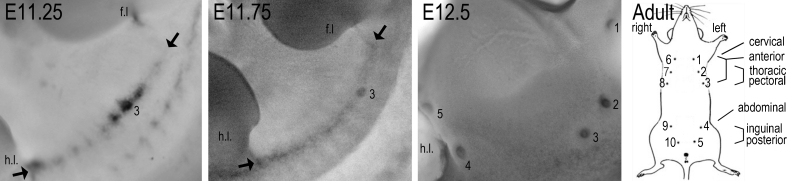
Fig. 1.
Mammary line formation and gland nomenclature. Images of flanks of wild-type (C57Bl6/J) mouse embryos hybridized with a Wnt10b probe showing the mammary line between arrows, at E11.25 visible as a fragmented expression pattern, and at E11.75 as a continuous line between forelimb (f.l.) and hindlimb (h.l.). The first mammary rudiment to appear is #3 (and #8 on the contralateral side, not shown). By E12.5, all five pairs are formed, shown here for the left side (#1–5) after removal of the limbs. Mammary rudiments and glands are numbered in anterior-posterior sequence, 1–5 on the left side and 6–10 on the right side, as shown in the cartoon. Corresponding but variable other nomenclature in the literature is also indicated
Studies in the rabbit led to the contention that mammary line formation is completed prior to mammary placode formation, per flank providing a shared ‘anatomical’ origin for all mammary glands arising on that flank. Moreover, other repetitive structures along the anterior-posterior (AP) body axis, such as limbs, are induced by the same molecular mechanism. Based on those two contentions, one might presume that all MRs are induced via shared molecular mechanisms.
However, we now know that MRs arise prior to completion of mammary line formation in the mouse, and not in sequence along the AP-axis [3], which suggests that the presumption that the mammary line is a pre-existing structure from which the mammary glands subsequently derive by a shared mechanism, is incorrect, at least for mice [4]. Moreover, defects in the formation and morphogenesis of select mammary gland pairs in various genetically modified mice show that different signaling networks induce and maintain mammary gland development at different positions along the AP-axis. Furthermore, the reciprocal tissue interactions governing mammary gland development [5] may co-vary with the high internal left-right (LR) asymmetry of the trunk. Indeed, LR-asymmetry between counterparts of mammary gland pairs has also become evident.
Thus, each mammary gland has an individual identity. Future studies on normal and pathological mammary gland development will benefit greatly from consistent documentation and analysis of data relative to mammary gland position. Accordingly, we will refer to them by number 1–10, as opposed to pair (1–5) or position (Fig. 1), unless the cited literature refers to mammary glands as clusters (e.g. anterior, posterior) without explicit specification of which glands were subject to examination.
Molecular Variation in the Induction of a Mammary Cell Fate Along the Mammary Lines
What determines the number and position of the individual pairs of mammary glands? The 2:1 correlation between the number of mammary glands and average litter size in most species [6] and co-incidence of supernumerary nipples with more frequent twinning as reported for isolated groups of sheep and macaques [7, 8] suggests considerable genetic plasticity that is molded by evolutionary pressure. Indeed, supernumerary breasts (polymastia) or nipples (polythelia) are an inherited trait in some humans [9], and pigs and sheep can be selectively bred for increased teat number [7, 10], indicating the involvement of a genetic component. Accordingly, several genetically modified mice have been identified with mammary induction defects leading to numerical aberrancies (Table 1). They can be organized into four categories depending on the subset of MRs affected, and loss or gain of MRs.
-
I
Loss of subset, linear sequence. The only example in this category is the transcription factor Hoxc6. Its loss occasionally leads to smaller thoracic buds at E12.5, and consistently to reduced fat pad development and branching morphogenesis by birth. These defects correspond to the Hoxc6 expression domain in the anterior part of the body, and are consistent with the general functions of Hox genes in providing and interpreting AP-positional information.
-
II
Loss of subset, non-linear sequence. This cluster contains a variety of transcription (Gli3, Pax3, Tbx2, Tbx3) and secreted factors (Fgf10, Nrg3). Interestingly, their expression in the somites or somite-derived dermal mesenchyme, and in particular somitic Fgf10 expression finally gave an identity to so-called ‘dermal factors’, which had long before been postulated to confer the mammary gland-inducing potential of the dermis [11, 12]. The dissimilarities in phenotypes of the mouse mutants in this category indicate that different molecular mechanisms for mammary induction exist along the AP-axis, and that the five mammary gland pairs develop independently of each other.
-
III
Loss of all MRs (with variable penetrance). Most genes in this category (Dkk1, Lef1, Pygo2, Lrp4-6, Wise) participate in the initial ectodermal Wnt signaling. The phenotypic clustering of their mutations suggests that the various mesodermal mammary inducers all converge on the same signaling cascade(s) in the ectoderm.
-
IV
Supernumerary MR formation. Increased ectodermal Ectodysplasin-A1 or Wnt signaling leads to formation of supernumerary glands along the mammary line, showing that the mammogenic capacity is maintained along the entire mammary line. Moreover, increased Wnt signaling leads to larger MRs, and in the case of reduced Wise/Sostdc1 function, a reduced spacing between MR2/7 and MR3/8 as well. The latter can be attributed to more ectodermal cells adopting a mammary fate and consequently losing proliferative activity that is required to fill the space between these MR pairs. A supernumerary MR slightly dorsal to the normal MR4 or MR9 is transiently present in some wild-type embryos of mixed genetic backgrounds but persists in some Nrg3Ska mutants, showing that mammogenic potential also exists at atypical positions outside the line.
Table 1.
Position-dependent mammary induction defects as established at E11.5 or E12.5
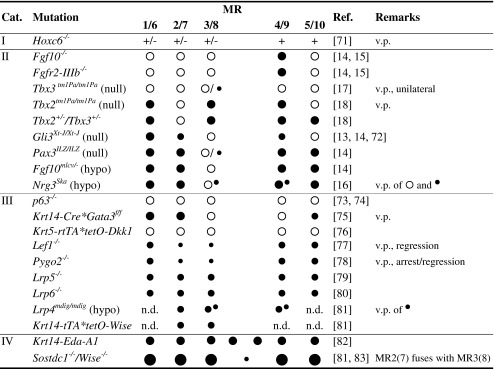
Legend: MR absent;
MR absent;  MR present without reported anomaly;
MR present without reported anomaly;  MR hypoplastic;
MR hypoplastic;  MR severely hypoplastic;
MR severely hypoplastic;  MR forms late and hypoplastic;
MR forms late and hypoplastic;  small supernumerary MR close to normal MR;
small supernumerary MR close to normal MR;  large MR. The region between MR3/8 and MR4/9 is prone to formation of supernumerary MRs of variable size (category IV)
large MR. The region between MR3/8 and MR4/9 is prone to formation of supernumerary MRs of variable size (category IV)
hypo hypomorphic mutant; n.d not determined; v.p. variable penetrance for presence, size and timing of formation
The various mesenchymal inductive mechanisms seem to converge on shared epithelial regulators, whose extent of activation appears to regulate the number of induced MRs. It is conceivable then that the variable inductive signals bestow the MR cells with unique properties. Indeed, preliminary gene profiling data suggest that each of the five paired mammary placode epithelia express unique subsets of genes (Sun and Veltmaat, http://www.veltmaatlab.net/research.html). While the functional relevance of these genes remains to be determined, mammary development proceeds differently in all MRs, as evidenced by the different growth rate and morphogenesis of the MRs [13], culminating in rudimentary glandular trees before birth that differ in size and number of branches (Fig. 2). These differences may very well underlie some of the differential attributes of adult MGs (discussed below).
