Abstract
The purpose of this study was to identify key genetic pathways involved in non-small cell lung cancer (NSCLC) and understand their role in tumor progression. We performed a genome wide scanning using paired tumors and corresponding 16 mucosal biopsies from four follow-up lung cancer patients on Affymetrix 250K-NSpI array platform. We found that a single gene SH3GL2 located on human chromosome 9p22 was most frequently deleted in all the tumors and corresponding mucosal biopsies. We further validated the alteration pattern of SH3GL2 in a substantial number of primary NSCLC tumors at DNA and protein level. We also overexpressed wild-type SH3GL2 in three NSCLC cell lines to understand its role in NSCLC progression. Validation in 116 primary NSCLC tumors confirmed frequent loss of heterozygosity of SH3GL2 in overall 51 % (49/97) of the informative cases. We found significantly low (p=0.0015) SH3GL2 protein expression in 71 % (43/60) primary tumors. Forced over-expression of wild-type (wt) SH3GL2 in three NSCLC cell lines resulted in a marked reduction of active epidermal growth factor receptor (EGFR) expression and an increase in EGFR internalization and degradation. Significantly decreased in vitro (p=0.0015–0.030) and in vivo (p=0.016) cellular growth, invasion (p=0.029–0.049), and colony formation (p=0.023–0.039) were also evident in the wt-SH3GL2-transfected cells accompanied by markedly low expression of activated AKT(Ser473), STAT3 (Tyr705), and PI3K. Downregulation of SH3GL2 interactor USP9X and activated β-catenin was also evident in the SH3GL2-transfected cells. Our results indicate that SH3GL2 is frequently deleted in NSCLC and regulates cellular growth and invasion by modulating EGFR function.
Keywords: Single nucleotide polymorphism array, Lung cancer, SH3GL2, Deletion
Introduction
Lung cancer kills more than one million people worldwide with smoking being the most important risk factor [ 1 ]. In the USA, the estimated number of lung cancer cases is 226,160 and predicted deaths of 160,340 in 2012 [2]. Approximately 85 % of these patients have non-small cell lung cancer (NSCLC) and two thirds are diagnosed at advanced stage with mean survival of 1 year receiving traditional treatment [3]. Eighty-five percent of lung cancers occur among the tobacco smokers and rest among the never-smokers [4], Despite a significant improvement in therapeutic modalities including surgery, platinum-based chemotherapy, and radiotherapy alone or in combination, the overall 5-year survival rate is only 15 % [5]. Lung cancer has a high morbidity because of difficulty in early detection and resistance to chemotherapy and radiotherapy [1]. Moreover, affected patients remain at significant risk for the development of secondary primary tumors throughout their lifetime.
Molecular genetic studies of LC have identified several genetic and epigenetic abnormalities, including DNA sequence/copy number changes and aberrant promoter hyper-methylation [1, 5–9]. Frequent allelic losses on chromosomes lp, 3p, 4q, 5q, 8p, 9p, 10p, 11p, 13q, and 17p and gain on chromosomes 1q, 3q, 5p, and 8q have been reported in lung cancer [1]. In total, these alterations result in the activation of oncogenes and inactivation of tumor suppressor genes [1]. Aberrant KRAS signaling among the smokers and epidermal growth factor receptor (EGFR) signaling among the never-smokers were also discovered as the predominant alteration pathways [4]. So far, other than the above pathways, only a few common inactivated target genes have been associated with lung carcinomas such as CDKN2A (9p), TP53 (17p), STK11 (19p), and BRF2 (8pl2) [9, 10]. Accordingly, further identification of the key oncogenes and tumor suppressor genes involved in lung tumorigenesis and interacting with the KRAS and EGFR pathways is necessary. Discovery of the key target genes underlying NSCLC development and progression requires high-resolution molecular characterization followed by appropriate validation platforms [9, 11]. In this context, high-density single nucleotide polymorphism (SNP) array is a feasible means for conducting a genome wide screen to identify novel targets of inactivation [9, 11]. Further understanding of the pivotal role of the altered genes involved in the initiation and progression of NSCLC will allow us to comprehend their role in lung tumorigenesis.
