Abstract
Background: Colorectal cancer metastasis is a multistep process involving degradation of extracellular matrix components by proteolytic enzymes. Among them, matrix metalloproteinases (MMPs) are the principal degrading enzymes and their expressions/activities are also correlated with survival. Much research has showed the associations between genetic polymorphisms in MMPs and risk of colorectal cancer; however, their prognostic significance has not been well determined.
Methods: We selected and genotyped 4 cancer-associated single nucleotide polymorphisms (SNPs) in a cohort of 282 colorectal cancer patients. The associations of these SNPs with distant metastasis-free survival and overall survival were evaluated by Kaplan-Meier analysis, Cox regression model, and survival tree analysis.
Results: The relative risks of developing distant metastasis after curative surgery were higher in individuals with minor homozygote AA genotype than in those with GG/GA genotypes at MMP2 rs243866 (P = 0.012). Survival tree analysis also identified a higher-order genetic interaction profile consisting of MMP2 rs243866 and MMP2 rs2285053 that was significantly associated with distant metastasis-free survival (Ptrend = 0.016). After adjusting for possible confounders, the genetic interaction profile remained significant (Ptrend = 0.050).
Conclusions: These results suggest that genetic variations in the MMP2 might be potential predictors of distant metastasis-free survival after curative surgery.
Keywords: colorectal cancer, matrix metalloproteinases, single nucleotide polymorphism, metastasis, survival.
Introduction
Colorectal cancer remains one of the leading causes of cancer death worldwide and in Taiwan. The 5-year survival rates are approximately 90% for early stage colorectal cancer patients, but decreasing to less than 10% in patients with distant metastases 1. In addition, about 25 to 40 percent of patients who undergo curative surgery for localized colorectal cancer eventually recur and develop metastases, but current clinicopathologic indicators do not predict the outcomes 2. Therefore, there is a need to identify and select appropriate adjuvant therapy for patients at high risk in development of metastases.
Cancer metastasis is a complex multifactorial and multistep process, involving invasion of stroma, intravasation into the circulation from the primary site, extravasation at the secondary site, and outgrowth of new tumors. These processes require degradation of the extracellular matrix components by proteolytic enzymes. The most important proteinases capable of creating a pathway for tumor cell migration are the matrix metalloproteinases (MMPs). One particular group of MMPs, the gelatinases A and B also known as MMP2 and MMP9, play a crucial role during colorectal cancer invasion and metastasis. The levels of gene expression and enzymatic activity of MMP2 and MMP9 were higher in tumors compared with normal mucosa, and their overexpression is associated with a poor prognosis in colorectal cancer patients 3-5.
Lots of molecular epidemiological studies have focused on the associations between MMP polymorphisms and the risk of colorectal cancer 6-10, but little is known about their relationship to cancer prognosis. Thus, the aim of the present study was to assess the association of MMP2 and MMP9 single-nucleotide polymorphisms (SNPs) with distant metastasis-free and overall survival in a cohort of 282 colorectal cancer patients.
Materials and Methods
Patient recruitment and data collection
Two hundred and eighty-two colorectal cancer patients were retrospectively examined the records in the China Medical University Hospital, Taiwan between 2001 and 2007. All cases were histopathologically confirmed, and the clinical data and outcomes were obtained from patients' clinical records and pathological reports. Among patients receiving curative surgery (stage I-III, n = 233), distant metastasis-free survival was defined as the time from surgery to the date of distant metastases or when censored at the latest date. Overall survival was defined as the time from diagnosis (n = 282) to the date of death from any cause or when censored at the latest date if patients were still alive. The survival data were updated most recently in 2010. This study was approved by the Institutional Review Board of the China Medical University Hospital and written informed consent was obtained from all patients.
SNP selection and genotyping
Four SNPs in MMP2 and MMP9 were selected based on the evidence of association with cancers 11-13. The MMP2 rs243866 (-1575) located immediately 5' to the potential estrogen receptor binding site and functioned as an enhancer 14. The MMP2 rs2285053 (-735) have been shown to affect MMP2 expression by lowering transcriptional activity 15, 16. Several studies have evaluated the association of MMP9 rs3787268 and rs17577 with cancer susceptibility, such as breast 12, prostate 13, and colorectal 17 cancers, but their clinical impact on cancer prognosis and outcomes has not been well evaluated. The MMP9 rs17577 (rs2274756, Arg668Gln) is in the hemopexin-like domain and probably functions in substrate binding, since removal of this domain disables the cleavage of collagen 18.
Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) and stored at -80°C until the time of study. Genotyping was performed as described previously 19-24 using Sequenom iPLEX matrix-assisted laser desorption/ionization-time of flight mass spectrometry technology at the National Center for Genome Medicine, Academia Sinica, Taiwan. The SNPs were all in Hardy-Weinberg equilibrium (P > 0.05), and the average genotype call rate for these SNPs was 99.5%.
Statistical analysis
Patient clinicopathologic characteristics were summarized as number and percentage of patients or median and interquartile range (IQR) of values. Age was dichotomized at the median value within the cohort. Carcinoembryonic antigen (CEA) level was dichotomized at 5 μg/L because of its correlation with an increasing stage of the colorectal cancer 25. The associations of 4 MMP SNPs and clinical characteristics with distant metastasis-free and overall survival were assessed using Cox proportional hazards model and Kaplan-Meier analysis with log-rank test. We tested different genetic models, dominant, recessive, and additive models for each SNP. The model with the most significant P value was considered the best-fitting model. Higher order SNP-SNP interactions were evaluated using survival tree analysis by STREE software (http://c2s2.yale.edu/software/stree/), which uses recursive partitioning to identify subgroups of individuals with similar risk 26. Patients were then categorized into low-, medium-, and high-risk groups based on the survival tree analysis. Multivariate analyses to determine the interdependency of genotypes and other known prognostic factors, such as age at diagnosis, gender, CEA levels, tumor differentiation, stage, lymphovascular invasion, perineural invasion, and lymph node involvement, were carried out using Cox proportional hazards model. Statistical Package for the Social Sciences software version 19.0.0 (IBM, Armonk, NY) was used for other statistical analyses. A two-sided P value of ≤ 0.05 was considered statistically significant.
Results
DNA was extracted for analysis from the blood samples of 282 colorectal cancer patients. Of the 233 patients receiving curative surgery (stage I-III), 30 (12.9%) developed distant metastasis during the median follow-up of 54.5 months (Table 1). Among the 282 patients diagnosed with colorectal cancer, 92 (32.6%) died after a median follow-up of 50.0 months. Stage, lymphovascular invasion, and lymph node involvement were significantly associated with both distant metastasis-free and overall survival. Gender, CEA levels, tumor differentiation, and perineural invasion were only associated with overall survival, and age was only associated with distant metastasis-free survival.
Table 1.
Demographic and clinical characteristics of colorectal cancer patients.
| Characteristics | Distant metastasis-free survival | Overall survival | ||||||
|---|---|---|---|---|---|---|---|---|
| n (%)* | n of events* | HR (95% CI) | P | n (%)* | n of events* | HR (95% CI) | P | |
| All patients | 233 | 30 | 282 | 92 | ||||
| Age, years | ||||||||
| Median (IQR) | 66 (56-73) | 65 (54-73) | ||||||
| <65 | 106 (45.5) | 19 | 1.00 | 133 (47.2) | 42 | 1.00 | ||
| ≥65 | 127 (54.5) | 11 | 0.48 (0.23-1.00) | 0.049 | 149 (52.8) | 50 | 1.09 (0.72-1.64) | 0.692 |
| Gender | ||||||||
| Male | 119 (51.1) | 17 | 1.00 | 148 (52.5) | 57 | 1.00 | ||
| Female | 114 (48.9) | 13 | 0.78 (0.38-1.61) | 0.508 | 134 (47.5) | 35 | 0.65 (0.43-0.99) | 0.045 |
| CEA, μg/L | ||||||||
| Median (IQR) | 3.5 (1.7-10.2) | 4.4 (2.0-14.9) | ||||||
| <5 | 129 (59.7) | 14 | 1.00 | 136 (52.3) | 23 | 1.00 | ||