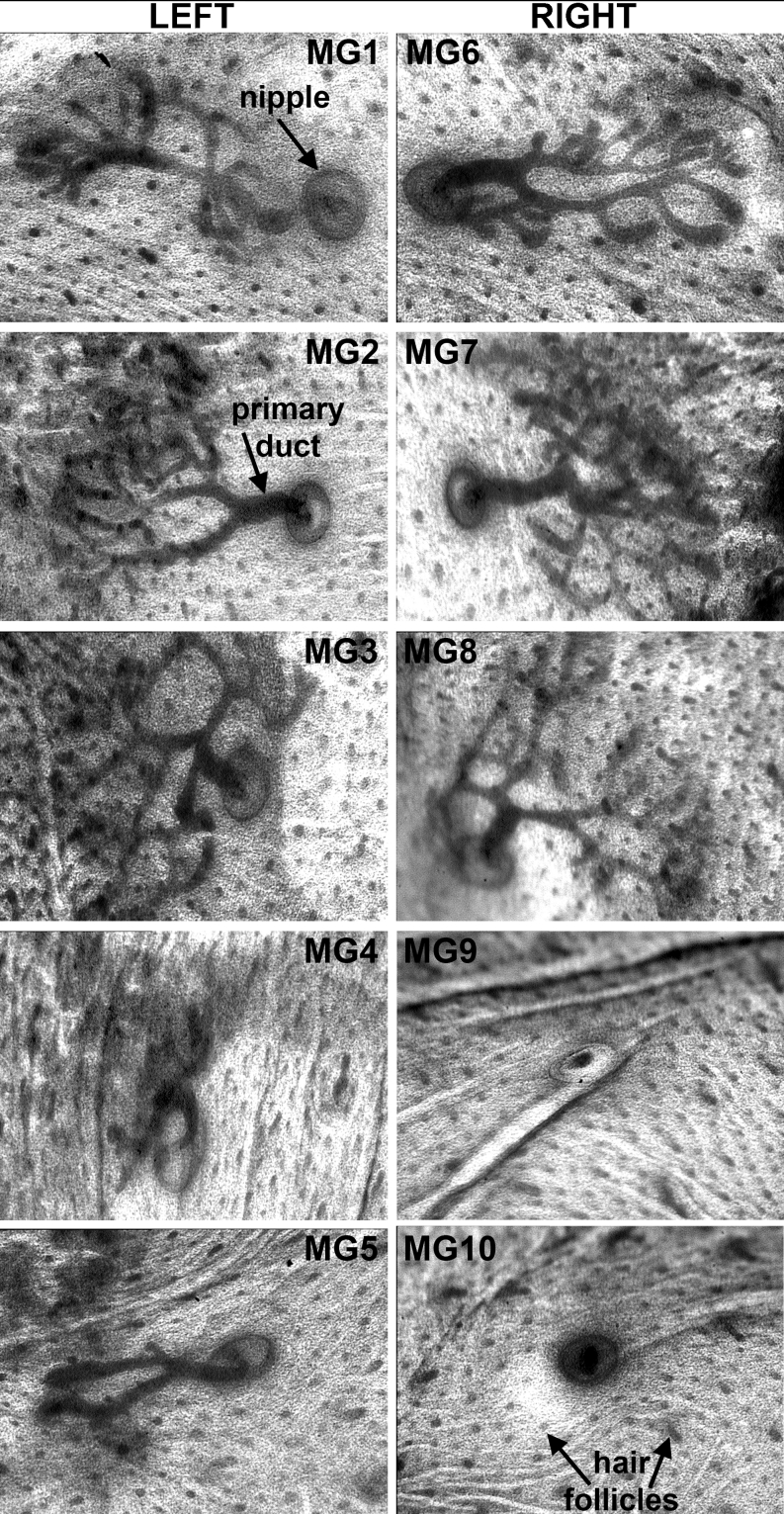
Fig. 2.
Position-dependent differences in mammary morphogenesis. Images of all mammary rudiments of a carmine-red stained skin of an E18.5 wild-type (C57Bl6/J) mouse embryo showing AP-axial differences in extent of outgrowth and branching of the mammary rudiments. Note that branching morphogenesis is not stereotypical, and left-right counterparts of a pair have no mirror-imaged symmetric morphology. MG9 and MG10 were broken off during dissection. MG: Mammary gland; hf: hair follicle
Why do mesodermal genes act differently at different positions along the mammary line? The answer may be found in their expression pattern, which is in part reflected in the temporal development of the murine mammary line. This line is actually composed of three initially unconnected streaks of Wnt10b expression [3]. As concluded from the phenotypes summarized in Table 1, these streaks form independently of each other.
The first streak forms on the flank, between the limbs. Its initially fragmented Wnt10b expression pattern in wild-type embryos (Fig. 1) and dorsalized position upon hypaxial truncation of the somites, as in Pax3 null mutants, indicate somitic involvement in the formation of this streak [14]. Fgf10 is expressed highly in the hypaxial dermomyotomal tips of somites #12–18, which underlie the region where MR2/7 and MR3/8 form [14, 15]. FGF10 activates its main receptor FGFR2-IIIb, expressed throughout the overlying surface ectoderm. Partial reduction of somitic Fgf10 expression, as in Gli3Xt-J/Xt-J null or hypomorphic Fgf10mlcv/- mutants, allows formation of a mammary line and all MRs except MR3/8, indicating that at least this pair requires somitic Fgf10 and, contrary to MR2/7, is susceptible to reduced levels of Fgf10 [14]. Fgf10 is not expressed in somites 19–24, and neither Wnt10b expression in this region nor induction of MR4/9 (above #24 somites) requires FGF10/FGFR2b signaling. These observations suggest that the anterior and posterior regions of this streak are two separate entities which develop by independent mechanisms, and that another yet to be identified somitic molecule(s) may be responsible for the initiation of mammogenesis in the posterior region, at the posterior edge of which arises MR4 or MR9. Interestingly, the boundary between the Fgf10-dependent and Fgf10-independent region at the level of #18 somites is home to a small mesenchymal Nrg3 expression domain, which seems a prerequisite for normal formation of MR3/8 [16].
