Abstract
Neutrophil-activating protein (NAP) is a major virulence factor expressed by Helicobacter pylori isolates associated with severe chronic gastroduodenal inflammation and peptic ulcers. NAP is one of the main protective antigens and a target for vaccine development against Helicobacter infection. In addition, NAP is a potent immune stimulator with potential application as a general vaccine adjuvant and in treatment of allergic diseases and cancer immunotherapy. NAP-specific immunoassays are needed for both H. pylori diagnostics and characterization of NAP-based vaccines and immunomodulatory preparations. We generated a panel of NAP-specific monoclonal antibodies (MAbs) by immunization of BALB/c mice with synthetic NAP peptides. The antibody reactivity against recombinant or native NAP antigen was characterized by enzyme-linked immunosorbent assay (ELISA), immunoblotting and immunofluorescence. A sensitive capture ELISA was developed using MAbs 23C8 and 16F4 (directed against different NAP epitopes) for detection of native or measles virus (MV) vector-expressed recombinant NAP in a concentration range of 4 ng/ml to 2000 ng/ml. MAb 23C8 antigen-binding depends on Tyr101 in a variable amino acid sequence of the NAP molecule, indicating the existence of antigenic variants among H. pylori strains. MAb 16F4 reacted with NAP from different H. pylori strains and was a sensitive tool for detection of small amounts of isolated NAP antigen or whole bacteria by immunoblotting or immunofluorescence. In conclusion, MAb-based immunoassays are highly specific and sensitive for detection of native NAP antigen and recombinant NAP immunostimulatory transgenes expressed by replication competent virus vectors.
Keywords: Monoclonal antibody, Immunoassays, Helicobacter pylori, NAP
1. Introduction
Helicobacter pylori neutrophil-activating protein (NAP) is a key virulence factor responsible for recruitment and activation of immune cells and induction of a robust chronic inflammatory reaction in gastroduodenal mucosa (D'Elios et al., 2007a; Evans et al., 1995; Montecucco and de Bernard, 2003).
NAP is a small (144 amino acid) iron-binding protein structurally similar to the DNA protecting (Dps) proteins described in Enterobacteriaceae (Tonello et al., 1999). NAP released by mucosa colonizing bacteria is transported via transcytosis at the luminal surface of endothelial cells, where it attracts neutrophils and mononuclear cells and triggers robust production of reactive oxygen species and secretion of pro-inflammatory cytokines (D'Elios et al., 2007a; Kottakis et al., 2009; Polenghi et al., 2007; Wang et al., 2008). NAP acts as a Toll-like receptor 2 (TLR-2) ligand and potent immunomodulator inducing strong interleukin 12 (IL-12) expression and Th1-biased polarization of the immune response (Amedei et al., 2006). NAP has been identified as one of the main H. pylori protective antigens (Satin et al., 2000) and it has been proposed as a component of recombinant protein vaccines for H. pylori immunoprophylaxis in humans (Malfertheiner et al., 2008). Recombinant NAP encoded by live attenuated viral vectors is highly immunogenic inducing both humoral and cellular immunity (Iankov et al., 2011). On the other hand, NAP is an attractive immune adjuvant and potent inducer of Th1 immunity with possible application in the treatment of allergic diseases and cancer immunotherapy. Co-administration of purified NAP can reverse the Th2 type immune response to ovalbumin and Trichinella spiralis allergens in animal models (Codolo et al., 2008; Del Prete et al., 2008). Recent reports demonstrated that local treatment with recombinant NAP significantly reduced tumor burden and tumor vascularization in an animal model of bladder cancer (Codolo et al., 2012). Another major advantage is that NAP can be successfully inserted in vector platforms and expressed in biologically active form by vector-transduced mammalian cells. Our recent data demonstrated that an attenuated measles virus (MV) strain engineered to encode secretory NAP forms expressed large amount of the NAP transgene and did not negatively impact viral propagation in vitro and development of anti-measles immunity in vaccinated animals (Iankov et al., 2011). We confirmed that NAP secreted by infected cells was biologically active and capable of inducing inflammatory cytokine production by monocytic cells. These results suggest that vector encoded NAP is a powerful immunomodulator that potentially can enhance the immunogenicity of vaccines and augment the therapeutic effect of oncolytic viruses in cancer therapy.
Characterization of recombinant NAP vaccines or replication competent vectors engineered to express NAP requires precise measurement of NAP concentration in the vaccine preparations or by vector-transduced cells in vitro and in vivo. Here, we present the generation of a panel of monoclonal antibodies (MAbs) against synthetic NAP peptides and their application in diagnostic immunological assays for detection of both bacteria-derived and vector-expressed NAP antigen. Capture ELISA, immunoblotting and immunofluorescent test developed with these antibodies demonstrated high sensitivity in detection of recombinant NAP. These assays can have an important application in characterization and quality control of H. pylori vaccines, NAP-containing immunomodulatory preparations and recombinant viral or bacterial vectors encoding NAP as therapeutic transgene.
2. Materials and methods
2.1. Recombinant NAP, H. pylori strains, MV strains encoding secretory NAP
Recombinant 6-histitidine-tagged NAP from H. pylori strains 26695 (NAP-26695) and 43504 (NAP-43504) was produced in Escherichia coli BL21 Star (DE3) cells (Invitrogen) and purified using Ni-NTA 6-his-tagged protein purification system (Qiagen) as described previously (Iankov et al., 2011). Purity of the recombinant NAP was verified by sodium-dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent SimplyBlue SafeStain staining (Invitrogen). Protein concentration was determined using a BCA protein assay kit (Pierce). Fresh bacterial culture of H. pylori strain 26695 (from ATCC) propagated on solid media was used for immunoblotting and IFT.