| ≥5 | 87 (40.3) | 15 | 1.61 (0.78-3.34) | 0.198 | 124 (47.7) | 58 | 3.35 (2.07-5.44) | <0.001 |
| Differentiation | ||||||||
| Well | 43 (20.0) | 5 | 1.00 | 51 (19.4) | 11 | 1.00 | ||
| Moderate | 153 (70.8) | 17 | 0.95 (0.35-2.56) | 0.912 | 185 (70.3) | 62 | 1.64 (0.86-3.12) | 0.131 |
| Poor | 19 (9.2) | 6 | 3.17 (0.97-10.4) | 0.057 | 27 (10.3) | 15 | 3.59 (1.65-7.83) | 0.001 |
| Stage† | ||||||||
| I-II | 150 (64.7) | 12 | 1.00 | - | ||||
| III | 82 (35.3) | 18 | 2.92 (1.41-6.06) | 0.004 | - | |||
| I-III | - | 232 (82.6) | 54 | 1.00 | ||||
| IV | - | 49 (17.4) | 38 | 5.19 (3.40-7.92) | <0.001 | |||
| Lymphovascular invasion | ||||||||
| Negative | 179 (76.8) | 14 | 1.00 | 201 (71.3) | 48 | 1.00 | ||
| Positive | 54 (23.2) | 16 | 4.13 (2.02-8.47) | <0.001 | 81 (28.7) | 44 | 2.78 (1.84-4.19) | <0.001 |
| Perineural invasion | ||||||||
| Negative | 202 (86.7) | 23 | 1.00 | 234 (83.0) | 65 | 1.00 | ||
| Positive | 31 (13.3) | 7 | 2.01 (0.86-4.67) | 0.107 | 48 (17.0) | 27 | 2.39 (1.53-3.75) | <0.001 |
| Lymph node involvement | ||||||||
| Negative | 143 (64.1) | 10 | 1.00 | 154 (56.8) | 33 | 1.00 | ||
| Positive | 80 (35.9) | 17 | 3.24 (1.48-7.08) | 0.003 | 117 (43.2) | 56 | 2.76 (1.79-4.24) | <0.001 |
Abbreviations: HR, hazard ratio; CI, confidence interval; CEA, carcinoembryonic antigen; IQR, interquartile range. *Column subtotals do not sum to n of patients and n of events due to missing data.†According to the American Joint Committee on Cancer - Cancer Staging Manual (version 6.0). P ≤ 0.05 are in boldface.
We assessed the association of 4 MMP SNPs with distant metastasis-free and overall survival under dominant, recessive, and additive genetic models (Table 2). We defined the model with the most significant P value as the best model. Patients carrying AA genotype, minor allele homozygote, of MMP2 rs243866 had a significantly increased risk of developing distant metastasis following curative surgery compared with those with at least one G, major allele [hazard ratio (HR): 6.32, 95% confidence interval (CI): 1.50-26.7, P = 0.012]. We did not observe statistically significant relations between other MMP SNPs and distant metastasis-free and overall survival. We also found no association between the 4 MMP SNPs with clinicopathologic features listed in Table 1, except for the MMP2 rs2285053 showing a significant association with CEA levels (P = 0.030, data not shown).
Table 2.
Association of MMP gene polymorphisms with distant metastasis-free survival and overall survival.
| Gene | SNP ID | Position | Location | Allele | Genotype Distribution* | Best-fitting genetic models | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Distant metastasis-free survival | Overall survival | ||||||||||
| Model | HR (95% CI) | P | Model | HR (95% CI) | P | ||||||
| MMP2 | rs243866 | Chr16:55511537 | 5' upstream | G>A | 223/53/4 | Recessive | 6.32 (1.50-26.7) | 0.012 | Additive | 0.91 (0.78-1.45) | 0.696 |
| MMP2 | rs2285053 | Chr16:55512377 | 5' upstream | C>T | 144/121/16 | Dominant | 1.59 (0.77-3.30) | 0.213 | Recessive | 0.79 (0.29-2.16) | 0.646 |
| MMP9 | rs3787268 | Chr20:44641731 | Intron 8 | G>A | 93/138/49 | Recessive | 1.55 (0.66-3.60) | 0.314 | Additive | 1.18 (0.88-1.59) | 0.260 |
| MMP9 | rs17577 | Chr20:44643111 | Exon 12, Arg668Gln | G>A | 213/64/4 | Recessive | 2.34 (0.32-17.1) | 0.404 | Additive | 0.76 (0.47-1.22) | 0.255 |
Abbreviations: SNP, single nucleotide polymorphism; HR, hazard ratio; CI, confidence interval; MMP, matrix metalloproteinase. *The number represents major allele homozygotes, heterozygotes, and minor allele homozygotes, respectively. P ≤ 0.05 are in boldface.