The two other mammary streaks are located in the axilla and inguen, where they give rise to MR1/6 and MR5/10, respectively. They gradually elongate dorsally until they connect to the first streak on the flank [3]. Defects in these streaks or their MRs often coincide with defects in limb induction or specification in the absence of limb mesenchymal factors like Tbx2, Tbx3, Gli3 and Fgf10 [13, 14, 17, 18]. It remains to be determined whether MR1/6 and MR5/10 require such factors or the limb mesenchyme directly, or indirectly via formation of the limbs as new signaling centers. Perhaps MR2/7, that form at the junction of the first and axillary streak, and depend less than MR3/8 on somitic signals [14], similarly benefit from signals that originate in the limb. In conclusion, MRs develop due to in part variable mesodermal inductive signals along the AP-axis that converge on common ectodermal executive pathways. Thus, the different glands experience unique developmental histories and, furthermore, are embedded and surrounded by tissue that exhibits region-specific properties.
Left-Right Asymmetry During Embryonic Mammary Development
The embryonic flanks develop in the trunk as two broad lateral regions where the dorsal paraxial mesoderm intermingles with the ventral lateral plate mesoderm (LPM). They establish new, ventro-lateral boundaries with the ventrum, which serve as bilateral platforms for mammary line formation. Externally, these presumptive mammary forming regions look left-right (LR) symmetric. But looks can be deceiving! LR asymmetries during mammogenesis become evident at the level of individual mammary gland pairs: The size and branching pattern differ between contralateral glands; one counterpart of a normal or supernumerary pair may be missing or shifted from its symmetric position, shown for mouse in Figs. 2 and 3. In humans with Poland syndrome, one breast (most often the right) may remain hypoplastic [19]. Interestingly, formation of the mammary line and MRs is slightly advanced on the left side (Fig. 3) and the left side seems to have a greater mammogenic potential. For example, unilateral polythelia or polymastia occurs most frequently on the left side in humans [20] and mice [21].
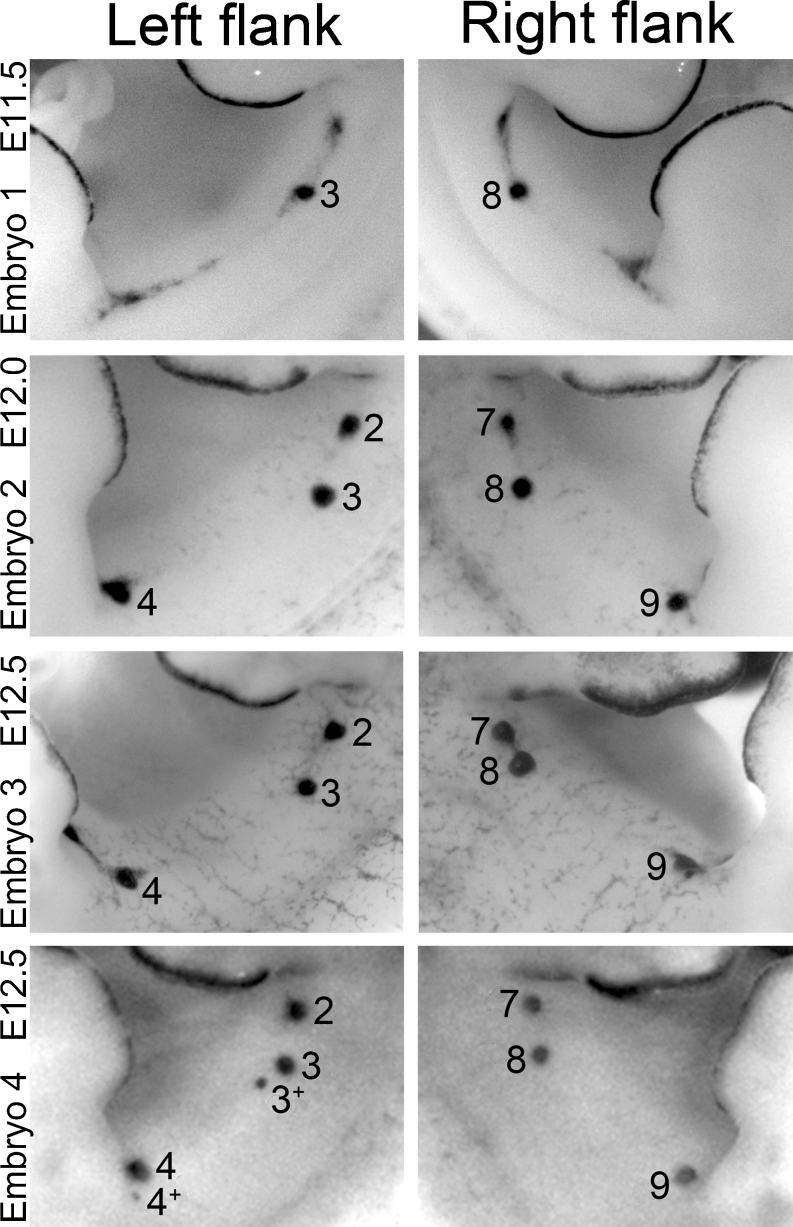
Fig. 3.
Examples of left-right asymmetry in embryonic mammary gland development. Images of flanks of Lac-Z stained mice (mixed genetic backgrounds) expressing Topgal as a marker for mammary line and rudiment formation. Top four panels show left-right asynchrony in formation of the mammary line and mammary rudiment (MR) pair 2 and 7. Lower four panels show anterior shift of MR 8 and formation of supernumerary MRs (indicated as 3+ and 4+ near the normal MR3 and MR4)
The basis for LR asymmetric mammary gland development is likely rooted in embryonic LR patterning: During vertebrate embryogenesis, the LR axis establishes the positions of the heart and visceral organs and promotes structural asymmetries that are necessary for their functions. The driving force for LR patterning is unilateral activity of molecules such as the secreted factor nodal, which is expressed by the left half of the node, left-sided LPM and overlying ectoderm [22]. In the left-sided LPM, nodal initiates the expression of the transcription factor Pitx2, an essential “interpreter” of LR axial patterning during organogenesis [reviewed by 23]. Thus, as ectodermal appendages that are induced at the ventro-lateral boundaries, i.e. at the fronts of somitic penetration into two molecularly different regions of left and right LPM [14], the left and right mammary glands have distinct molecular “histories” from their inception!