In the present study, we examined tumor and corresponding mucosal biopsy specimens from four follow-up lung cancer patients using a high-resolution SNP array platform (NSpI-250K, Affymetrix). We identified that a single gene SH3GL2 on chromosome 9p22 as the most frequently deleted in all the tumors and corresponding histologically normal-appearing mucosal biopsies. These observations were further validated in a large number of primary NSCLC tumors. Forced overexpression of SH3GL2 in NSCLC cells resulted in decreased in vitro and in vivo growth and invasion with concomitant decrease in the expression level of activated EGFR and its interacting molecules PI3K, AKT, STAT3, USP9X, and beta-catenin.
Materials and methods
Tissue specimens
Matched tumor and tumor-free normal lymph nodes were collected from four primary NSCLC tumors as described earlier [12]. All were squamous cell carcinomas (SCCs). In addition, five postoperative mucosal biopsies each from two patients (patients 1 and 2) and three biopsies each from the other two patients (patients 3 and 4) were also collected (total of 16 biopsies) [12]. Although all the follow-up biopsy specimens were histologically diagnosed as normal, however, in the context of multistage carcinogenesis, they will be regarded as “normal appearing.” All specimens were procured following an IRB-approved protocol at Johns Hopkins University. We also procured two additional sets of matched normal and tumor tissues from 116 NSCLC cases at various stages and histologic grades from University of British Columbia (S.L., UBC, 60 cases) and Mayo Clinic (MC, 56 cases, Y.P and J.E.Y.) as per respective IRB-approved protocols. Most of the patients were adenocarcinomas (ADCs). The available demographic information for these two cohorts of patients with alteration status of SH3GL2 and/or EGFR and KRAS was provided in Table 1 and Table 2 and supplementary Table S1, supplementary Table S2 (JHU), and 3 (MC).
Table 1.
Alteration pattern of SH3GL2 in the lung cancer patients carrying EGFR/KRAS mutation
| Smoking status | No. of patientsa | LOH of SH3GL2b | EGFR mutation | KRAS mutation | Simultaneous SH3GL2 loss/EGFR mutationb |
Simultaneous SH3GL2 loss/KRAS mutationb |
|---|---|---|---|---|---|---|
| Never-smoker | 30 | 16/26 (62 %) | 17/30 (57 %) | 1/30 (3 %) | 11/26 (43 %) | N |
| Current smoker | 30 | 13/25 (52 %) | 0/30 (0 %) | 19/30 (64 %) | N | 6/25 (24 %) |
N none detected
These 60 primary lung adenocarcinomas were obtained from University of British Columbia
Only informative cases were considered for determining LOH and simultaneous gene alteration pattern
Table 2.
The alteration pattern of SH3GL2 in the lung cancer patients obtained from Mayo Clinic
| Smoking status | No. of patientsa | LOH of SH3GL2b | EGFR mutation | KRAS mutation | Simultaneous SH3GL2 loss/EGFR mutationb |
Simultaneous SH3GL2 loss/KRAS mutationb |
|---|---|---|---|---|---|---|
| Never-smoker | 56 | 20/46 (44 %) | ND | ND | ND | ND |
ND not determined
These 56 primary lung adenocarcinomas were obtained from Mayo Clinic
Only informative cases were considered for determining LOH and simultaneous gene alteration pattern
Cell lines and reagents
All the lung cancer cell lines were purchased from American Type Culture Collection (ATCC) and cultured in ATCC-recommended medium. The cell lines were periodically checked for Mycoplasma contamination using Mycoplasma detection kit from Sigma (MP-0025). All tissue culture medium and reagents were purchased either from ATCC or Invitrogen.