Engineering and characterization of MV strains expressing secretory NAP forms have been recently reported (Iankov et al., 2011). In the MV-lambda-NAP construct H. pylori NAP from strain 26695 was inserted in the human lambda immunoglobulin light chain replacing the variable domain of the molecule. MV-s-NAP is a truncated variant of the lambda-NAP protein and encodes the N-terminus with the immunoglobulin leader peptide but without the constant domain of lambda chain (see Suppl. Fig. 1). The presence of immunoglobulin leader peptide facilitates extracellular secretion of the chimeric NAP proteins by mammalian cells. MV expressing green fluorescent protein (MV-GFP) was used as control virus strain (Duprex et al., 1999).
2.2. Cell culture and MV propagation
African green monkey kidney Vero cells (ATCC) were grown in DMEM cell culture medium (HyClone) supplemented with 10% fetal bovine serum (FBS) and antibiotics (Invitrogen). NAP-expressing recombinant MV strains and MV-GFP were grown and viral stocks were titrated on Vero cells as previously described (Duprex et al., 2000; Iankov et al., 2011).
2.3. Production of NAP-specific MAbs
Synthetic peptides corresponding to amino acid residues 70–90 and 97–119 of H. pylori 26695 NAP molecule conjugated to keyhole limpet hemocyanin (KLH) were used for immunization of BALB/c mice. Peptide sequences were designed and synthesized in the peptide synthesis facility of the Proteomics Research Center, Mayo Clinic, Rochester, MN. Female 5–6-week old animals were immunized by intraperitoneal injection of 50 μg of the NAP-70–90 or NAP-97–119 peptide conjugates mixed 1/1 with Inject Alum adjuvant (Pierce). Mice with the highest serum titer in antigen-mediated ELISA were boosted intravenously with 25 μg of the corresponding antigen and 72 h later spleen cells were collected and fused with myeloma line Sp2/0-Ag14 (ATCC) using polyethylene glycol 4000 (EMD Chemicals) as previously described (Campbell, 1991; Köhler and Milstein, 1975). Hybridomas were grown in 24-well plates and on day 14 were screened for specific reactivity against homologous NAP antigen in antigen-mediated ELISA. Positive (for NAP-specific MAbs) wells were cloned from a single cell in 96-well plates in the presence of 5 ng/ml human recombinant IL-6 (Novus Biologicals). Hybridoma clones were further characterized for specific reactivity against NAP antigen in antigen-mediated ELISA and immunoblotting. Immunoglobulin isotype of NAP-specific MAbs was determined by IsoStrip Monoclonal Antibody Isotyping kit (Santa Cruz Biotechnology).
Characterization of MAb 16F4 generated using bacterial cell derived whole NAP molecule has been reported recently (Iankov et al., 2011).
2.4. Purification of MAbs
Exponentially grown hybridoma cells were washed and resuspended in serum-free culture medium Hybridoma-SFM medium (Invitrogen) and cultured for 72–96 h. Supernatants were harvested and MAbs were purified by affinity chromatography on Protein G columns according to the manufacturer's protocol (Pierce). MAbs were dialyzed overnight against PBS and protein concentration was measured using a BCA kit (Pierce). Purified MAbs were biotinylated or directly conjugated to horse-radish peroxidase (HRP) using Lightning-Link conjugation kits following the manufacturer's protocol (Innova Biosciences, UK).
2.5. Antigen-mediated ELISA
ELISA plates (Nunc) were coated overnight with 0.3 μg/well of purified NAP antigen in carbonate–bicarbonate buffer (CBB) pH 9.6. Plates were blocked with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 2 h at room temperature. Serum samples from the immunized BALB/c mice, hybridoma culture supernatants and purified MAbs were diluted in PBS containing 0.05% Tween 20 (PBS/T) and 1% BSA was added to the wells and incubated for 1–2 h at room temperature. Plates were washed 3 times in PBS/T and incubated for 1 h with a secondary anti-mouse polyvalent immunoglobulin (G, A, M) HRP-conjugated antibody (Sigma) diluted 1/2000 in 1% BSA in PBS/T. TMB peroxidase substrate (Bethyl Laboratories) was used for enzyme reaction development.
Antigen-mediated ELISA for epitope mapping of MAb 23C8 was performed using ELISA plates coated with KLH-conjugated NAP-97–119 synthetic peptides that correspond to the sequence of different H. pylori strains.
2.6. Inhibition ELISA
In order to determine a possible cross-reactivity and concurrence for epitope binding between MAb 16F4 and the panel of MAbs against NAP-97–119 amino acid residues, recombinant NAP-26695-coated plates were pre-incubated for 1 h with 10 μg/ml of either MAb 16F4 or MAb 23C8. Next, HRP-conjugated MAb 16F4 or MAb 23C8 (in 4-fold serial dilutions starting from 2 μg/ml) was added to the wells and plates were incubated for additional 1 h. The reaction was developed as described above. Wells pre-incubated with dilution of the same MAb (self-inhibition test) served as positive controls in the assay.
2.7. Quantitative capture ELISA
Quantitative sandwich ELISA for detection of NAP antigen was developed using MAb 16F4 as capturing antibody and HRP-conjugated MAb 23C8 as detection antibody. ELISA plates were coated with 0.2–2 μg/well of purified MAb 16F4 diluted in CBB. After overnight incubation at either 4 °C or room temperature, plates were blocked using 1% BSA (Sigma), 1% Casein (Pierce) or Synblock solution (Novus Biologicals). Serially diluted purified recombinant NAP-26695 or chimeric lambda-NAP protein from supernatants of MV-lambda-infected Vero cells was used as standards. Concentration of lambda-NAP in supernatants from MV-lambda-NAP-infected Vero cells was measured by human lambda immunoglobulin specific quantitative ELISA according to the manufacturer's protocol (Bethyl Laboratories). To optimize reaction conditions, the plates were incubated for different time periods (from 1 h at room temperature to overnight at 4 °C) with dilutions of recombinant NAP or lambda-NAP standards. Secondary HRP-conjugated MAb 23C8 (diluted in 1% BSA in PBS/T) was added at a concentration of ~1 μg/ml for 1–4-h incubation. Reaction was developed as described for antigen-mediated ELISA. Detection sensitivity was determined based on the highest dilution of the standards with an absorbance reading significantly different (mean±4×SD) than the wells incubated without (standard=0 ng/ml) NAP antigen.