Moreover, and despite their morphological bilateral symmetry, the left and right somites also differ from each other with significant LR differences in gene expression [24, 25]. Of interest is perhaps heparin-binding EGF-like growth factor [25], which is transiently elevated on the left side, temporally overlapping with formation of MR3/8. Furthermore, retinoic acid (RA) signaling promotes LR asymmetry in the somites, as demonstrated by one-sided delay in somite formation in chick and zebrafish embryos, caused by pharmacological inhibition of RA production [26, 27]. Several modulators of RA signaling and synthesis, including Raldh2, are expressed in the somites proper, and the RA receptor RARβ is expressed in the flank mesenchyme underlying MR2/7, 3/8, 4/9 [28]. Notably, perturbation of RA-signaling affects MR3/8 formation ex vivo [28]. Unfortunately, laterality in effects of RA signaling on mammary gland development was not tested. This will be difficult to do, due to lethality of RA-deprived mouse models prior to mammary gland formation, and due to artifacts introduced during MR development ex vivo under otherwise normal conditions, but especially under experimental conditions such as pharmological modulation of RA-signaling. However, it is of interest that RA-deficient Raldh2−/− mouse embryos exhibit transiently delayed right-sided development and segmentation of the paraxial mesoderm into somites #8–15 [29]. With MR2/7 and MR3/8 forming above the #12 and #15 somites respectively, it is thus conceivable that RA signaling may be a pivotal link between somitic laterality and asynchrony and asymmetry in development of LR counterparts of particularly these MR pairs. Moreover, the already known requirement of somitic signals for induction of MR3/8 [14], provides strong support for the hypothesis that this mammary gland pair—despite its seemingly morphological symmetry - may be lateralized, either directly or indirectly via inductive interaction with lateralized aspects of somites, including effects of RA-signaling.
Finally, the curvature of the AP-body axis may also be involved in promoting subtle asymmetry of mammogenesis: Most often the AP-axis is curved to the right (tail over right shoulder), leading to a ‘compressed’ right flank and ‘stretched’ left flank. If molecular gradients are involved in early mammary gland formation, which is the case at least for somitic Fgf10 [14], their shape may be different between left and right flanks, which could possibly explain the slight LR asynchrony in length of the mammary lines and mis-alignment between paired glands that is sometimes observed even in wild-type embryos (Fig. 3).
Left-Right Asymmetry in Breast Cancer
A striking but still unexplained feature of breast cancer is a higher tumor incidence in the left breast. Although modest with a ~1.10 left/right incidence ratio, such laterality was consistently found in several epidemiological studies [30–36] regardless of gender and stage of disease. Even in bilateral breast cancer cases, the left breast is more frequently affected first, or with a larger tumor [37].
Survival rates also appear to reflect LR differences: A breast cancer study among 590 Russian patients reported a statistically significant higher survival rate of patients with right-sided, advanced stage disease [38]. In contrast, a study of 163 Norwegian patients associated longer survival with left-sided disease, but did not relate this finding to disease stage [39]. In a third study of 10,702 Israeli women, no laterality was detected for overall patient survival from years 1–15 [40], but trends of a higher 5–10 year survival rate of left-sided later stage disease, and a higher 10–15 year survival rate of right-sided early stage disease, call for statistical analysis of the data within such smaller time periods, which had not been done. The most recent study of 384 Pakistani breast cancer patients indicates that although left-sided tumors are larger and more prevalent, when standardized for tumor size, right-sided tumors are more prone to metastasis and are associated with lower survival rates [36]. Although the results of these studies appear discordant with each other, they certainly attest to the need for well-defined parameters to better assess laterality. Furthermore, these available data contest, at least in part, the assumption that asymmetry in survival may be due to radiation-induced heart damage, which is greater with left-side targeted therapy. Instead, the data suggest that the LR differences may be due to lateralized tumor biology, similar to LR asymmetry in incidence and patient survival observed with cancers of other paired organs, including kidney cancer, which does not entail cardiac complications following side-specific radiation therapy [33, 41].
Why do tumors form with unequal probability and perhaps outcome in left versus right members of organ pairs? A variety of epidemiological correlates have been investigated, including age, race, ethnicity, sex, breast size, stage of disease at diagnosis, histological tumor type, and even left- versus right-handedness. None of these factors emerged as a consistently significant variable for the increased left-sided incidence [30–32, 37]. By contrast, laterality in outcome may in part be explained by laterality of axillary lymph node metastasis. A higher mean number of lymph nodes with metastases in breast cancer patients has been reported for right-sided tumors, consistent with more lymph nodes present on the right side [43, 44]. However, when axillary lymph node involvement was analyzed with respect to the total number of affected nodes, the right-sided increase was present only in patients with 1–3 positive nodes [45]. For patients with >3 positive nodes (a characteristic of locally advanced or higher stage disease) left-sided tumors were associated with more affected nodes despite the LR asymmetry in overall node number [45]. Thus, metastasis to lymph nodes seems to be a lateralized disease feature, and may indicate LR differences in tumor biology, e.g. greater metastatic potential of left-sided tumors, or lateralized features of the lymph nodes themselves. In general, attempts to identify lateralized features may be confounded by the profound heterogeneity of breast cancer and could possibly be improved by inclusion of more detailed cancer subtype-specific information such as genomic classifiers. Indeed, an interesting correlate in laterality of breast tumors is the expression of Her-2 and perhaps also hormone receptors, although the sidedness may co-vary with other parameters: Among 166 Kuwaiti patients spanning all disease stages at diagnosis, right-sided tumors are significantly associated with amplified Her-2 expression, without lateralized differences for hormone receptor expression [46], whereas among 384 Pakistani breast cancer patients with bone metastasis, right-sided tumors were markedly less prone to express Her-2, estrogen receptor, or progesterone receptor, either individually or in combination [36].
It is tempting to speculate that the LR differences in tumor incidence and patient survival may be related to expression of genes that govern LR axial patterning during embryonic development, like Nodal and Pitx2 [reviewed by 47]. Increased expression of Nodal and methylation of Pitx2 DNA correlates with poor prognostic outcome in breast cancer patients [48–50]. Overexpression of another nodal pathway component, the co-receptor Cripto-1, is also associated with decreased patient survival [reviewed by 49]. Consistent with these findings, nodal pathway signaling promotes an invasive phenotype in breast cancer cells and induces tumor vascularization, which has implications for disease progression and relapse [51]. Whether nodal or nodal pathway members are expressed asymmetrically during mammary gland development remains to be investigated, as does the question of whether such putative LR differences are recapitulated in breast tumors.