Preparation of genomic DNA and SNP 250K-NSpI array analysis
Genomic DNA was extracted from microdissected tumor and undissected normal-appearing tissue samples using a standard laboratory protocol [13]. We used the 250K-Nsp I array platform from the Affymetrix 500K array set for genotyping each sample (www.affymetrix.com). This array platform contains ∼262,000 SNPs and utilizes Nsp I restriction enzyme. Two hundred fifty nanograms of genomic DNA was digested with Nsp I and ligated to adaptors that recognize the cohesive four base pair (bp) overhangs. All fragments resulting from restriction enzyme digestion was then ligated using T4 ligase and PCR-amplified using an Applied Biosystems Thermal Cycler I (9700) to achieve a size range of 200–1,100 bp. Amplified DNA was then pooled, concentrated, and cleaned up. The amplified and cleaned product was fragmented using DNaseI (Affymetrix Inc.) and subsequently labeled, denatured, and hybridized to arrays. Hybridized arrays were scanned using the GeneChip Scanner 3000 7G (Affymetrix Inc). The genotype call was automatically detected by the Affymetrix Genotype Analysis Software. All the experimental steps were followed as per the manufacturers' specification.
Determination of loss of heterozygosity and visualization of the results
The resulting data were analyzed using both dChip (www.dchip.org) and Nexus software package (http://www.snp-nexus.org) for the determination of loss of heterozygosity (LOH). Intensity (.CEL) files were normalized and modeled using the PM-MM difference modeling method [8, 9]. Array normalization and quality control were performed as per the Affymetrix protocol and requirement. A region of “LOH” was designated when there was a consecutive loss of at least 10 SNPs. Subsequently, utilizing the mapping information of the SNP tags based on the Affymetrix annotation, the specific regions of losses/gains and the genes were identified utilizing the UCSC Genome Browser (http://genome.ucsc.edu/) as well as the NCBI data base (http://www.ncbi.nlm.nih.gov/SNP/). All SNP data files were submitted to gene expression omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29365).
Loss of heterozygosity analysis by gene-specific microsatellite markers
For validation purpose, gene-specific LOH analysis (for SH3GL2, 9p22) was performed using Gene Marker version 1.91 (Soft Genetics, State College, PA). Briefly, we designed two STS markers (UniSTS: 28906 and 12760) in D9S157 based on the UniSTS database and synthesized to 6-FAM-labeled primers (Operon Biotechnology, Huntsville, AL). PCR primers used for the UniSTS 28906 and UniSTS 12760 were 5’-6-FAM CATTTCATCTGGTAGACCCA-3’ (forward) and 5’-TTTGATTGGCTGGAAGTAGA-3’ (reverse) and 5’-6-FAM AGCAAGGCAAGCCACATTTC-3’ (forward) and 5’-TGGGGATGCCCAGATAACTATATC-3’ (reverse), respectively. PCR was carried out at 95 °C for 4 min, followed by 35 °C cycles of 15 s at 95 °C; 30 s at 59 °C for UniSTS 28906 and 30 s at 61 °C for UniSTS 12760; 1 min at 72 °C; and a final elongation at 72 °C for 7 min. The PCR products were loaded with LIZ 500 size standard (Applied Biosystems) onto ABI PRISM 3730XL DNA Analyzer (Applied Biosystems) for capillary electrophoresis. LOH was determined as a more than 25 % change in the ratio of allele peak size compared to the paired normal lung and tumor tissue at both marker loci. Analysis of EGFR and KRAS gene mutation in NSCLC samples was described earlier [14].
FISH analysis
For analysis of gene-specific copy number loss, dual color fluorescence in situ hybridization (FISH) was performed on tissue microarrays containing 25 cases of lung adenocarcinoma, corresponding lung tissue adjacent to adenocarcinoma, and normal lung parenchyma. Bacterial artificial chromosome-derived test probe targeting SH3GL2 (9p22, contig of RP11-98 M23) (BACPAC Resources Center, Oakland, CA) and CTD-2522C6 (Invitrogen, Carlsbad, CA) was labeled with Spectrum Orange. This was paired for dual-target hybridization diluted 1:50 in Den-Hyb hybridization buffer (Insitus Laboratories, Albuquerque, NM) with Spectrum Green-labeled control probe CEP9 (control probe targeting centromeric region of chromosome 9). Ten microliters of the resultant hybridization mix was applied to the tissue microarray sections, with simultaneous denaturing of probe and target at 80 °C for 2 min and 50 °C for 45 min. Overnight hybridization was carried out at 37 °C which occurred in a humidified chamber, followed by post-hybridization washes with 50 % formamide/l× SSC (5 min) and 2× SSC (5 min). DAPI (0.125 ng/ml) (Abbott Laboratories) served as a nuclear counter stain. Sections showing sufficient hybridization efficiency (majority of nuclei with signals) were considered informative and were scored. Corresponding non-neoplastic lung specimens served as the controls in each case. Deletion for SH3GL2 was subsequently defined by a SH3GL2/CEP9 ratio of <0.80 (mean+3 standard deviations in non-neoplastic controls).
