Abstract
Background: Previously, we identified a sequence variant (N375S) of c-Met gene, however, its association with lung cancer risk and prognosis remain undefined.
Patients and Methods: We investigated the genotype distribution of the c-Met-N375S sequence variant in 206 lung cancer patients and 207 non-cancer controls in the Taiwanese population by DNA sequencing.
Results: Lung cancer patients with variant A/G and G/G genotypes showed 1.08-fold increased cancer risk when compared to patients with the wild-type A/A genotype (95% CI, 0.60-1.91). There were no significant differences in postoperative survival between c-Met-N375S and wild-type patients. In the cell model, the c-Met-N375S cells showed a decrease in cell death upon treatment with MET inhibitor SU11274 compared to wild-type cells.
Conclusion: Our data suggest that the c-Met-N375S sequence variant may not play a significant role in cancer susceptibility and the prognosis of lung cancer patients. The correlation with chemoresponse of c-Met-N375S is worth further investigation in patients receiving MET therapy.
Keywords: c-Met, N375S, cancer risk, prognosis, lung cancer.
Introduction
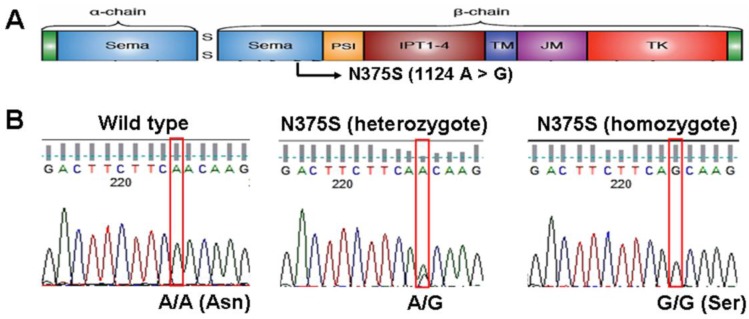
Over the past decade, lung cancer has become the leading cause of cancer death in worldwide. According to the new statistics, more than 150,000 people died from lung cancer in US in 2010 and more than 7,000 people died from lung cancer in Taiwan in 2010 1, 2. Uncontrolled activation of MET, the tyrosine kinase receptor for hepatocyte growth factor (HGF), is oncogenic and has been implicated in the growth, invasion and metastasis in a variety of tumors including lung cancer 3. The gene for c-Met, a transmembrane receptor tyrosine kinase, is located on chromosome 7q21-q31, and it is 120 kb in length with 21 exons and 20 introns 4. The primary transcript produces a 150 kDa polypeptide that is partially glycosylated to produce a 170 kDa precursor protein. The precursor protein is further cleaved to produce a 50 kDa α chain and a 140 kDa β chain, which are linked by disulfide bonds 5. The functional structures and domains of c-Met are shown in Figure 1A, including the sema (semaphorin domain), PSI (found in Plexins, Semaphorins, Integrins), four IPT repeats (found in Immunoglobulins, Plexins, Transcription factors), TM (transmembrane), JM (juxtamembrane) and TK (tyrosine kinase) domains.
Figure 1.
Sequence analysis reveals c-Met-N375S polymorphism in human NSCLC. (A): c-Met-N375S polymorphism site is illustrated schematically in the context of functional domains of Met. (B): The representative sequencing results of c-Met-N375S polymorphism are shown. Left: wild type (A/A). Middle: heterozygous change (A/G). Right: homozygous change (G/G). The nucleotide position of N375 (1124) was assessed with respect to full-length Met gene sequence (Ensemble transcript ID: ENST00000318493).
In our previous study, we found a sequence variant of c-Met (N375S, rs33917957) while screening for mutations in the exons coding domains of c-Met in tumor tissues from non-small cell lung cancer (NSCLC) from patients of East Asian, African-American, and Caucasian ethnicities 6. This c-Met-N375S sequence variant was also reported in a previous study by Tengs et al in 22 NSCLC patients and 24 healthy individuals, indicating that it is a germline polymorphism 7. N375S sequence variant was located in sema domain (1124A > G, Figure 1A) and occurred at a higher frequency in East Asians compared with Caucasians, and was absent in African Americans. Molecular modeling study suggested that replacement of asparagine at 375 by a serine resulted in the loss of a hydrogen bond between the altered residue and the adjacent arginine residue at 280, which may possibly disrupt HGF ligand binding 6. Therefore, an investigation of the genotype distribution of the c-Met-N375S polymorphism in more samples is important in order to understand the possible involvement of the c-Met-N375S polymorphism in NSCLC tumorigenesis. In addition, no association between the c-Met-N375S polymorphism and cancer risk and prognosis was reported. The purpose of this study, therefore, is to investigate the genotypic frequency of the c-Met-N375S polymorphism in NSCLC patients in Taiwan, and to examine the association of this polymorphism with lung cancer risk and prognosis.