To evaluate the effect of serum on the assay accuracy we employed a spike recovery sample test. Briefly, a known concentration of lambda-NAP was added (spiked) to human AB blood group serum (from Sigma) or serum from BALB/c mice. Samples were then diluted 1/4 (final dilution of the serum) and run using lambda-NAP standard dilutions for calculation of the concentration.
2.8. SDS-PAGE and immunoblotting
Diluted proteins or suspension of whole bacteria was mixed 1/1 vol./vol. with sample buffer (Bio-Rad) and resolved on 15% criterion gels using criterion electrophoresis system (Bio-Rad). Pre-stained marker (Bio-Rad) was used to show the molecular weight (MW) of the fractionated proteins. Resolved proteins were then blotted on PVDF membranes (Bio-Rad) using semi-dry transfer system (Bio-Rad). Membranes were blocked in 10% non-fat dry milk (Bio-Rad) in PBS/T. Hybridoma culture supernatants were diluted in 10% non-fat dry milk in PBS/T and incubated with the membranes for 2–4 h. Mouse specific polyvalent immunoglobulin (G, A, M) HRP conjugate (Sigma) was used as secondary antibody and reaction was visualized using chemiluminescent substrate (Pierce).
2.9. Indirect immunofluorescent test (IFT)
H. pylori strain 26695 resuspended in PBS was fixed on the microscope slides by incubation in methanol for 10 min. E. coli TOP10 cells (Invitrogen) used for cloning and amplification of the NAP-encoding plasmids were used as negative control. Slides were blocked in 10% FBS and incubated with biotin-conjugated MAbs for 1 h at room temperature. Slides were washed in PBS and incubated for 30 min with DyLight™594-streptavidin conjugate diluted 1/250 in PBS. Samples were washed 5× in PBS, counterstained with DAPI (Santa Cruz Biotechnology) to visualize all bacterial cells and examined by fluorescent microscopy using Axiovert 200M microscope (Zeiss).
3. Results
3.1. Generation of NAP-specific MAbs using synthetic peptides as immunogen
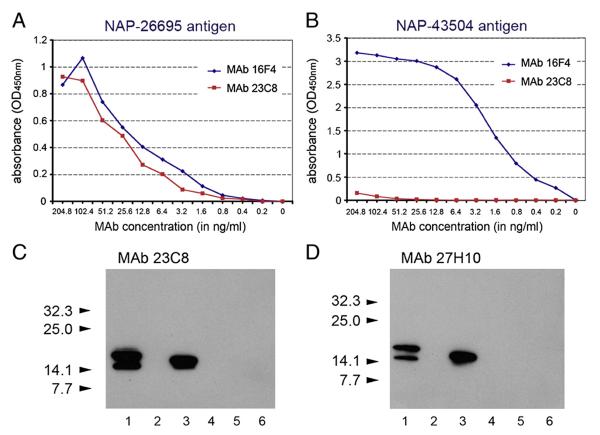
BALB/c mice developed a strong antibody response against the peptide corresponding to NAP-97–119 amino acid residues. Fusion between spleen cells isolated from the animal with the highest serum anti-NAP titer and Sp2/0-Ag14 myeloma gave rise to multiple NAP-specific hybridomas. Five hybridomas were selected, cloned from a single cell and further characterized in immunological assays. Clones 23C8 and 27H10 (both of IgG1 isotype) reacting strongly with the homologous NAP in ELISA and immunoblot (Table 1 and Fig. 1) were selected for affinity chromatography purification and development of NAP quantitative ELISA. MAb 23C8 reached high concentrations in serum-free supernatants and was purified on protein G column with yield of >80%. The final high purity preparations of the antibody had concentrations of 1–1.8 mg/ml. Purified preparations of MAb 16F4 (Iankov et al., 2011) had concentrations of approximately 700 μg/ml. Purity of both MAbs was confirmed by SDS-PAGE (not shown). In contrast to the previously characterized MAb 16F4, both MAb clone 23C8 and clone 27H10 reacted strongly with homologous NAP-26695 but not with NAP antigen from strain 43504 (see Table 1 and Fig. 1). These results indicate the existence of serological differences in antigenic determinants among the different H. pylori strains located between 97 and 119 amino acid residues of NAP molecule.
Table 1.
Reaction of MAbs 23C8 and 27H10 hybridoma supernatants in antigen-mediated ELISA against homologous (H. pylori strain 26695) and non-homologous (H. pylori strain 43504) NAP antigen.
| Supernatant |
MAb 23C8 |
MAb27H10 |
||
|---|---|---|---|---|
| Dilution | NAP-26695 | NAP-43504 | NAP-26695 | NAP-43504 |
| 1/20 | 1.389 | 0.044 | 1.256 | 0.039 |
| 1/80 | 0.934 | 0.025 | 0.278 | 0.050 |
| 1/320 | 0.376 | 0.052 | 0.106 | 0.043 |
Results are presented as absorbance reading (OD450 nm).
Fig. 1.
Characterization of MAbs against NAP-97–119 peptide. Comparison of NAP-97–119-specific MAb 23C8 and MAb 16F4 reactivity against NAP-26695 (A) and NAP-43504 (B) antigens in antigen-mediated ELISA. Purified MAbs with known protein concentration were incubated in 2-fold serial dilutions with the antigen. Results are presented as absorbance (OD450 nm). Both NAP-97–119-specific MAbs clone 23C8 and clone 27H10 reacted in immunoblotting (C and D) with homologous secretory NAP (H. pylori strain 26695) antigen from MV-s-NAP-infected Vero cells (lane 1) and E. coli expressed recombinant NAP-26695 antigen (lane 3) but not with non-homologous NAP-43504 recombinant antigen (lane 4). MV-GFP-infected Vero cells (lane 2) or protein extracts from E. coli BL21 Star (DE3) cells transformed with pET-28 empty expression plasmid were used as controls (lanes 5 and 6). Arrows indicate MW and position the pre-stained protein standards (C and D).