While the laterality in breast cancer incidence and outcome seems subtle, evidence for its existence is compelling. We may be unaware of more significant aspects of laterality because situs is often not a factor in molecular and clinical correlations, and experimental data in mouse models typically are generated from a single side, or if from both sides, may be combined for analyses, obscuring potential LR differences. To our knowledge, laterality has not been investigated systematically in any mouse tumor mammary model to date. Better insights into laterality of breast cancer incidence, biology, and outcome may lead to a better understanding of the disease(s) and have prognostic value.
“Breast” Cancer in Mouse: Location, Location, Location!
The mouse is widely used as a breast cancer model. The axial position of the human breast approximates the 5th thoracic somite or its derived rib. Taking into account the differences in somite number between mouse and human, this area corresponds best to the area between MG2/7 and 3/8 in the mouse. However, the inguinal mouse MG4/9 are generally used for molecular and histological analyses. Does it matter? As described above, the five pairs of mammary glands in the mouse are each induced by unique inductive signaling networks. Yet surprisingly little information exists on differences between the five pairs of glands in adult mice, both at the level of normal physiology and with respect to response to tumor-initiating events. However, several studies report significantly differential tumor incidence or biology depending on the position along the AP axis. So the answer is almost certainly: Yes!
Examples for AP variations in tumor-incidence and biology come from endogenous tumor agents, chemical carcinogenesis, various treatments, and transgenic approaches. Integration of mouse mammary tumor viruses (MMTV) in the C3H mouse strain causes mammary tumors most often in thoracic glands. While tumor incidences are equal among the gland pairs after correction for total wet tissue weight [52], the relative ratios of epithelial to stromal/adipose tissue may not correspond to wet weights and could confound the analysis. Indeed, increased incidence of DMBA-induced tumors in the thoracic mammary glands of rats was correlated to asynchronous development of the glands. Differences in the number of terminal end buds (TEBs) and alveolar buds per gland results in variable abundance of target cells for chemical carcinogens depending on treatment age [53]. Likewise, murine MG2/7 and MG3/8 exhibit a greater number and larger TEBs than MG4/9 (see also Fig. 2); and MG2/7 specifically undergoes a reduction in TEBs from 5 to 6 weeks of age [54]. The observed asynchrony in postnatal development can also result in differential responses to hormones [53]. These AP-specific developmental features are a plausible basis for increased tumor incidence in thoracic glands. Interestingly, exposure to power frequency magnetic fields (MF) increases DMBA-induced mammary tumorigenesis in SD1 rats, particularly of MG1/6 and 2/7 in correlation with more significant MF-induced increases in ornithine decarboxylase activity—a possible indicator for stem cell proliferation [55]. However, in F433 Fisher rats, MF exposure increased the incidence of DMBA-induced adenocarcinoma specifically in the 5th of the six pairs of glands in rats, but latency was reduced most in the 3rd pair [56]. Thus, region-specific effects on tumorigenesis may vary even between strains of the same species.
Transgenic MMTV-Polyoma middle T antigen (PyMT) causes the largest tumor burden with shortest latency in MG1/6 of C57BL/6 mice [57]. Tumor latency is more similar among all other glands, with MG4/9 harboring the second largest tumor burden. Introduction of an iNOS (Nos2) null mutation increases tumor latency and reduces tumor burden most in MG4/9 and least in MG1/6 [57]. Overexpressed Erbb2 in MMTV-cNeu mice [58] induces tumors preferentially in MG2/7 + 3/8 (Table 2) and tumor incidence is comparatively low in MG4/9/5/10. Transgenic C3(1)-SV40-T/t-antigen causes tumor formation first in MG1/6 and MG2/7 with the highest total tumor burden in MG2/7 [59]. Differential activity of transgene promoters is one possible explanation for variations in transgene-initiated mammary tumorigenesis. Even then, such mechanism(s) would serve to highlight the significance of biological differences between pairs of glands. What is clear from these data, though, is that these transgenic models generate most tumors in specific anterior mammary glands, raising the question if other transgenic models exhibit similar or other unique gland-specific incidences along the AP-axis.
Table 2.
Regional distribution of mammary tumors in MMTV-cNeu transgenic mice
| Cohort \ Glands | MG1/6 | MG2/7+3/8 | MG4/9+5/10 |
|---|---|---|---|
| A | 4 (15) | 19 (73)** | 3 (12) |
| B | 11 (27) | 26 (63)** | 4 (10) |
| Weight (%) | (18) | (46) | (36) |
(A) Number of palpable tumors observed in 21 tumor-bearing transgenic mice at the age of 9 months (% of 26 tumors total), and (B) number of tumors as assessed by necropsy in 22 mice at tumor endpoint (% of 41 tumors total). The relative wet weight of the specified glands in % is quoted from [52] and was not specifically determined for this strain. **P < 0.0001 compared to MG4/9 + 5/10 and to MG1/6 by mixed-effects Poisson regression analysis
Transplantation experiments also provide clues to the existence of AP-axial effects both in the tumor cells and environment. PyMT tumor cells isolated from anterior tumors proliferate faster than cells from posterior tumors, independent of implantation site. Significant differences in gene expression exist between anterior and posterior tumors despite equivalent histology, and the difference in proliferative potential was maintained over transplantation passages [60]. Moreover, the higher take rate of teratocarcinoma cell grafts at lumbar subcutaneous sites despite their faster growth rate at thoracic sites [61] provides evidence for unique positional features. Likewise, MDA-MB-435 tumor cell xenografts in thoracic MFPs grow better in response to high fat diet but not when in inguinal MFPs [62]. Thus, along the AP-axis, both the mammary epithelium and stroma harbor positional variations that influence tumor growth.
Finally, modality of disease progression may also depend on positional differences: Tumors of C3HxVY-Avy mice are more likely to metastasize from thoracic glands than from abdominal glands [63]. MDA-MB-435 cell implants into MFPs of athymic nude mice showed the same tendency and moreover, thoracic implants spread via the vascular system, while inguinal implants spread via the lymphatic system [62].
What could be the cause of these positional variations? A comprehensive review article on wound healing, immune responsiveness, and mammary tumorigenesis in mice and rats, concluded that not only tumor growth, but also wound healing responses are more pronounced in anterior regions [64]. The prominent role of inflammatory signaling in breast cancer suggests that this AP-gradient may influence murine mammary tumor biology. The AP-gradient of the nervous system and vascularization, resulting in reduced blood flow in the posterior body, provides perhaps the likeliest explanation for the region-specific variations in tumor biology [64]. Such a gradient has also been confirmed for blood flow in pectoral versus inguinal mammary glands [65]. Consequently, one can hypothesize that posterior glands exhibit reduced oxygen tension and lower concentrations of systemic factors, which may modulate tumor incidence. Alternatively, varying relative positions of lymph nodes may alter lymph drainage of the tissue and consequently tumor biology.