Patients and Methods
Study population
A total of 206 patients with NSCLC who were admitted to China Medical University Hospital, Taichung, and Veterans General Hospital, Taichung and Taipei, Taiwan were recruited for this study after obtaining appropriate institutional review board permission and informed consent from the patients. The histological determinations, including tumor type and disease stage, were performed according to the World Health Organization classification and the TNM classification system, respectively. Information on the sex, age and smoking history of the patients were obtained from hospital records. Among this cohort, 177 patients had completed the follow-up of post-operative survival. The follow-up was performed at 2-month intervals in the first year after surgery and at 3-month intervals thereafter at out-patient clinics or by routine phone calls. The end of the follow-up period was September 2012. The mean follow-up period for all patients was 38 months (range 1-74 months). For the 101 patients who survived the follow-up period (censored patients), the mean follow-up time was 49 months. For the 76 patients who died during the follow-up period, the mean follow-up period was 24 months. The control group comprised of 207 unrelated, age-matched and sex-matched, cancer-free individuals recruited from Veterans General Hospital, Taichung. Informed consent from each participant was obtained before the study. Tumor typing and disease staging were performed according to the World Health Organization classification and the TNM classification system, respectively. Information on the sex, age, and smoking history of the patients were obtained from hospital records. Patients were categorized as non-smokers (never smokers and ex-smokers) and smokers (including regular smokers and continuously occasional smokers).
Sequence variant analysis
Blood samples (5-10 ml) were obtained and genomic DNA was extracted from the peripheral lymphocytes using standard methods. Purified genomic DNA was amplified by PCR using the primers: sense, 5'GCA TTC CTA CAT GGA AAT GCC TCT GGA GTG; antisense, 5'CCT ATT AAA GCA GTG CTC ATG ATT GGG TCC G. To determine Met genotype at N375, PCR-amplified DNA samples were examined by direct DNA sequencing using sense primer as sequencing primer.
Functional analysis of c-Met-N375S variant
c-Met-N375S mutant and its wild-type (WT) counterpart in pIRES2-EGFP vector were created as described previously 8, 9. The recombinant vector and empty vector were transfected into A549 lung cancer cells using TurboFectTM transfection reagent (Thermo Scientific Inc.). The transfected cells were treated with a MET-specific inhibitor SU11274 (5 μM, Sigma-Aldrich) and assayed for cell viability by trypan blue staining and cell counting at 48 h and 72 h post-treatment. Anti-phosphatidylserine antibody (#05-719, Millipore) was used for detection of apoptosis. Cells were then photographed under an OLYMPUS FV1000 confocal microscope. Western blot was performed to measure protein expression level using c-Met antibody (#3127, Cell Signaling).
Statistical analysis
The SPSS program (SPSS Inc. Headquarters Chicago, Illinois) was used for all statistical analysis. Statistical modeling with logistic regression was used to calculate the relative risk (odds ratio) of genotypes for the case/control study. ORs were expressed together with 95% confidence intervals (95% CIs). Multivariate logistic regression analysis was adjusted for age and sex. The Pearson X2 test was used to analyze the correlation between c-Met-N375S distribution and clinicopathological parameters in NSCLC patients. Survival curves were calculated according to the Kaplan-Meier method, and comparison was performed using the log-rank test.
Results
Effect of c-Met-N375S sequence variant on lung cancer risk
The genotype of c-Met-N375S was determined by direct DNA sequencing of blood samples from 206 Taiwanese NSCLC patients and 207 Taiwanese cancer-free individuals matched with age and sex distribution. Figure 1B shows the representative DNA sequencing data for c-Met-WT (A/A) or heterozygous (A/G) and homozygous (G/G) c-Met-N375S genotypes. Frequencies of the c-Met-N375S genotypes A/A, A/G and G/G were 85.5%, 14.0%, and 0.5%, respectively in Taiwanese cancer-free individuals; and 85.9%, 13.0%, and 1.1%, in Taiwanese NSCLC patients. Lung cancer patients with variant A/G and G/G genotypes showed a non-significant 1.08-fold increased risk of lung cancer when compared to individuals with the WT A/A genotype (95% CI, 0.60-1.91 by multiple logistic regression analysis) (Table 1). In addition, the distribution of c-Met-N375S genotypes in relation to patients' clinicopathological parameters was examined to assess the lung cancer susceptibility of the variant allele (A/G and G/G genotypes) (Table 2). Data analyzed using a different model of inheritance, i.e. G/G genotype vs. AG and AA genotypes did not show any significant difference in cancer risk or association with clinicopathological parameters. The results indicated that higher odds ratios for lung cancer were not seen for N375S A/G or G/G genotypes in case/control study and in the subgroup according to clinicopathological parameters of lung cancer patients.