In contrast to NAP-97–119 peptide immunization, response in NAP-70–90-immunized mice was poor and we were not able to isolate hybridoma clones directed against this NAP protein sequence.
3.2. MAb 23C8 epitope mapping
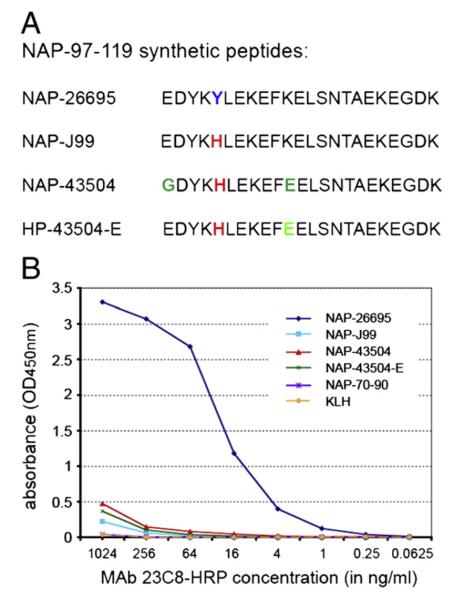
KLH-conjugated synthetic NAP-97–119 peptides containing variable residues observed among the NAP sequence of different strains were used for plate coating in antigen-mediated ELISA. Data analysis demonstrated that MAb 23C8 recognizes an epitope located within NAP-97–119 with specificity restricted to the NAP sequence of H. pylori strain 26695 (Fig. 2). Synthetic peptide combinations used in the assay showed that tyrosine in position 101 is critical for the formation of MAb 23C8 epitope (Fig. 2B). A single amino acid change Tyr→His alone in the NAP-97–119 peptide, corresponding to NAP from H. pylori strain J99, abolished the antibody binding. The same alteration at Tyr101 is present in strain 43504 and that can also explain the lack of reactivity of MAb 23C8 against NAP-43504 antigen.
Fig. 2.
Synthetic NAP-97–119 peptide sequences (A) and reactivity of purified and HRP-conjugated MAb 23C8 against KLH-conjugated NAP-97–119 peptides in antigen mediated ELISA (B). Synthetic peptides correspond to NAP-97–119 sequence of H. pylori strains 26695 (NAP-26695), J99 (NAP-J99) and 43504 (NAP-43504). In NAP-43504-E glycine-97 is substituted with glutamic acid, such as in strains 26695 and J99. KLH and KLH-conjugated NAP-70–90 peptide were used as control antigens in antigen-mediated ELISA (B).
3.3. MAb 16F4 and 23C8 reacted with different NAP epitopes
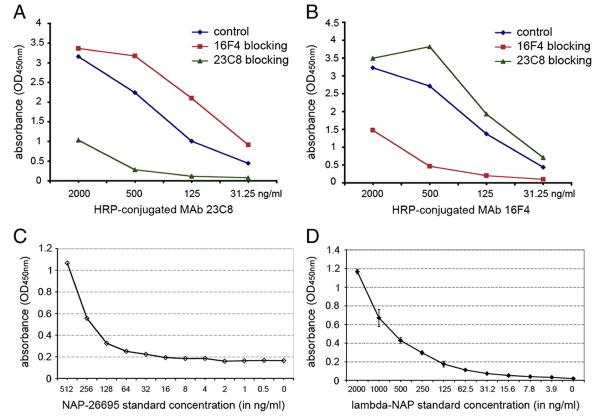
Antigen-mediated inhibition ELISA was used to test a possible cross-reactivity between the MAbs selected for development of NAP quantitative immunoassay. Pre-incubation of the NAP-26695-coated plates with MAb 16F4 had no any effect on MAb 23C8 reactivity with the antigen (Fig. 3A). The same results were obtained when MAb 23C8 was pre-incubated prior to reaction with HRP-conjugated MAb 16F4. MAb 23C8 did not inhibit MAb 16F4 binding to NAP (Fig. 3B). In contrast, pre-incubation with the same unconjugated MAb significantly inhibited reaction in concentration dependent way (Fig. 3A,B). This data indicated that the MAb 16F4 and MAb 23C8 recognize distinct NAP epitopes without interference for binding.
Fig. 3.
Inhibition ELISA demonstrated the absence of cross-reactivity between MAb 23C8 and MAb 16F4. Pre-incubation with different concentration of MAb 16F4 did not inhibit HRP-conjugated MAb 23C8 binding to NAP-26695 antigen (A). Similar results were obtained after pre-incubation with MAb 23C8 and HRP-conjugated MAb 16F4 (B). In contrast HRP-conjugated antibody binding was inhibited when the same MAb was used for pre-incubation. Standard curves from representative capture ELISA tests using bacterial derived recombinant NAP-26695 (C) or lambda-NAP from MV-lambda-NAP-infected Vero cells (D) as assay standards, demonstrate the higher sensitivity of the assay variant with mammalian cell expressed lambda-NAP standard.