The role of embryonic signaling pathways in promoting cancer cell stemness and metastasis [66] may suggest that differential intrinsic features of tumor cells and their microenvironment could possibly be traced back to the differential developmental signals along the mammary line of the mouse. For example, by interpretation of regional cues, Hox transcription factors lay down positional memory that results in region-of-origin specific cell biology, as shown for dermal fibroblasts [67]. By analogy, mammary stroma may secrete unique factors depending on their position along the AP axis. Aberrant Hox gene activity has also been implicated in breast cancer [68] and in altered Wnt signaling [69, 70]. Given the prominent role of the Wnt pathway in embryonic MG development and in breast tumor biology, it is conceivable that the Hox and Wnt pathways affect tumors differentially along the AP axis.
Conclusions and Perspectives
The mouse is an invaluable model system to study mammalian development, physiology and cancer. Here, we reviewed evidence that each of the ten mammary glands in mouse is unique, from inception to maintenance, throughout normal and tumor development as well as metastasis. Mechanistic insights into causes for variations in prenatal development of the mammary glands begin to emerge. One can presently only speculate about the cause(s) for regional variations in tumor incidence and biology, which may in part stem from intrinsic molecular differences during embryogenesis, and from variations in stromal factors including vascularization and lymph drainage. However, this phenomenon can possibly be exploited to gain insights into factors that modulate tumorigenesis. Clearly, future studies on normal and pathological mammary gland development will benefit greatly from improved documentation of positional variations. In particular, laterality should be treated as a possible variable in studies of human patients to discern why tumor incidence and progression are lateralized disease features and to assess the potential implications for the clinic. Inbred mouse strains and engineered mouse models provide unique opportunities to study positional variations from both left-right and anterior-posterior axis effects. Invaluable information may be lost when samples/data from different locations are combined before analyses, or confounded when phenotypic aspects of one gland are correlated to molecular data from another. Moreover, experimental design and analysis of specific cancer models may improve when regional effects are taken into account, e.g. by tailoring treatments to developmental asynchrony and by making guided choices such as using inguinal glands for studies of lymphatic metastasis. Ultimately, such consideration of positional variations in mammary gland development and tumorigenesis should improve the predictive value of studies in mouse for various aspects of human breast cancer.
Acknowledgments
We are indebted to many colleagues for suggestions and discussion. We thank Glenn Summers, Robert Leighty, Thu Duong, May Yin Lee and Li Sun for expert assistance. Research was supported by A*STAR (J.M.V.), National Institute of Health (NIH) HD068993 (A.F.R.) and the Intramural Research Program of the NIH in part with Federal Funds under contract no.HHSN261200800001E (E.S).
Abbreviations
- AP
anterior-posterior
- DMBA
7,12-demethylbenz-alpha-anthracene
- DV
dorso-ventral
- LR
left-right
- LPM
lateral plate mesoderm
- MFP
mammary fat pad
- MG
mammary gland
- MR
mammary rudiment
- RA
retinoic acid
- TEB
terminal end bud
Contributor Information
Jacqueline M. Veltmaat, Email: jveltmaat.sc@gmail.com
Ann F. Ramsdell, Email: ramsdell@musc.edu
Esta Sterneck, Email: sternecg@mail.nih.gov.
References
- 1.Bresslau E. The structure and development of the mammary apparatus in the marsupialia., in The Mammary Apparatus of the Mammalia—in the light of ontogenesis and phylogenesis. London: Methuen and Co. Ltd; 1920. pp. 44–95. [Google Scholar]
- 2.Propper AY. Wandering epithelial cells in the rabbit embryo milk line. A preliminary scanning electron microscope study. Dev Biol. 1978;67(1):225–31. doi: 10.1016/0012-1606(78)90311-1. [DOI] [PubMed] [Google Scholar]
- 3.Veltmaat JM, et al. Identification of the mammary line in mouse by Wnt10b expression. Dev Dyn. 2004;229(2):349–56. doi: 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]
- 4.Propper AY, Howard BA, Veltmaat J.M. Prenatal morphogenesis of mammary glands in mouse and rabbit. J Mammary Gland Biol Neoplasia. 2013. doi:dx.doi.org/10.1007/s10911-013-9298-0. [DOI] [PMC free article] [PubMed]
- 5.Robinson GW, Karpf AB, Kratochwil K. Regulation of mammary gland development by tissue interaction. J Mammary Gland Biol Neoplasia. 1999;4(1):9–19. doi: 10.1023/A:1018748418447. [DOI] [PubMed] [Google Scholar]
- 6.Gilbert AN. Mammary number and litter size in Rodentia: the “one-half rule”. Proc Natl Acad Sci U S A. 1986;83(13):4828–30. doi: 10.1073/pnas.83.13.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell A. Bell on the development by selection of supernumerary mammae in sheep. Science. 1899;9(227):637–9. doi: 10.1126/science.9.227.637. [DOI] [PubMed] [Google Scholar]
- 8.Hsu MJ, et al. High incidence of supernumerary nipples and twins in formosan macaques (Macaca cyclopis) at Mt. Longevity, Taiwan. Am J Primatol. 2000;52(4):199–205. doi: 10.1002/1098-2345(200012)52:4<199::AID-AJP4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 9.Cellini A, Offidani A. Familial supernumerary nipples and breasts. Dermatology. 1992;185(1):56–8. doi: 10.1159/000247404. [DOI] [PubMed] [Google Scholar]
- 10.Hirooka H, et al. A whole-genome scan for quantitative trait loci affecting teat number in pigs. J Anim Sci. 2001;79(9):2320–6. doi: 10.2527/2001.7992320x. [DOI] [PubMed] [Google Scholar]
- 11.Kratochwil K. Organ specificity in mesenchymal induction demonstrated in the embryonic development of the mammary gland of the mouse. Dev Biol. 1969;20(1):46–71. doi: 10.1016/0012-1606(69)90004-9. [DOI] [PubMed] [Google Scholar]
- 12.Propper A, Gomot L. Tissue interactions during organogenesis of the mammary gland in the rabbit embryo. C R Acad Sci Hebd Seances Acad Sci D. 1967;264(22):2573–5. [PubMed] [Google Scholar]
- 13.Lee MY, et al. Ectodermal influx and cell hypertrophy provide early growth for all murine mammary rudiments, and are differentially regulated among them by Gli3. PLoS One. 2011;6(10):e26242. doi: 10.1371/journal.pone.0026242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veltmaat JM, et al. Gli3-mediated somitic Fgf10 expression gradients are required for the induction and patterning of mammary epithelium along the embryonic axes. Development. 2006;133(12):2325–35. doi: 10.1242/dev.02394. [DOI] [PubMed] [Google Scholar]
- 15.Mailleux AA, et al. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129(1):53–60. doi: 10.1242/dev.129.1.53. [DOI] [PubMed] [Google Scholar]
- 16.Howard B, et al. Identification of the scaramanga gene implicates Neuregulin3 in mammary gland specification. Genes Dev. 2005;19(17):2078–90. doi: 10.1101/gad.338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130(10):2263–73. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 18.Jerome-Majewska LA, et al. Tbx3, the ulnar-mammary syndrome gene, and Tbx2 interact in mammary gland development through a p19Arf/p53-independent pathway. Dev Dyn. 2005;234(4):922–33. doi: 10.1002/dvdy.20575. [DOI] [PubMed] [Google Scholar]
- 19.Shermak MA. Congenital and developmental abnormalities of the breast, in Management of Breast Diseases, Jatoi I, Kaufman M, editors. 2010, Springer. p. 37–51.