Table 1.
Distribution of the c-Met-N375S genotypes in case/control studies justified by age, gender and lifestyle.
| Characteristics | Genotypes | Logistic regression model | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A/A | (%) | A/G | (%) | G/G | (%) | Total | Crude OR | (95% CI) a | Adjusted OR | (95% CI) b | |||||||||
| Cancer free controls | 177 | (85.5) | 29 | (14.0) | 1 | (0.5) | 207 | 1.00 | 1.00 | ||||||||||
| NSCLC patients | 177 | (85.9) | 27 | (13.0) | 2 | (1.1) | 206 | 0.97 | (0.56-1.68)0.90 | 1.08 | (0.60-1.91)0.80 | ||||||||
| Male | |||||||||||||||||||
| Controls | 104 | (85.2) | 18 | (14.8) | 0 | (0.0) | 122 | 1.00 | 1.00 | ||||||||||
| Patients | 105 | (85.4) | 17 | (13.8) | 1 | (0.8) | 123 | 1.00 | (0.49-2.04)1.00 | 1.03 | (0.48-2.17)0.93 | ||||||||
| Female | |||||||||||||||||||
| Controls | 73 | (85.9) | 11 | (12.9) | 1 | (1.2) | 85 | 1.00 | 1.00 | ||||||||||
| Patients | 72 | (86.7) | 10 | (12.0) | 1 | (1.2) | 83 | 0.93 | (0.38-2.25)0.87 | 1.18 | (0.46-2.97)0.72 | ||||||||
| Non-smoking | |||||||||||||||||||
| Controls | 72 | (86.7) | 10 | (12.0) | 1 | (1.2) | 83 | 1.00 | 1.00 | ||||||||||
| Patients | 54 | (85.7) | 8 | (12.7) | 1 | (1.6) | 63 | 1.11 | (0.42-2.87)0.83 | 1.15 | (0.43-3.01)0.77 | ||||||||
| Smoking | |||||||||||||||||||
| Controls | 105 | (84.7) | 19 | (15.3) | 0 | (0.0) | 124 | 1.00 | 1.00 | ||||||||||
| Patients | 93 | (85.3) | 15 | (13.8) | 1 | (0.9) | 109 | 1.04 | (0.50-2.14)0.91 | 1.04 | (0.50-2.15)0.91 | ||||||||
| Age<70 | |||||||||||||||||||
| Controls | 88 | (86.3) | 14 | (13.7) | 0 | (0.0) | 102 | 1.00 | 1.00 | ||||||||||
| Patients | 96 | (88.1) | 12 | (11.0) | 1 | (0.9) | 109 | 0.87 | (0.38-1.96)0.73 | 0.99 | (0.42-2.27)0.98 | ||||||||
| Age ≥70 | |||||||||||||||||||
| Controls | 89 | (84.8) | 15 | (14.3) | 1 | (1.0) | 105 | 1.00 | 1.00 | ||||||||||
| Patients | 81 | (83.5) | 15 | (15.5) | 1 | (1.0) | 97 | 1.19 | (0.55-2.55)0.66 | 1.19 | (0.52-2.66)0.67 | ||||||||
a. Odds ratios (ORs) were calculated to measure the association of the A/G+G/G genotype with lung cancer risk, with the A/A genotype being referred to as 1. CI: confidence interval. P values with significance are labeled in superscript. b. Adjusted for age, sex, and smoking; for Sex, adjusted for age and smoking; for Age, adjusted for sex and smoking; for Smoking, adjusted for sex and age.
Table 2.