3.4. Quantitative capture ELISA
Two MAbs — clones 16F4 and 23C8 reacting with different NAP epitopes were selected for development of quantitative ELISA methods for detection of recombinant NAP or NAP-“tagged” antigens. The lack of concurrence between the two antibodies for antigen-binding (described above, see Fig. 3A,B) is important for the assay sensitivity. Different concentrations of the coating antibody, blocking reagents and incubation periods were explored in order to achieve optimal detection of NAP (data not shown). Both bacterial expression system-derived recombinant NAP and mammalian cell-expressed chimeric lambda-NAP proteins were used as standards. The combination of MAb 16F4 as a capture and HRP-conjugated MAb 23C8 as detection antibody had the highest sensitivity and lowest background of all conditions explored. The optimal blocking effect was achieved using 1% BSA or Synblock blocking reagent. The ELISA sensitivity using bacteria-derived recombinant NAP-26695 standard was determined to be 16–64 ng/ml based on standard curves obtained in different experiments (Fig. 3C). The assay variant with mammalian cell-expressed lambda-NAP standard showed higher sensitivity (3.9 ng/ml) and lower background using 1% BSA as blocking solution (Fig. 3D). The lambda-NAP supernatant standard was produced by MV-lambda-NAP infected Vero cells and the concentration was determined using lambda chain specific ELISA. Stability of the lambda-NAP standard was checked after repeated freeze–thaw and storage at −20 °C. Recombinant NAP from the bacterial expression system tended to form aggregates after thawing and required careful pre-warming at 37 °C and vortexing before use. Repeated freeze–thaw affected its stability causing formation of larger aggregates that resulted in lower sensitivity (data not shown). In contrast, the lambda-NAP standard demonstrated excellent stability after freeze–thaw and storage at −20 °C. In addition, the lambda-NAP standard concentration can be measured and controlled by lambda-specific ELISA. These results illustrated the significant advantage of employing a chimeric lambda-NAP standard, as compared to bacterial expression system, in the development of sensitive and reproducible immunoassays for precise NAP measurement.
NAP has been proposed as an immunoadjuvant and a vector-encoded immunomodulatory transgene (D'Elios et al., 2007b; Iankov et al., 2011). Thus, detection of NAP in biological fluids is an important consideration. We evaluated the possible inhibitory effect of serum on the quantitative ELISA accuracy. Known amounts of chimeric lambda-NAP were mixed with serum and concentration was measured based on the lambda-NAP standard curve. Correction coefficient (CC) to obtain the actual NAP concentration in the sample was calculated using the formula: CC=(known concentration)/(measured concentration). Results indicated that both human and mouse serum at dilution of 1/4 affect the precise measurement of lambda-NAP. CC for human and mouse serum in this experiment were calculated to be 1.88 and 2.23 respectively. Thus, the accurate calculation of the actual NAP concentration in biological specimens requires spike recovery test for each sample.
3.5. Diagnostic evaluation of the MAbs in immunoblotting and IFT
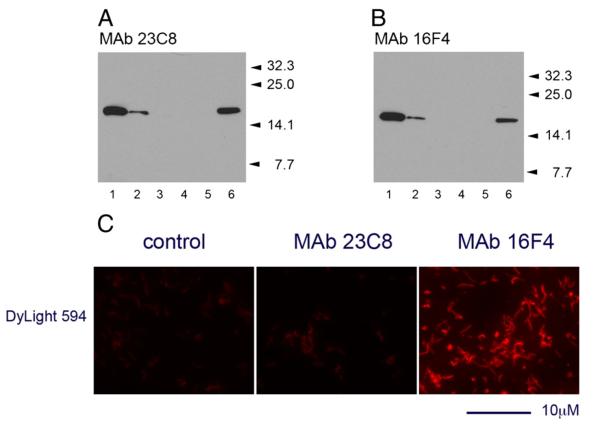
Immunoblotting analyses demonstrated the high sensitivity of MAbs in detection of NAP antigen in whole bacterial cell lysates (5–10×106 cells per lane loaded on the gel). Both MAb 23C8 and MAb 16F4 reacted with a single protein band corresponding to H. pylori NAP antigen (Fig. 4A,B).
Fig. 4.
Reaction of MAb 23C8 (A) and MAb 16F4 (B) against H. pylori strain 26695 whole bacteria in immunoblotting. Approximately 107 bacteria undiluted (lane 1), diluted 1/4 (lane 2) or 1/16 (lane 3) were mixed 1/1 with sample buffer and loaded on criterion gel. Undiluted (lane 4) or 1/4 diluted (lane 5) E. coli suspension was used as control. Purified recombinant NAP-26695 (40 ng/lane) was used as positive control for the reaction (lane 6). Positions of the MW standards are indicated by arrows. In contrast to MAb 23C8 directed against NAP-97–119 peptide, broadly reactive MAb 16F4 reacted with the H. pylori strain 26695 in IFT (C). Incubation with streptavidin-DyLight594 conjugated alone was used as control. IFT analysis was performed using a 100× oil-immersion objective on Axiovert 200M fluorescent microscope. The scale bar indicates 10 μm length.
The potential diagnostic application of the NAP-specific MAbs in detection of H. pylori directly in samples was evaluated in indirect IFT. Reaction was developed by incubation of the biotinylated MAbs with methanol-fixed bacteria in the primary step and DyLight594™-steptavidin as secondary reagent. Bright red-fluorescent bacterial cells were detected by fluorescent microscopy following incubation of the MAb 16F4 (Fig. 4C). Under the same conditions MAb 23C8 showed weak reactivity against the homologous H. pylori strain 26695. Fluorescent intensity was difficult to distinguish from that of control samples incubated with the secondary reagent alone, thus the test for MAb 23C8 was considered negative.
4. Discussion
H. pylori is a major cause of chronic gastritis and peptic ulcer in humans and is associated with increased risk of gastric cancer and primary mucosal lymphoma development. It is one of the highest prevalence pathogens, with approximately 50% of the human population being chronic H. pylori carriers (D'Elios and Andersen, 2007; Khalifa et al., 2010). Routine microbiology diagnostics includes invasive (requiring upper gastrointestinal tract endoscopy) and non-invasive techniques for detection of primary Helicobacter infection or post-treatment evaluation (Cirak et al., 2007; Guarner et al., 2010; Hino et al., 2004; Hooton et al., 2006; Perri et al., 2005). Bacterial culture or direct detection of the pathogen is used for invasively collected biological specimens. Non-invasive methods include detection of major virulence factors such as urease (13C-urea breath test), detection of H. pylori antigens in stool samples or serological tests for detection of specific antibody response (Adachi et al., 2002; Mégraud and Lehours, 2007).