DNA methylation analysis
DNA methylation status of SH3GL2 (refSeq NM003026.2) was examined using specific primers (MePh 12267-2A, SA Bioscience) as per the manufacturer's protocol and data analysis tool.
Immunohistochemistry
Immunohistochemistry (IHC) was performed using specific antibody in paired lung cancer specimens at their different stages as described earlier [15]. Each experiment was repeated at least two times. For counting of the positively stained cells or intensity measurement, Meta-morph software (Universal Imaging, Downington, PA) was used. At least 10 fields were chosen at random for counting and data were expressed as mean±SE. For intensity measurement, lowest value (≤50.00) was represented by a single + sign and each fold increase was represented by additional + sign [15]. We have procured SH3GL2 antibody for IHC analysis from Abnova Corporation (#H00006456-MOl).
Construction and cloning of wild-type SH3GL2
The wild-type (wt) SH3GL2 sequence (gene ID: 6456) was subcloned into SalI and NotI sites of the phosphorylated cytomegalovirus pCMV/myc/cyto plasmid. The resultant plasmids were resequenced using the ABI Big Dye cycle sequencing kit (Applied Biosystems) for verification of the insert sequences. In transfections, H1299, H1437, and H1944 NSCLC cells were transfected with wt-SH3GL2 plasmids in the presence of the FuGene 6 transfection regent. An empty vector was used as control. Transfected cells were maintained in 300 μg/ml of G418. The expression of exogenous SH3GL2 was confirmed by western blot analysis using cmyc antibody in the stable clones. Three different clones were examined for all the subsequent experiments.
BrdU cell proliferation assay
The proliferation capabilities of the transfected cells were determined by BrdU incorporation assay kit (Roche Diagnostics) as per the manufacturer's protocol. Briefly, the cells were incubated at 37 °C, 5 % CO2 for about 15–60 min in BrdU-labeling medium followed by fixation in ethanol fixative for 20 min at −20 °C. The cells were then treated with anti-BrdU reagent for 30 min at 37 °C followed by incubation for 30 min at 37 °C with anti-mouse-Ig-AP solution. Color substrate solution (NBT+BCIP) was then added to the cells. After 15–30 min of incubation at room temperature, the cells were analyzed directly under a light microscope. At least 10 fields were randomly selected for counting the positive cells. Data are represented as mean±SE of duplicate experiments.
Soft agar colony formation assay
In vitro colony-forming ability of the cells was examined by soft agar assay as described [16]. Briefly, for base agar, an equal volume of medium and 1 % agarose was mixed and layered in 2-ml volume in well of a six-well tissue culture plate. For the top layer, an equal volume of medium and 0.5 % agarose was mixed and layered on the base agar as above containing 1 × 104 cells. The cells were cultured for 14 days until colonies are visible, stained with 0.5 % crystal violet, and photographed. The numbers of colonies bigger than 2 mm were counted in each well of triplicate wells per group. Data were represented as mean±SE of duplicate experiments.
Cell invasion assay
Cell invasion capacity was assessed using the Cell Invasion Assay Kit (BD Biosciences) [16]. The assay was performed in triplicate. At least 10 fields were randomly selected for counting cells that invaded through the membrane from each group. Data were represented as mean±SE of duplicate experiments.