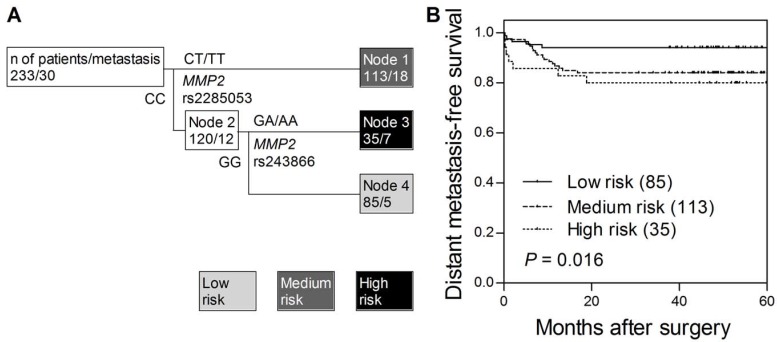
We noted that the association of MMP2 rs243866 with distant metastasis-free survival was based on a very low number of minor allele homozygous patients (n = 4) and required more cautious interpretation. Thus, we further used survival tree analysis to explored higher order SNP-SNP interactions to evaluate whether the interactions among MMP SNPs could determine the clinical outcomes. The tree structure resulted in 3 terminal nodes, ranging from low, medium, and high risk in developing distant metastasis after curative surgery (Fig. 1A). When using low risk node 4 as the reference group (wild-type homozygous genotypes of MMP2 rs2285053 and MMP2 rs243866), the HR was 2.79 (95% CI, 1.04-7.53, Table 3) for medium risk node 1, and 3.70 (95% CI, 1.18-11.7) for high risk node 3 (Ptrend = 0.017). The time to develop distant metastasis decreased as the increase in risk classification by MMP2 SNPs (log-rank P = 0.016, Fig. 1B). However, no genetic interaction was observed for overall survival.
Figure 1.
Potential higher order SNP-SNP interactions between MMP2 gene polymorphisms. (A) Survival tree analysis identifies the interactions between MMP2 gene polymorphisms. (B) Kaplan-Meier curves of distant metastasis-free survival based on the survival tree analysis. Numbers in parentheses indicate the number of patients.
Table 3.
Cox proportional hazards analysis of MMP2 genetic interaction profiles and distant metastasis-free survival.
| Variables | n (%) | n of events | Univariate analysis | Multivariate analysis* | ||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |||
| MMP2 gene polymorphisms | ||||||
| Low risk | 85 (36.5) | 5 | 1.00 | 1.00 | ||
| Medium risk | 113 (48.5) | 18 | 2.79 (1.04-7.53) | 0.042 | 3.73 (1.18-11.8) | 0.025 |
| High risk | 35 (15.0) | 7 | 3.70 (1.18-11.7) | 0.025 | 3.37 (0.86-13.2) | 0.082 |
| Trend | 1.87 (1.12-3.12) | 0.017 | 1.77 (1.00-3.15) | 0.050 | ||
Abbreviations: HR, hazard ratio; CI, confidence interval; MMP, matrix metalloproteinase. *Age, gender, CEA levels, tumor differentiation, stage, lymphovascular invasion, perineural invasion, lymph node involvement, and genetic risk classification by MMP2 polymorphisms were included in the multivariate analysis. P ≤ 0.05 are in boldface.
To adjust for potential confounding between the genetic interactions of MMP2 SNPs and distant metastasis-free survival, a Cox proportional hazards model was constructed. The multivariable model was adjusted for age, gender, CEA levels, tumor differentiation, stage, lymphovascular invasion, perineural invasion, and lymph node involvement as covariates. The genetic interaction profile between MMP2 rs2285053 and MMP2 rs243866 remained significantly associated with distant metastasis-free survival (HR: 1.77, 95% CI: 1.00-3.15, adjusted Ptrend = 0.050, Table 3).