There is no diagnostic test, however, for NAP antigen detection. NAP is one of the genes that determine the H. pylori virulence and studies showed that it is highly expressed by all clinical isolates (Momynaliev et al., 2005). Higher expression levels of NAP are associated with more severe H. pylori infection and serological tests confirm significantly higher rate of serum antibody response against NAP antigen in patients with peptic ulcers and gastric cancer (Long et al., 2009). Thus, detection of NAP antigen could be a valuable diagnostic marker in detection of H. pylori infection in humans. Proposed clinical application of NAP as potent immunoadjuvant, vaccine candidate and therapeutic transgene (Amedei et al., 2010; Codolo et al., 2012; Del Giudice et al., 2001; Iankov et al., 2011; Malfertheiner et al., 2008) also requires development of accurate quantitative assay for measurement of NAP concentrations.
We generated a panel of MAbs directed against the highly immunogenic synthetic NAP-97–119 peptide and evaluate their possible application as diagnostic tools. This fragment of NAP molecule carried strain specific variances in the protein sequence. It is of note that in contrast to MAb 16F4 raised against whole NAP (Iankov et al., 2011), all of the anti-NAP-97–119 MAbs showed a strain-restricted reactivity. Further investigation using a combination of synthetic NAP-97–119 variants revealed the existence of Tyr101-dependent antigenic specificity among H. pylori strains. Although NAP is highly conserved differences in the immunodominant epitopes of the protein can be involved in evasion of the protective antibodies and can have an important implication in serological diagnostics. The immunostimulatory property of NAP resides at the carboxy terminal region of the molecule (Kottakis et al., 2008). This was one of the reasons to target this sequence with MAbs. Homology 3D models of dodecameric NAP analysis revealed the positions of amino acids involved in iron coordination and localization of the surface domains of the molecule (Tonello et al., 1999; Zanotti et al., 2002). Additional modeling and structure/function experiments indicate that NAP-mediated neutrophil activation does not require dodecamer formation and two sequences (69–75 and 88–113) exposed on the molecule surface are recognized as potential immune receptor binding sites (Kottakis et al., 2008). Full-length NAP or only the portion carrying immunoadjuvant activity can be cloned in chimeric “NAP-tagged” proteins. Thus, MAbs specific for C-terminus of the NAP molecule may be useful reagents for detection of immunostimulatory transgenes expressing truncated NAP. NAP-97–119 peptide used for immunization and MAb 23C8 epitope overlap with one of the NAP molecule regions responsible for immune cell activation.
Based on their specificity and the absence of concurrence for epitope-binding, two MAbs: clone 23C8 (specific for NAP-97–119 sequence of H. pylori strain 26695) and clone 16F4 (Iankov et al., 2011) were selected for development of quantitative capture ELISA for detection of NAP antigen. Since MAbs are monoepitope specific, capture ELISA requires the capture and detection antibodies to recognize different antigenic determinants on the molecule. Although, a single antibody could be used in the case of repeated epitopes on molecule, such as polysaccharide antigens or protein antigens that forms oligomers. Naturally produced NAP forms dodecamers (Tonello et al., 1999; Zanotti et al., 2002). Characterization of MAb 16F4 showed that in capture ELISA it could detect as low as 1 ng/ml of bacterial-derived oligomeric NAP-43504 (MAb 16F4 homologous NAP antigen) but not monomeric NAP constructs, such as lambda-NAP (Iankov et al., 2011). However, further experiments with MAb 16F4 showed a variation in sensitivity, which depends on strain (homologous or non-homologous NAP), protein source (soluble or insoluble inclusion body fraction) and purification steps of the NAP standards. These data suggested that the development of highly sensitive and reproducible assays for detection of both monomeric and oligomeric NAP could be accomplished by a sandwich ELISA using MAbs that recognized distinct NAP epitopes. We tried to produce MAbs against another NAP determinant (NAP-70–90) but were unsuccessful because of the poor immune response against this peptide in BALB/c mice. Although the MAb 16F4 specific epitope is still unknown we found that it is distinct from the MAb 23C8 epitope. Thus, a capture ELISA was developed using MAb 16F4 as the capture antibody and MAb 23C8 as the detection antibody. The optimal blocking conditions, low background, assay reproducibility and sensitivity of less than 10 ng/ml were achieved with 1% BSA as blocker and mammalian cell-expressed lambda-NAP standards. The assay allows accurate measurement of NAP concentration in control of vaccine preparations with a recombinant NAP component as well as detection of vector-encoded NAP transgene expression. However, the presence of high serum concentrations can affect the reading accuracy and precise concentration calculation requires a correction factor determined by spike recovery tests for individual samples. Since MAb 23C8 has strain specific reactivity dependent upon Tyr101, this variant of sandwich ELISA is restricted to detection of NAP antigen with sequence similar to H. pylori 26695. We are currently working on generation of MAbs that react with His101 substituted NAP-97–119 peptide. This could allow detection of NAP antigen based on serological differences among the H. pylori strains.
Further characterization of NAP-specific MAbs confirmed that immunoblotting is a highly specific and sensitive immunoassay for detection of both bacterial cell-derived and vector-encoded NAP. All MAbs were able to detect nanogram quantities of the homologous antigen in samples loaded on the gel. In contrast to MAb 16F4, clones against NAP-97–119 sequence (similar to the ELISA results) showed strain specific reactivity, confirming the presence of Tyr101-dependent linear epitope on NAP molecule.
In addition to its potent pro-inflammatory effect NAP can act as adhesin promoting H. pylori attachment to the epithelial cell surface via interaction with sulfated carbohydrates on mucins, including Lewis blood group antigen (Namavar et al., 1998; Teneberg et al., 1997). NAP localized on bacterial surface can be detected by immunostaining methods without needing of a cell-permeabilization step. Indirect IFT using methanol-fixed bacteria demonstrated the diagnostic value of the anti-NAP MAb 16F4 in detection of the pathogen.