Treatment of the transfected cells with epidermal growth factor
Stably transfected cells with wt-SH3GL2 or empty vector were serum starved for 12 h and treated with 100 ng/ ml of epidermal growth factor (EGF) for 30 min in a tissue culture incubator [17] followed by immunofluorescence analysis using anti-EGFR antibody as described below.
Immunofluorescence
EGF-treated cells were fixed in 4 % para-formaldehyde and stained with anti-EGFR primary antibody overnight at 4 °C followed by staining with FITC-labeled secondary antibody for 30 min [18]. Cells were then washed with PBS and examined under a fluorescent microscope for examining the distribution pattern of EGFR molecules. Each experiment was repeated two times.
Western blot analysis
Preparation of whole cell lysates and western blot analysis was performed following standard protocol as described earlier [18]. Antibodies were purchased from Cell Signaling or Abnova Corporation.
Animal model and tumor growth analysis
For tumor growth, 1 × 106 cells in 100 μl mixture of PBS and basement membrane extract (R&D systems) were injected subcutaneously at the left flank of athymic, 4-to 6-week-old female nude (CD-NUBR) mice (Charles River) as described earlier [16]. All experiments were performed in accordance with the Animal Care and Use Committee guidelines at the Johns Hopkins University. Each group was consisted of six mice and each experiment was repeated two times. Tumor growth was monitored every day, and mice showing signs of morbidity were immediately sacrificed according to guidelines. After 4 weeks, mice were sacrificed and gross tumor weight was taken. Data were represented as mean±SE of duplicate experiments.
Co-immunoprecipitation
The co-immunoprecipitation analysis was performed using Dynabead Protein G kit (#100.07D) and DynaMag-2 system (#123.21D) and following protocols from Invitrogen Corporation. Antibodies for SH3GL2 and USP9X were purchased from Abnova Corporation. Appropriate IgG control antibody was used in each case.
Statistical analysis
Chi-square test, Fisher's exact test, or Student's t test was used as appropriate. All p values will be two-sided and all confidence intervals were at the 95 % level. Computation for all the analysis was performed using the Statistical Analysis System.
Results
Identification of frequent loss of SH3GL2 in the primary lung tumors and corresponding normal-appearing mucosa by SNP array
In this pilot study, we first examined primary tumors and 16 corresponding normal-appearing mucosal biopsies from four lung cancer patients on the Affymetrix 250K-Nsp I array platform. In each case, corresponding tumor-free normal lymph nodes were used as controls. Primarily, we focused on identifying the most frequently deleted genes that were simultaneously altered in the primary tumors and the normal-appearing biopsies. We found that a single gene SH3GL2 located on human chromosome 9p22 was most frequently deleted in the all the four tumors and corresponding normal-appearing biopsies.
Validation of SH3GL2 deletion status in primary NSCLC tumors
We further validated the deletion status of SH3GL2 in an additional set of 116 primary NSCLC tumors. We examined LOH at SH3GL2 locus (D9S157) utilizing two intragenic microsatellite markers (Fig. 1a). Allelic loss at both the marker loci was only considered for determining LOH. Of the 116 cases analyzed, first cohort comprised 60 primary tumors from JHU (Table 1). Among these, 30 were never-smoker with predominant EGFR gene mutation (Table 1 and Table S1) as reported earlier [14]. In this group, four cases were non-informative and the LOH of SH3GL2 was detected among 62 % (16/26) of the informative cases (Table 1 and Table S1). No association between SH3GL2 loss with stage (p=0.17), age (p=0.85), or gender (p= 1.00) was found. However, SH3GL2 loss was significantly higher (p=0.03) among the never-smoker Asian compared to the Caucasian patient population (Table S1). The other 30 cases were current smokers with predominant KRAS gene mutation (Table 1 and Table S2). In this group, five cases were non-informative and LOH of SH3GL2 was detected among 52 % (13/25) of the informative cases (Table 1 and Table S2). No significant association between SH3GL2 loss with stage (p=0.57), age, (p= 0.45), gender (p=0.54), and ethnicity (p=1.00) was found in this group. No association was also found between SH3GL2 loss and smoking habit (p=0.58).