Distribution of the c-Met-N375S genotypes in relation to clinicopathological parameters in NSCLC patients. a
| Characteristics | c-Met-N375S genotypes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | A/A | A/G+G/G c | |||||||
| n | N (%) | n | (%) | ||||||
| Overall b 206 | 177 | (85.9) | 29 | (14.1) | |||||
| Clinicopathological parameters | |||||||||
| Age | <70 | 109 | 96 | (88.1) | 13 | (11.9) | |||
| ≧70 | 97 | 81 | (83.5) | 16 | (16.5) | ||||
| Sex | Male | 123 | 105 | (85.4) | 18 | (14.6) | |||
| Female | 83 | 72 | (86.7) | 11 | (13.3) | ||||
| Smoker | Yes | 109 | 93 | (85.3) | 16 | (14.7) | |||
| No | 63 | 54 | (85.7) | 9 | (14.3) | ||||
| Tumor type d | |||||||||
| ADC | 133 | 117 | (88.0) | 16 | (12.0) | ||||
| SCC | 61 | 50 | (82.0) | 11 | (18.0) | ||||
| LCC | 12 | 10 | (83.3) | 2 | (16.7) | ||||
| Tumor stage | |||||||||
| I + II | 88 | 75 | (85.2) | 13 | (14.8) | ||||
| III + IV | 67 | 61 | (91.0) | 6 | (9.0) | ||||
a. The P value was analyzed by Pearson χ2 test and all P values were > 0.05. b. Total number of sample in some categories is less than the overall number analyzed because clinical data were not available for these patients. c. The risk genotype was defined as A/G+G/G genotypes. d. ADC: adenocarcinoma, SCC, squamous cell carcinoma, LCC, large cell carcinoma.
Effect of c-Met-N375S polymorphism on prognosis of lung cancer patients
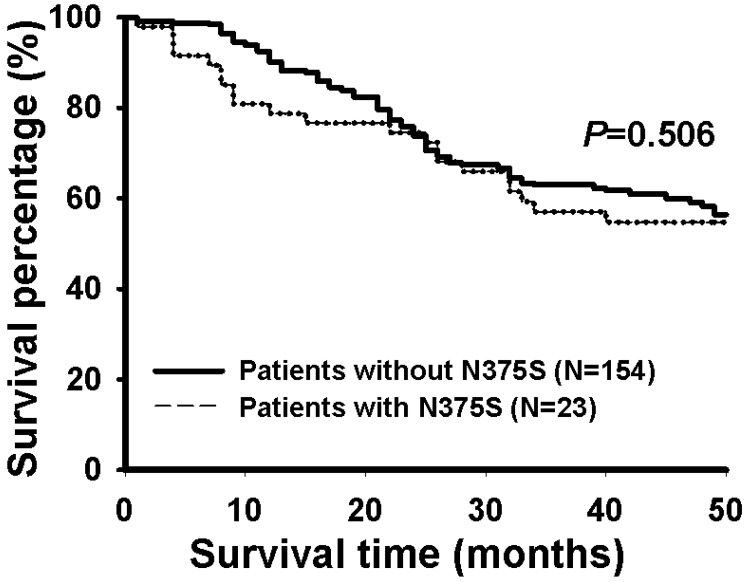
Survival was assessed using the Kaplan-Meier method by comparing patients with WT and N375S polymorphism of Met gene in 177 Taiwanese NSCLC patients whose survival data were available. The data indicated that there were no significant differences in post-operative survival between these two groups of patients (P=0.506, Figure 2).
Fig 2.
The Kaplan-Meier survival curves for patients with or without N375S polymorphism of the c-Met gene in lung cancer patients. Patients with N375S polymorphism of the c-Met gene showed similar post-operative survival rate as patients without N375S polymorphism. A total of 177 patients were analyzed.