In conclusion, we developed a sensitive capture ELISA for detection of NAP antigen and vector-expressed recombinant NAP proteins based on the NAP-specific MAb clones 23C8 and 16F4. MAb 23C8 helped to identify previously unknown antigenic specificity among different H. pylori strains, strictly dependent on Tyr101 of NAP molecule. MAb 16F4, which reacts with NAP antigen from different strains, is a powerful diagnostic tool for detection of NAP antigen expression in isolated bacteria by immunoblotting or direct detection of H. pylori by immunofluorescence. NAP-specific immunoassays will be important in characterization and quantification of NAP antigen as a vaccine component or vector-expressed recombinant NAP transgene products.
Supplementary Material
Acknowledgments
We wish to thank Dr. D. McCormick and Mayo Proteomics Research Center, Mayo Clinic, Rochester MN for the help with design, synthesis and KLH conjugation of the NAP peptides.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jim.2012.06.010.
References
- Adachi K, Kawamura A, Ono M, Masuzaki K, Takashima T, Yuki M, Fujishiro H, Ishihara S, Kinoshita Y. Comparative evaluation of urine-based and other minimally invasive methods for the diagnosis of Helicobacter pylori infection. J. Gastroenterol. 2002;37:703. doi: 10.1007/s005350200115. [DOI] [PubMed] [Google Scholar]
- Amedei A, Cappon A, Codolo G, Cabrelle A, Polenghi A, Benagiano M, Tasca E, Azzurri A, D'Elios MM, Del Prete G, de Bernard M. The neutrophil-activating protein of Helicobacter pylori promotes Th1 immune responses. J. Clin. Invest. 2006;116:1092. doi: 10.1172/JCI27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amedei A, Codolo G, Del Prete G, de Bernard M, D'Elios MM. The effect of Helicobacter pylori on asthma and allergy. J. Asthma Allergy. 2010;3:139. doi: 10.2147/JAA.S8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AM. Laboratory Techniques in Biochemistry and Molecular Biology. vol. 23. Elsevier; Amsterdam: 1991. Monoclonal antibody and immunosensor technology. [Google Scholar]
- Cirak MY, Akyön Y, Mégraud F. Diagnosis of Helicobacter pylori. Helicobacter. 2007;12(Suppl. 1):4. doi: 10.1111/j.1523-5378.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- Codolo G, Mazzi P, Amedei A, Del Prete G, Berton G, D'Elios MM, de Bernard M. The neutrophil-activating protein of Helicobacter pylori down-modulates Th2 inflammation in ovalbumin-induced allergic asthma. Cell. Microbiol. 2008;10:2355. doi: 10.1111/j.1462-5822.2008.01217.x. [DOI] [PubMed] [Google Scholar]
- Codolo G, Fassan M, Munari F, Volpe A, Bassi P, Rugge M, Pagano F, D'Elios MM, de Bernard M. HP-NAP inhibits the growth of bladder cancer in mice by activating a cytotoxic Th1 response. Cancer Immunol. Immunother. 2012;61:31. doi: 10.1007/s00262-011-1087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giudice G, Covacci A, Telford JL, Montecucco C, Rappuoli R. The design of vaccines against Helicobacter pylori and their development. Annu. Rev. Immunol. 2001;19:523. doi: 10.1146/annurev.immunol.19.1.523. [DOI] [PubMed] [Google Scholar]
- Del Prete G, Chiumiento L, Amedei A, Piazza M, D'Elios MM, Codolo G, de Bernard M, Masetti M, Bruschi F. Immunosuppression of TH2 responses in Trichinella spiralis infection by Helicobacter pylori neutrophil-activating protein. J. Allergy Clin. Immunol. 2008;122:908. doi: 10.1016/j.jaci.2008.08.016. [DOI] [PubMed] [Google Scholar]
- D'Elios MM, Andersen LP. Helicobacter pylori inflammation, immunity, and vaccines. Helicobacter. 2007;12(Suppl. 1):15. doi: 10.1111/j.1523-5378.2007.00530.x. [DOI] [PubMed] [Google Scholar]
- D'Elios MM, Montecucco C, de Bernard M. VacA and HP-NAP, Ying and Yang of Helicobacter pylori-associated gastric inflammation. Clin. Chim. Acta. 2007a;381:32. doi: 10.1016/j.cca.2007.02.026. [DOI] [PubMed] [Google Scholar]
- D'Elios MM, Amedei A, Cappon A, Del Prete G, de Bernard M. The neutrophil-activating protein of Helicobacter pylori (HP-NAP) as an immune modulating agent. FEMS Immunol. Med. Microbiol. 2007b;50:157. doi: 10.1111/j.1574-695X.2007.00258.x. [DOI] [PubMed] [Google Scholar]
- Duprex WP, McQuaid S, Hangartner L, Billeter MA, Rima BK. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J. Virol. 1999;73:9568. doi: 10.1128/jvi.73.11.9568-9575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprex WP, McQuaid S, Roscic-Mrkic B, Cattaneo R, McCallister C, Rima BK. In vitro and in vivo infection of neural cells by a recombinant measles virus expressing enhanced green fluorescent protein. J. Virol. 2000;74:7972. doi: 10.1128/jvi.74.17.7972-7979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DJ, Jr., Evans DG, Takemura T, Nakano H, Lampert HC, Graham DY, Granger DN, Kvietys PR. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 1995;63:2213. doi: 10.1128/iai.63.6.2213-2220.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner J, Kalach N, Elitsur Y, Koletzko S. Helicobacter pylori diagnostic tests in children: review of the literature from 1999 to 2009. Eur. J. Pediatr. 2010;169:15. doi: 10.1007/s00431-009-1033-x. [DOI] [PubMed] [Google Scholar]
- Hino B, Eliakim R, Levine A, Sprecher H, Berkowitz D, Hartman C, Eshach-Adiv O, Shamir R. Comparison of invasive and non-invasive tests diagnosis and monitoring of Helicobacter pylori infection in children. J. Pediatr. Gastroenterol. Nutr. 2004;39:519. doi: 10.1097/00005176-200411000-00013. [DOI] [PubMed] [Google Scholar]