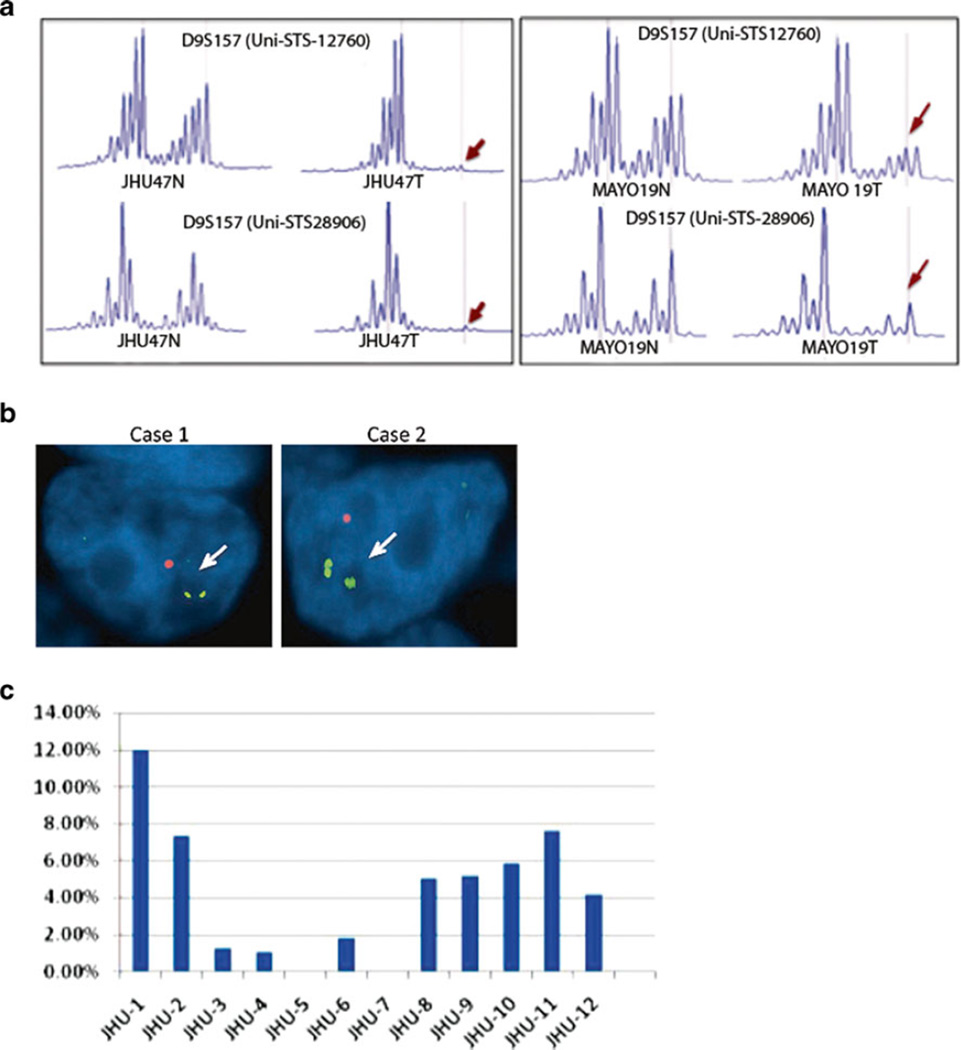
Fig. 1.
LOH, copy number loss, and hypermethylation of SH3GL2 in primary NSCLC tumors. a Two sets of primers (UniSTS 12760 and 28906) from the intragenic D9S157 locus of the SH3GL2 gene were examined. LOH occurs when allelic loss was observed at both marker loci (indicated by arrows) in the tumor (T) compared to the corresponding normal (N). b FISH analysis revealed loss of SH3GL2 copy (orange red, arrows) on 9p22 compared to control 9p centromeric probe CEP9 (green). c No significant changes were observed in the methylation pattern of SH3GL2 in the 12 primary NSCLC tumors analyzed compared to corresponding control tissues
SH3GL2 status in primary lung tumors with EGFR and KRAS gene mutation
As shown in Table 1 (bold italics), loss of SH3GL2 was higher in the primary tumors carrying EGFR gene mutation compared to the tumors carrying KRAS gene mutation (43 vs. 24 %). However, it did not reach a statistically significant level (p=0.24).