- 20.Schmidt H. Supernumerary nipples: prevalence, size, sex and side predilection—a prospective clinical study. Eur J Pediatr. 1998;157(10):821–3. doi: 10.1007/s004310050944. [DOI] [PubMed] [Google Scholar]
- 21.Little CC, McDonald H. Abnormalities of the mammae in the house mouse. J Hered. 1945;36:285–288. [Google Scholar]
- 22.Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133(11):2095–104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 23.Ramsdell AF. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol. 2005;288(1):1–20. doi: 10.1016/j.ydbio.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 24.Golding JP, et al. Mouse myotomes pairs exhibit left-right asymmetric expression of MLC3F and alpha-skeletal actin. Dev Dyn. 2004;231(4):795–800. doi: 10.1002/dvdy.20176. [DOI] [PubMed] [Google Scholar]
- 25.Golding JP, et al. Heparin-binding EGF-like growth factor shows transient left-right asymmetrical expression in mouse myotome pairs. Gene Expr Patterns. 2004;5(1):3–9. doi: 10.1016/j.modgep.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Kawakami Y, et al. Retinoic acid signalling links left-right asymmetric patterning and bilaterally symmetric somitogenesis in the zebrafish embryo. Nature. 2005;435(7039):165–71. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- 27.Vermot J, Pourquie O. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature. 2005;435(7039):215–220. doi: 10.1038/nature03488. [DOI] [PubMed] [Google Scholar]
- 28.Cho KW, et al. Retinoic acid signaling and the initiation of mammary gland development. Dev Biol. 2012;365(1):259–66. doi: 10.1016/j.ydbio.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Vermot J, et al. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005;308(5721):563–6. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- 30.Blot W, Fraumeni J, Young J. Left-sided breast cancer. Lancet. 1977;2:762–763. doi: 10.1016/S0140-6736(77)90266-5. [DOI] [PubMed] [Google Scholar]
- 31.Ekbom A, et al. Epidemiologic correlates of breast cancer laterality. Cancer Causes Control. 1994;7:539–543. doi: 10.1007/BF01831378. [DOI] [PubMed] [Google Scholar]
- 32.Garinkel L, Craig L, Seidman H. An appraisal of left and right breast cancer. J Natl Cancer Inst. 1959;23:617–631. [PubMed] [Google Scholar]
- 33.Roychoudhuri R, Putcha V, Møller H. Cancer and laterality: a study of the five major paired organs (UK) Cancer Causes and Control. 2006;17(5):655–662. doi: 10.1007/s10552-005-0615-9. [DOI] [PubMed] [Google Scholar]
- 34.Tulinius H, et al. Tumors in Iceland: 10. Malignant tumours of the breast. A histological classification, laterlaity, survival and epidemiological considerations. Acta path Microbiol Immunol Scand. 1988;96:229–238. doi: 10.1111/j.1699-0463.1988.tb05296.x. [DOI] [PubMed] [Google Scholar]
- 35.Weiss H, Devesa S, Brinton L. Laterality of breast cancer. Cancer Causes Control. 1996;7:539–543. doi: 10.1007/BF00051887. [DOI] [PubMed] [Google Scholar]
- 36.Fatima N, et al. Lower incidence but more aggressive behavior of right sided breast cancer in pakistani women: does right deserve more respect? Asian Pac J Cancer Prev. 2013;14(1):43–5. doi: 10.7314/APJCP.2013.14.1.43. [DOI] [PubMed] [Google Scholar]
- 37.Senie R, et al. Epidemiology of breast carcinoma II: factors related to the predominance of left-sided disease. Cancer Causes and Control. 1980;46:1705–1713. doi: 10.1002/1097-0142(19801001)46:7<1705::aid-cncr2820460734>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 38.Borisenkov MF and Bazhenov SM. Survival in human breast cancer: effects of tumor laterality and the time of year of surgery. Human Physiology. 2001;27(5):631-34.