c-Met-N375S variant and its effect on susceptibility to MET inhibitor SU11274
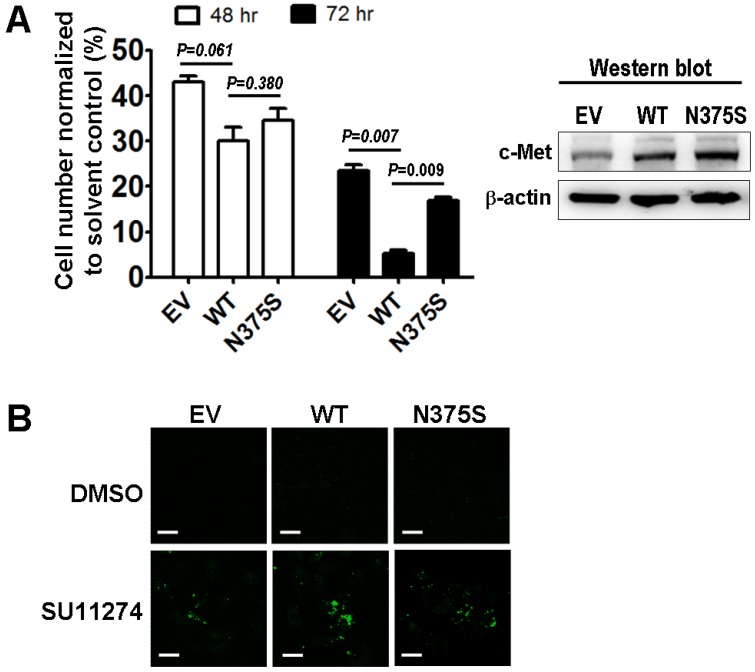
To understand the role of the c-Met-N375S sequence variant in modulating sensitivity to MET inhibitor, we measured the viability of A549 lung cancer cells transfected with the recombinant c-Met-WT and c-Met-N375S constructs after treatment with the preclinical MET inhibitor SU11274 8, 10, 11. The relative levels of c-Met-WT and c-Met-N375S protein expressed were comparable. However, t the c-Met-N375S cells showed a decrease in SU11274-induced cell death compared with cells that expressed c-Met-WT using cell viability and phosphatidylserine (PS) apoptosis assays (Figure 3).
Fig 3.
The functional significance of c-Met-N375S in response to SU11274. (A): Trypan blue exclusion assays of A549 lung cancer cells expressing empty vector (EV), Met-wild-type (WT) or Met-N375S (N375S) upon MET inhibitor SU11274 treatment was measured at 48 and 72 hours (left). Immunoblots of c-Met in cells expressing EV, WT, or N375S vector are shown (fight). (B): PS apoptosis assay was analyzed upon treatment with solvent control (DMSO) or SU11274 for 24 hours in A549 lung cancer cells expressing EV, WT, or N375S vector.
Discussion
Our results showed that c-Met-N375S sequence variant was not associated with lung cancer susceptibility and prognosis. The distribution of the c-Met-N375S genotypes was similar in both cases and controls (Table 1), and no significant association between the c-Met-N375S sequence variant and clinicopathological parameters (Table 2) and prognosis (Figure 2) of lung cancer was observed. Lack of association of c-Met-N375S genotypes with lung cancer susceptibility and prognosis is consistent with the observation by Tengs et al that c-Met-N375S may not represent a true cancer-causing genotype 7. Notably, the c-Met-N375S cells showed a decrease in SU11274-induced cell death compared with c-Met-WT cells (Figure 3).
The c-Met sema domain harbors the ligand binding site, and based on results from our and other laboratories, it seems to be a key region that is targeted for polymorphic mutations in lung cancer 8, 9. Our previous data suggested that the amount of HGF bound to c-Met-N375S was much lower when compared with c-Met-WT, thereby demonstrating a loss of affinity for HGF in the case of c-Met-N375S mutant 6. Our previous study also demonstrated that the COS-7 cell expressing c-Met-N375S mutation was less sensitive to specific MET kinase inhibitors such as SU11274 compared to cell expressing c-Met wild-type 12. The current study confirmed that the c-Met-N375S cells showed a decrease in cell death upon treatment of MET inhibitor SU11274 compared to wild-type cells in the A549 lung cancer cell model.
Since the NSCLC cohort recruited in this study did not receive MET therapy, the correlation with chemoresponse and prognostic effects of c-Met-N375S sequence variant needs to be further investigated in larger number of patients who receive MET therapy. In light of the reports of c-Met signaling as the target for lung cancer treatment 13-16, it is of interest to study whether c-Met-N375S cells are less sensitive to SU11275 in the presence of HGF. It is also important to determine the genotype of important sequence variants in addition to HGF expression in lung cancer response to MET therapy. Although more patients are needed to confirm the risk with a minor allele frequency of 0.074 such as c-Met-N375S sequence variant in populations with different ethnic backgrounds, our data provide the first clinical evidence of the effects of c-Met-N375S polymorphism on lung cancer risk and prognosis.
Acknowledgments
This work was supported in part by grant NSC99-2698-B-006-004-MY3 from the National Science Council, grant DOH98-TD-G-111-024 from the Department of Health (The Executive Yuan, Republic of China) and grant CMNCKU10002 from the Chi Mei Medical Center, Taiwan.