The remaining 56 primary NSCLC tumors were from Mayo Clinic. All the tumors were histologically ADCs and never-smokers. Of the 56 cases, 10 were non-informative and the LOH of SH3GL2 was detected among 44 % (20/46) of the informative cases (Table 2 and Table S3). We did not observe any significant correlation between SH3GL2 loss and stage (p=0.62), age (p=0.24), gender (p=0.64), or follow-up status (p=1.00). The status of EGFR/KRAS gene mutation in these samples was not available. Representative example of LOH in JHU and MC samples was shown in Fig. 1a.
Copy number loss and promoter methylation analysis of SH3GL2 in primary NSCLC tumors
We performed FISH analysis on 25, paired NSCLC tumors (tissue microarray) to identify copy number loss at SH3GL2 locus on 9p22. All samples were ADC and we observed similar rate of copy number loss among 56 % (14/25) cases analyzed (Fig. 1b). We further examined 12 primary NSCLC tumors based on DNA availability from the never-smoker group for promoter hypermethylation at the SH3GL2 locus using methylation-specific primers (MePh 12267-2A, SA Bioscience). Seven of these 12 cases had LOH at SH3GL2 locus, 3 were non-informative, and 2 had no LOH (Table S2, cases 07L6-07L38). We did not find significant DNA methylation of the SH3GL2 promoter in these tumors (Fig. 1c).
Expression of SH3GL2 protein in primary NSCLC tumors
Since we have observed a high frequency of allelic loss at SH3GL2 locus in the primary NSCLC tumors, we further analyzed primary tumors and adjacent normal from 60 NSCLC cases with both squamous cell carcinoma (15 cases of SCC) and adenocarcinoma histology (45 cases of ADC) for determining the expression of SH3GL2 at the protein level by IHC. For IHC analysis, all tissue samples were run in duplicates using appropriate isotype controls and scored by two independent investigators. Expression of SH3GL2 was detectable mostly in the cytoplasm and was significantly lower (+++ vs. +;p=0.0015) in 71 % cases (43/60; Fig. 2a). Of the 60 cases, 15 were SCCs and 73 % (11/15) had significantly low expression of SH3GL2. The rest of the 45 cases were ADCs and 71 % (32/45) had significantly low expression of SH3GL2 compared to the corresponding normal. Thus, a marked loss of SH3GL2 protein expression was evident in the primary NSCLC tumors. We did not find any significant correlation between SH3GL2 protein expression and clinical stage, histological grade, age, and gender in these primary tumors (data not shown).