Discussion
The expression levels of MMP2 and MMP9 have been shown to be correlated with colorectal cancer initiation, progression, angiogenesis, metastasis, and recurrence 27-29. Many studies have also proved that MMP SNPs have associations with cancer susceptibility including colorectal cancer 6-10, but little is known about their prognostic significance. In the present study, we found MMP2 rs243866 and a higher-order genetic interaction profile between MMP2 rs243866 and MMP2 rs2285053 were significantly associated with distant metastasis-free survival in colorectal cancer patients receiving curative surgery. Moreover, the association between the genetic interaction profile and distant metastasis-free survival persisted despite controlling for known clinical prognostic factors, including age, gender, CEA levels, tumor differentiation, stage, lymphovascular invasion, perineural invasion, and lymph node involvement.
The MMP2 rs2285053 (-735) C to T transition has been previously reported to affect MMP2 transcription due to the ablation of specificity protein (Sp) 1 transcription factor binding 15, 16. Similarly, it has also been reported that the MMP2 rs243866 G allele located in a potential estrogen receptor binding site and functioned as an enhancer, whereas the A allele showed reduced transcriptional activity in estrogen receptor-positive breast cancer cell line MCF-7 cells 14. In this study, we identified that colorectal cancer patients carrying AA genotype at MMP2 rs243866 had a 6.32-fold higher risk of developing distant metastasis after curative surgery compared with those with at least one G. These results are in contrast to the previous notion that high expression levels of MMP2 in rs243866 GG genotype carriers are associated with an invasive phenotype of cancer. However, our observation coincides with a previous study showing that the MMP2 rs243866 GG genotype was associated with a reduced risk of colorectal cancer 30. There are several possible explanations for this discrepancy. First, the MMP2 rs243866 was also reported to be not functional when tested within estrogen receptor-negative cells 15, suggesting that the functional effect varies depending on cell type. Second, it is important to consider that the activity of MMP2 is influenced not only by the level of its expression but also by its activators and inhibitors. Finally, although the original notion that MMPs act as metastasis-promoting enzymes by clearing a pathway for cancer cell invasion, more recent evidence indicates that some MMPs have anti-metastatic functions 31. Mounting evidence demonstrates that MMPs not only regulate the metastatic potential of cancer cells but also regulate cell proliferation, apoptosis, epithelial-mesenchymal transition, angiogenesis, and inflammatory response by targeting non-matrix proteins such as growth factors and their receptors, chemokines, and adhesive molecules. The complexity of the pro- and anti-tumorigenic and -metastatic functions might at least partly explain why MMP inhibitors failed in clinical trials. Together, these observations indicate that the balance between the pro- and anti-metastatic actions of MMPs is critical in determining their overall impact on cancer progression, and this paradox needs further investigation.
There are several strengths in this study. The patients analyzed in this study were enrolled from a single hospital, so patient characteristics and treatments are highly homogenous. We chose to investigate common MMP SNPs for which an association with cancer is biologically plausible and has been previously reported. However, some limitations in this study should also be considered when interpreting our results. The sample size is small and we had relatively few cases with distant metastasis. In addition, our homogeneous Chinese Han population might make our findings less generalizable to other ethnic groups. Additional studies in other populations are necessary to validate our findings.
In conclusion, our findings suggest that genetic variants in MMP2 might modulate colorectal cancer metastasis. We are able to identify subgroups of patients with different genetic profiles at MMP2 and predict their distant metastasis-free survival after curative surgery.
Acknowledgments
This work was supported by the National Science Council (NSC), Taiwan (grant number: grants NSC-98-2320-B-039-019-MY3 and NSC-100-2314-B-039-009-MY3), and China Medical University (grant number: CMU98-N1-21 and CMU98-C-12). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the National Center for Genome Medicine at Academia Sinica, Taiwan, for genotyping/technical support.
Abbreviations
- MMP
matrix metalloproteinase
- SNP
single nucleotide polymorphism
- CEA
carcinoembryonic antigen
- IQR
interquartile range
- HR
hazard ratio
- CI
confidence interval.