Biographies
Dr. Yi-Ching Wang studies the molecular mechanisms involved in lung tumorigenesis. Dr. Wang investigates the etiological association of alterations in several tumor suppressor genes, oncogenes, and DNA repair genes with lung tumorigenesis. The alteration analyses include the following aspects: gene mutation and polymorphism, gene loss, hypermethylation of promoter, chromatin structure alteration of gene locus, mRNA alteration, and protein expression alteration. More recently, Dr. Wang has continued to do research on cancer genomics and epigenomics such as genome-wide search of loss of heterozygosity, and genome-scanning approaches of DNA methylation and chromatin alteration profiles for identification of new genes critical to lung tumorigenesis. In addition, several potential anti-cancer drugs are developing in her laboratory. Dr. Wang has published more than 60 SCI papers on lung cancer including prestigious journals such as J Clin Oncol, J Clin Invest, and Cancer Res. Dr. Wang is one of the recipients of the Excellent Research Award of Taiwan National Science Council.
Dr. Jiunn-Min Shieh is a physician scientist with 10 years of research experience in pulmonary and critical care medicine. Dr Shieh is a pioneer in pulmonary ultrasound diagnostics and therapeutics. His research group has discovered novel genes and sequence variants that are associated with lung cancer pathogenesis and progression. Dr Shieh is currently the Director of the Department of Chest Medicine, Chi Mei Medical Center, Tainan, Taiwan.
References
- 1.Department of Health. Health and Vital Statistics, Republic of China. R. O. C. Press, Taipei, pp 86-108; 2011. The Executive Yuan RoC. General Health Statistics, 2010. [Google Scholar]
- 2.Jemal A, Siegel R, Xu J. et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Blumenschein GR Jr, Mills GB, Gonzalez-Angulo AM. Targeting the hepatocyte growth factor-cMET axis in cancer therapy. J Clin Oncol. 2012;30:3287–96. doi: 10.1200/JCO.2011.40.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cipriani NA, Abidoye OO, Vokes E. et al. MET as a target for treatment of chest tumors. Lung Cancer. 2009;63:169–79. doi: 10.1016/j.lungcan.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y. The human hepatocyte growth factor receptor gene: complete structural organization and promoter characterization. Gene. 1998;215:159–69. doi: 10.1016/s0378-1119(98)00264-9. [DOI] [PubMed] [Google Scholar]
- 6.Krishnaswamy S, Kanteti R, Duke-Cohan JS. et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res. 2009;15:5714–23. doi: 10.1158/1078-0432.CCR-09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tengs T, Lee JC, Paez JG. et al. A transforming MET mutation discovered in non-small cell lung cancer using microarray-based resequencing. Cancer Lett. 2006;239:227–33. doi: 10.1016/j.canlet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Ma PC, Jagadeeswaran R, Jagadeesh S. et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–88. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- 9.Ma PC, Kijima T, Maulik G. et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–81. [PubMed] [Google Scholar]
- 10.Dulak AM, Gubish CT, Stabile LP. et al. HGF-independent potentiation of EGFR action by c-Met. Oncogene. 2011;30:3625–35. doi: 10.1038/onc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence RE, Salgia R. MET molecular mechanisms and therapies in lung cancer. Cell Adh Migr. 2010;4:146–52. doi: 10.4161/cam.4.1.10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berthou S, Aebersold DM, Schmidt LS. et al. The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants. Oncogene. 2004;23:5387–93. doi: 10.1038/sj.onc.1207691. [DOI] [PubMed] [Google Scholar]
- 13.Salgia R, Hensing T, Campbell N. et al. Personalized treatment of lung cancer. Semin Oncol. 2011;38:274–83. doi: 10.1053/j.seminoncol.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Stabile LP, Rothstein ME, Keohavong P. et al. Targeting of Both the c-Met and EGFR Pathways Results in Additive Inhibition of Lung Tumorigenesis in Transgenic Mice. Cancers (Basel) 2010;2:2153–70. doi: 10.3390/cancers2042153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Du R, Jiang S. et al. Dual MET-EGFR combinatorial inhibition against T790M-EGFR-mediated erlotinib-resistant lung cancer. Br J Cancer. 2008;99:911–22. doi: 10.1038/sj.bjc.6604559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap TA, Olmos D, Brunetto AT. et al. Phase I trial of a selective c-MET inhibitor ARQ 197 incorporating proof of mechanism pharmacodynamic studies. J Clin Oncol. 2011;29:1271–9. doi: 10.1200/JCO.2010.31.0367. [DOI] [PubMed] [Google Scholar]