- 39.Hartveit F, Tangen M, Hartveit E. Side and survival in breast cancer. Oncology. 1984;41(3):149–54. doi: 10.1159/000225812. [DOI] [PubMed] [Google Scholar]
- 40.Melnik Y, et al. Breast cancer in Israel: laterality and survival. J Cancer Res Clin Oncol. 1979;95(3):291–3. doi: 10.1007/BF00410651. [DOI] [PubMed] [Google Scholar]
- 41.Delahunt B, Bethwaite P, Nacey JN. Renal cell carcinoma in New Zealand: a national survival study. Urology. 1994;43(3):300–9. doi: 10.1016/0090-4295(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 42.Pece S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. cell. 140(1): 62–73. [DOI] [PubMed]
- 43.Cappello F, et al. Study of axillary lymph node asymmetry in a female population. J Anat. 2001;199(Pt 5):617–20. doi: 10.1017/S002187820100841X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dane S, et al. Asymmetries in breast cancer lateralization and both axillary lymph node number and metastatic involvement. Lymphology. 2008;41(2):75–9. [PubMed] [Google Scholar]
- 45.Nouh MA, et al. Lymph node metastasis in breast carcinoma: clinicopathological correlations in 3747 patients. J Egypt Natl Canc Inst. 2004;16(1):50–6. [PubMed] [Google Scholar]
- 46.Saleh F, Abdeen S. Pathobiological features of breast tumours in the State of Kuwait: a comprehensive analysis. J Carcinog. 2007;6:12. doi: 10.1186/1477-3163-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilting J, Hagedorn M. Left-right asymmetry in embryonic development and breast cancer: common molecular determinants? Curr Med Chem. 2011;18(36):5519–27. doi: 10.2174/092986711798347252. [DOI] [PubMed] [Google Scholar]
- 48.Kenney NJ, Adkins HB, Sanicola M. Nodal and Cripto-1: embryonic pattern formation genes involved in mammary gland development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2004;9(2):133–144. doi: 10.1023/B:JOMG.0000037158.91940.1c. [DOI] [PubMed] [Google Scholar]
- 49.Strizzi L, et al. Emerging roles of nodal and cripto-1: from embryogenesis to breast cancer progression. Breast Dis. 2008;29(−1):91–103. doi: 10.3233/bd-2008-29110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harbeck N, et al. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008;26(31):5036–42. doi: 10.1200/JCO.2007.14.1697. [DOI] [PubMed] [Google Scholar]
- 51.Quail DF, et al. Nodal signalling in embryogenesis and tumourigenesis. Int J Biochem Cell Biol. 2013;45(4):885–898. doi: 10.1016/j.biocel.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 52.Vaage J. Relationship between tumor growth characteristics and preferential sites of growth. J Natl Cancer Inst. 1984;72(5):1199–203. [PubMed] [Google Scholar]
- 53.Russo IH, Russo J. Hormone prevention of mammary carcinogenesis: a new approach in anticancer research. Anticancer Res. 1988;8(6 489011536):1247–1264. [PubMed] [Google Scholar]
- 54.Ball SM. The development of the terminal end bud in the prepubertal-pubertal mouse mammary gland. Anat Rec. 1998;250(4):459–464. doi: 10.1002/(SICI)1097-0185(199804)250:4<459::AID-AR9>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 55.Mevissen M, Haussler M, Loscher W. Alterations in ornithine decarboxylase activity in the rat mammary gland after different periods of 50 Hz magnetic field exposure. Bioelectromagnetics. 1999;20(6):338–46. doi: 10.1002/(SICI)1521-186X(199909)20:6<338::AID-BEM2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 56.Fedrowitz M, Loscher W. Exposure of Fischer 344 rats to a weak power frequency magnetic field facilitates mammary tumorigenesis in the DMBA model of breast cancer. Carcinogenesis. 2008;29(1):186–93. doi: 10.1093/carcin/bgm217. [DOI] [PubMed] [Google Scholar]
- 57.Davie SA, et al. Effects of FVB/NJ and C57Bl/6J strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res. 2007;16(2):193–201. doi: 10.1007/s11248-006-9056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89(22):10578–82. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Green JE, et al. 2-difluoromethylornithine and dehydroepiandrosterone inhibit mammary tumor progression but not mammary or prostate tumor initiation in C3(1)/SV40 T/t-antigen transgenic mice. Cancer Res. 2001;61(20):7449–55. [PubMed] [Google Scholar]
- 60.Varticovski L, et al. Accelerated preclinical testing using transplanted tumors from genetically engineered mouse breast cancer models. Clin Cancer Res. 2007;13(7):2168–77. doi: 10.1158/1078-0432.CCR-06-0918. [DOI] [PubMed] [Google Scholar]
- 61.Oosterhuis JW, et al. The effects of regional factors on the growth rate and the differentiation of mouse teratocarcinoma. Br J Cancer. 1983;47(3):407–411. doi: 10.1038/bjc.1983.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meschter CL, Connolly JM, Rose DP. Influence of regional location of the inoculation site and dietary fat on the pathology of MDA-MB-435 human breast cancer cell-derived tumors grown in nude mice. Clin Exp Metastasis. 1992;10(3 982934656):167–173. doi: 10.1007/BF00132748. [DOI] [PubMed] [Google Scholar]
- 63.Sheldon WG, et al. Distribution of mammary gland neoplasms and factors influencing metastases in hybrid mice. Lab Anim Sci. 1982;32(2):166–8. [PubMed] [Google Scholar]
- 64.Auerbach R, Auerbach W. Regional differences in the growth of normal and neoplastic cells. Science. 1982;215(4529):127–34. doi: 10.1126/science.7053564. [DOI] [PubMed] [Google Scholar]
- 65.Minasian H, Minassian D. The relationship between blood flow of mammary tissue and its overlying skin in the mouse. Lab Anim. 1983;17(4):321–3. doi: 10.1258/002367783781062352. [DOI] [PubMed] [Google Scholar]
- 66.Takebe N, Warren RQ, Ivy SP. Breast cancer growth and metastasis: interplay between cancer stem cells, embryonic signaling pathways and epithelial-to-mesenchymal transition. Breast Cancer Res. 2011;13(3):211. doi: 10.1186/bcr2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rinn JL, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22(3):303–7. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–71. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 69.Aulehla A, Pourquie O. Signaling gradients during paraxial mesoderm development. Cold Spring Harb Perspect Biol. 2010;2(2):a000869. doi: 10.1101/cshperspect.a000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bondos S. Variations on a theme: Hox and Wnt combinatorial regulation during animal development. Sci STKE. 2006;2006(355):pe38. doi: 10.1126/stke.3552006pe38. [DOI] [PubMed] [Google Scholar]
- 71.Garcia-Gasca A, Spyropoulos DD. Differential mammary morphogenesis along the anteroposterior axis in Hoxc6 gene targeted mice. Dev Dyn. 2000;219(2):261–76. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1048>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 72.Hatsell SJ, Cowin P. Gli3-mediated repression of Hedgehog targets is required for normal mammary development. Development. 2006;133(18):3661–70. doi: 10.1242/dev.02542. [DOI] [PubMed] [Google Scholar]
- 73.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 74.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 75.Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9(2):201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 76.Chu EY, et al. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004; 131(19): 4819–29. [DOI] [PubMed]
- 77.Boras-Granic K, et al. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295(1):219–31. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 78.Gu B, et al. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol. 2009;185(5):811–26. doi: 10.1083/jcb.200810133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lindvall C, et al. The Wnt signaling receptor Lrp5 is required for mammary ductal stem cell activity and Wnt1-induced tumorigenesis. J Biol Chem. 2006;281(46):35081–7. doi: 10.1074/jbc.M607571200. [DOI] [PubMed] [Google Scholar]
- 80.Lindvall C, et al. The Wnt co-receptor Lrp6 is required for normal mouse mammary gland development. PLoS One. 2009;4(6):e5813. doi: 10.1371/journal.pone.0005813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ahn Y, et al. Lrp4 and Wise interplay controls the formation and patterning of mammary and other skin appendage placodes by modulating Wnt signaling. Development. 2013;140(3):583–93. doi: 10.1242/dev.085118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mustonen T, et al. Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development. 2004;131(20):4907–19. doi: 10.1242/dev.01377. [DOI] [PubMed] [Google Scholar]
- 83.Narhi K, et al. Sostdc1 defines the size and number of skin appendage placodes. Dev Biol. 2012;364(2):149–61. doi: 10.1016/j.ydbio.2012.01.026. [DOI] [PubMed] [Google Scholar]