References
- 1.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–5. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 2.August DA, Ottow RT, Sugarbaker PH. Clinical perspective of human colorectal cancer metastasis. Cancer Metastasis Rev. 1984;3:303–24. doi: 10.1007/BF00051457. [DOI] [PubMed] [Google Scholar]
- 3.Langers AM, Sier CF, Hawinkels LJ. et al. MMP-2 geno-phenotype is prognostic for colorectal cancer survival, whereas MMP-9 is not. Br J Cancer. 2008;98:1820–3. doi: 10.1038/sj.bjc.6604380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Jagt MF, Sweep FC, Waas ET. et al. Correlation of reversion-inducing cysteine-rich protein with kazal motifs (RECK) and extracellular matrix metalloproteinase inducer (EMMPRIN), with MMP-2, MMP-9, and survival in colorectal cancer. Cancer Lett. 2006;237:289–97. doi: 10.1016/j.canlet.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Wilson S, Wakelam MJ, Hobbs RF. et al. Evaluation of the accuracy of serum MMP-9 as a test for colorectal cancer in a primary care population. BMC Cancer. 2006;6:258. doi: 10.1186/1471-2407-6-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elander N, Soderkvist P, Fransen K. Matrix metalloproteinase (MMP) -1, -2, -3 and -9 promoter polymorphisms in colorectal cancer. Anticancer Res. 2006;26:791–5. [PubMed] [Google Scholar]
- 7.Woo M, Park K, Nam J. et al. Clinical implications of matrix metalloproteinase-1, -3, -7, -9, -12, and plasminogen activator inhibitor-1 gene polymorphisms in colorectal cancer. J Gastroenterol Hepatol. 2007;22:1064–70. doi: 10.1111/j.1440-1746.2006.04424.x. [DOI] [PubMed] [Google Scholar]
- 8.Xing LL, Wang ZN, Jiang L. et al. Matrix metalloproteinase-9-1562C>T polymorphism may increase the risk of lymphatic metastasis of colorectal cancer. World J Gastroenterol. 2007;13:4626–9. doi: 10.3748/wjg.v13.i34.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu E, Lai M, Lv B. et al. A single nucleotide polymorphism in the matrix metalloproteinase-2 promoter is associated with colorectal cancer. Biochem Biophys Res Commun. 2004;324:999–1003. doi: 10.1016/j.bbrc.2004.09.150. [DOI] [PubMed] [Google Scholar]
- 10.Xu E, Xia X, Lu B. et al. Association of matrix metalloproteinase-2 and -9 promoter polymorphisms with colorectal cancer in Chinese. Mol Carcinog. 2007;46:924–9. doi: 10.1002/mc.20323. [DOI] [PubMed] [Google Scholar]
- 11.Beeghly-Fadiel A, Lu W, Long JR. et al. Matrix metalloproteinase-2 polymorphisms and breast cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 2009;18:1770–6. doi: 10.1158/1055-9965.EPI-09-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beeghly-Fadiel A, Lu W, Shu XO. et al. MMP9 polymorphisms and breast cancer risk: a report from the Shanghai Breast Cancer Genetics Study. Breast Cancer Res Treat. 2011;126:507–13. doi: 10.1007/s10549-010-1119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs EJ, Hsing AW, Bain EB. et al. Polymorphisms in angiogenesis-related genes and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:972–7. doi: 10.1158/1055-9965.EPI-07-2787. [DOI] [PubMed] [Google Scholar]
- 14.Harendza S, Lovett DH, Panzer U. et al. Linked common polymorphisms in the gelatinase a promoter are associated with diminished transcriptional response to estrogen and genetic fitness. J Biol Chem. 2003;278:20490–9. doi: 10.1074/jbc.M211536200. [DOI] [PubMed] [Google Scholar]
- 15.Price SJ, Greaves DR, Watkins H. Identification of novel, functional genetic variants in the human matrix metalloproteinase-2 gene: role of Sp1 in allele-specific transcriptional regulation. J Biol Chem. 2001;276:7549–58. doi: 10.1074/jbc.M010242200. [DOI] [PubMed] [Google Scholar]