- Hooton C, Keohane J, Clair J, Azam M, O'Mahony S, Crosbie O, Lucey B. Comparison of three stool antigen assays with the 13C-urea breath test for the primary diagnosis of Helicobacter pylori infection and monitoring treatment outcome. Eur. J. Gastroenterol. Hepatol. 2006;18:595. doi: 10.1097/00042737-200606000-00004. [DOI] [PubMed] [Google Scholar]
- Iankov ID, Haralambieva IH, Galanis E. Immunogenicity of attenuated measles virus engineered to express Helicobacter pylori neutrophil-activating protein. Vaccine. 2011;29:1710. doi: 10.1016/j.vaccine.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa MM, Sharaf RR, Aziz RK. Helicobacter pylori: a poor man's gut pathogen? Gut Pathog. 2010;2:2. doi: 10.1186/1757-4749-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Kottakis F, Papadopoulos G, Pappa EV, Cordopatis P, Pentas S, Choli-Papadopoulou T. Helicobacter pylori neutrophil-activating protein activates neutrophils by its C-terminal region even without dodecamer formation, which is a prerequisite for DNA protection — novel approaches against Helicobacter pylori inflammation. FEBS J. 2008;275:302. doi: 10.1111/j.1742-4658.2007.06201.x. [DOI] [PubMed] [Google Scholar]
- Kottakis F, Befani C, Asiminas A, Kontou M, Koliakos G, Choli-Papadopoulou T. The C-terminal region of HPNAP activates neutrophils and promotes their adhesion to endothelial cells. Helicobacter. 2009;14:177. doi: 10.1111/j.1523-5378.2009.00678.x. [DOI] [PubMed] [Google Scholar]
- Long M, Luo J, Li Y, Zeng FY, Li M. Detection and evaluation of antibodies against neutrophil-activating protein of Helicobacter pylori in patients with gastric cancer. World J. Gastroenterol. 2009;15:2381. doi: 10.3748/wjg.15.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfertheiner P, Schultze V, Rosenkranz B, Kaufmann SH, Ulrichs T, Novicki D, Norelli F, Contorni M, Peppoloni S, Berti D, Tornese D, Ganju J, Palla E, Rappuoli R, Scharschmidt BF, Del Giudice G. Safety and immunogenicity of an intramuscular Helicobacter pylori vaccine in noninfected volunteers: a phase I study. Gastroenterology. 2008;135:787. doi: 10.1053/j.gastro.2008.05.054. [DOI] [PubMed] [Google Scholar]
- Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin. Microbiol. Rev. 2007;20:280. doi: 10.1128/CMR.00033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momynaliev KT, Rogov SI, Selezneva OV, Chelysheva VV, Akopian TA, Govorun VM. Comparative analysis of transcription profiles of Helicobacter pylori clinical isolates. Biochemistry (Mosc.) 2005;70:383. doi: 10.1007/s10541-005-0129-9. [DOI] [PubMed] [Google Scholar]
- Montecucco C, de Bernard M. Molecular and cellular mechanisms of action of the vacuolating cytotoxin (VacA) and neutrophil-activating protein (HP-NAP) virulence factors of Helicobacter pylori. Microbes Infect. 2003;5:715. doi: 10.1016/s1286-4579(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Namavar F, Sparrius M, Veerman EC, Appelmelk BJ, Vandenbroucke-Grauls CM. Neutrophil-activating protein mediates adhesion of Helicobacter pylori to sulfated carbohydrates on high-molecular-weight salivary mucin. Infect. Immun. 1998;66:444. doi: 10.1128/iai.66.2.444-447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri F, Quitadamo M, Ricciardi R, Piepoli A, Cotugno R, Gentile A, Pilotto A, Andriulli A. Comparison of a monoclonal antigen stool test (Hp StAR) with the 13C-urea breath test in monitoring Helicobacter pylori eradication therapy. World J. Gastroenterol. 2005;11:5878. doi: 10.3748/wjg.v11.i37.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polenghi A, Bossi F, Fischetti F, Durigutto P, Cabrelle A, Tamassia N, Cassatella MA, Montecucco C, Tedesco F, de Bernard M. The neutrophil-activating protein of Helicobacter pylori crosses endothelia to promote neutrophil adhesion in vivo. J. Immunol. 2007;178:1312. doi: 10.4049/jimmunol.178.3.1312. [DOI] [PubMed] [Google Scholar]
- Satin B, Del Giudice G, Della Bianca V, Dusi S, Laudanna C, Tonello F, Kelleher D, Rappuoli R, Montecucco C, Rossi F. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 2000;191:1467. doi: 10.1084/jem.191.9.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teneberg S, Miller-Podraza H, Lampert HC, Evans DJ, Jr., Evans DG, Danielsson D, Karlsson KA. Carbohydrate binding specificity of the neutrophil-activating protein of Helicobacter pylori. J. Biol. Chem. 1997;272:19067. doi: 10.1074/jbc.272.30.19067. [DOI] [PubMed] [Google Scholar]
- Tonello F, Dundon WG, Satin B, Molinari M, Tognon G, Grandi G, Del Giudice G, Rappuoli R, Montecucco C. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 1999;34:238. doi: 10.1046/j.1365-2958.1999.01584.x. [DOI] [PubMed] [Google Scholar]
- Wang CA, Liu YC, Du SY, Lin CW, Fu HW. Helicobacter pylori neutrophil-activating protein promotes myeloperoxidase release from human neutrophils. Biochem. Biophys. Res. Commun. 2008;377:52. doi: 10.1016/j.bbrc.2008.09.072. [DOI] [PubMed] [Google Scholar]
- Zanotti G, Papinutto E, Dundon W, Battistutta R, Seveso M, Giudice G, Rappuoli R, Montecucco C. Structure of the neutrophil-activating protein from Helicobacter pylori. J. Mol. Biol. 2002;323:125. doi: 10.1016/s0022-2836(02)00879-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.