- 16.Yu C, Zhou Y, Miao X. et al. Functional haplotypes in the promoter of matrix metalloproteinase-2 predict risk of the occurrence and metastasis of esophageal cancer. Cancer Res. 2004;64:7622–8. doi: 10.1158/0008-5472.CAN-04-1521. [DOI] [PubMed] [Google Scholar]
- 17.Yang ZH, Li SN, Liu JX. et al. MMP-9 polymorphisms are related to serum lipids levels but not associated with colorectal cancer susceptibility in Chinese population. Mol Biol Rep. 2012;39:9399–404. doi: 10.1007/s11033-012-1804-8. [DOI] [PubMed] [Google Scholar]
- 18.Han YJ, Kim HN, Yoon JK. et al. Haplotype analysis of the matrix metalloproteinase-9 gene associated with advanced-stage endometriosis. Fertil Steril. 2009;91:2324–30. doi: 10.1016/j.fertnstert.2008.03.047. [DOI] [PubMed] [Google Scholar]
- 19.Bao BY, Pao JB, Huang CN. et al. Significant associations of prostate cancer susceptibility variants with survival in patients treated with androgen-deprivation therapy. Int J Cancer. 2012;130:876–84. doi: 10.1002/ijc.26091. [DOI] [PubMed] [Google Scholar]
- 20.Bao BY, Pao JB, Huang CN. et al. Polymorphisms inside microRNAs and microRNA target sites predict clinical outcomes in prostate cancer patients receiving androgen-deprivation therapy. Clin Cancer Res. 2011;17:928–36. doi: 10.1158/1078-0432.CCR-10-2648. [DOI] [PubMed] [Google Scholar]
- 21.Bao BY, Pao JB, Lin VC. et al. Individual and cumulative association of prostate cancer susceptibility variants with clinicopathologic characteristics of the disease. Clin Chim Acta. 2010;411:1232–7. doi: 10.1016/j.cca.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 22.Huang CN, Huang SP, Pao JB. et al. Genetic polymorphisms in androgen receptor-binding sites predict survival in prostate cancer patients receiving androgen-deprivation therapy. Ann Oncol. 2012;23:707–13. doi: 10.1093/annonc/mdr264. [DOI] [PubMed] [Google Scholar]
- 23.Huang CN, Huang SP, Pao JB. et al. Genetic polymorphisms in oestrogen receptor-binding sites affect clinical outcomes in patients with prostate cancer receiving androgen-deprivation therapy. J Intern Med. 2012;271:499–509. doi: 10.1111/j.1365-2796.2011.02449.x. [DOI] [PubMed] [Google Scholar]
- 24.Huang SP, Lan YH, Lu TL. et al. Clinical significance of runt-related transcription factor 1 polymorphism in prostate cancer. BJU Int. 2011;107:486–92. doi: 10.1111/j.1464-410X.2010.09512.x. [DOI] [PubMed] [Google Scholar]
- 25.Wanebo HJ, Rao B, Pinsky CM. et al. Preoperative carcinoembryonic antigen level as a prognostic indicator in colorectal cancer. N Engl J Med. 1978;299:448–51. doi: 10.1056/NEJM197808312990904. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HP, Singer B. Recursive partitioning and applications. New York: Springer; 2010. [Google Scholar]
- 27.Asano T, Tada M, Cheng S. et al. Prognostic values of matrix metalloproteinase family expression in human colorectal carcinoma. J Surg Res. 2008;146:32–42. doi: 10.1016/j.jss.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 28.van der Jagt MF, Wobbes T, Strobbe LJ. et al. Metalloproteinases and their regulators in colorectal cancer. J Surg Oncol. 2010;101:259–69. doi: 10.1002/jso.21462. [DOI] [PubMed] [Google Scholar]
- 29.Zucker S, Vacirca J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 2004;23:101–17. doi: 10.1023/a:1025867130437. [DOI] [PubMed] [Google Scholar]
- 30.Park KS, Kim SJ, Kim KH. et al. Clinical characteristics of TIMP2, MMP2, and MMP9 gene polymorphisms in colorectal cancer. J Gastroenterol Hepatol. 2011;26:391–7. doi: 10.1111/j.1440-1746.2010.06504.x. [DOI] [PubMed] [Google Scholar]
- 31.Decock J, Thirkettle S, Wagstaff L. et al. Matrix metalloproteinases: protective roles in cancer. J Cell Mol Med. 2011;15:1254–65. doi: 10.1111/j.1582-4934.2011.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]