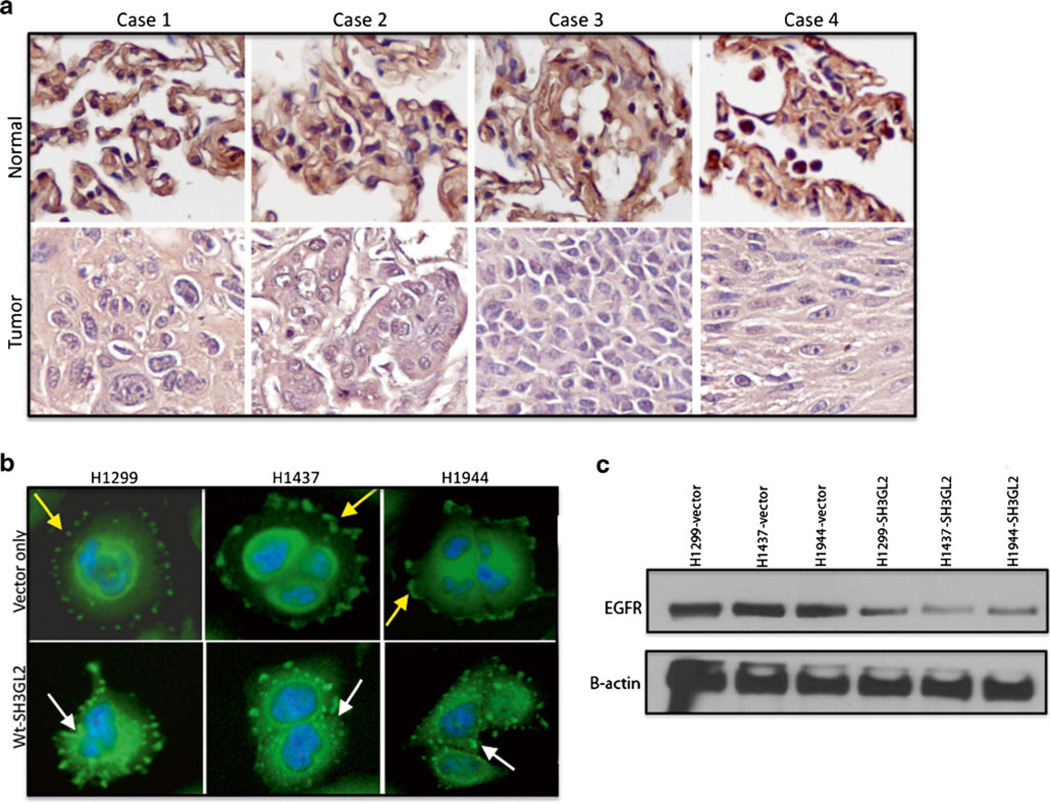
Fig. 2.
Pattern of SH3GL2 protein expression in paired primary NSCLC tissues. a Tissue sections were stained with a monoclonal anti-human SH3GL2 antibody. The expression of SH3GL2 was significantly lower (+++ vs. +; p=0.0015) in the tumors compared to the corresponding normal. Representative photomicrograph of two independent experiments is shown. Cases 1–3 were adenocarcinomas, and case 4 was squamous cell carcinoma. Magnification ×200. b Effect of EGF on EGFR internalization in the transfected cells. EGFR internalization markedly increased in the wt-SH3GL2-transfected groups as indicated (lower panel, white arrow) compared to the control vector-transfected groups (upper panel, yellow arrow). Representative photomicrograph of two independent experiments is shown. Magnification ×400. c Transfected cells were serum starved and treated with EGF for 6 h followed by western blot analysis. Markedly decreased expression of EGFR was observed in SH3GL2-transfected cell line as indicated
SH3GL2 induces EGFR internalization in the NSCLC cells
In the next step, we examined the impact of forced over-expression of wt-SH3GL2 on EGFR internalization in lung cancer cells. We treated the stably transfected and serum-starved cells with EGF followed by staining with EGFR antibody. As shown in Fig. 2b, EGFR internalization was markedly increased in the wt-SH3GL2-transfected cells (white arrows) compared to the control vector (yellow arrows). Thus, forced overexpression of wt-SH3GL2 resulted in increased internalization of EGFR in the lung cancer cells. To further elucidate whether the internalization was followed by degradation of EGFR or not, we treated the stably transfected cells with EGF for 6 h followed by western blot analysis. As shown in Fig. 2c, we have observed markedly low EGFR expression in the SH3GL2-transfected cells.
SH3GL2 modulates EGFR expression in the NSCLC cells
The SH3GL2 is a gene involved in controlling clathrin-mediated receptor endocytosis including EGFR in association with its constitutive binding partner CIN85 and Cbl [19]. Therefore, to understand the impact of SH3GL2 alteration on EGFR expression, we overexpressed the wild-type version of the SH3GL2 (wt-SH3GL2) construct in three NSCLC cell lines, which initially harbored low expression of SH3GL2 (Fig. 3a, left panel 1). The exogenously expressed SH3GL2 fusion protein was tagged and detectable by the myc antibody in the transfected cells (Fig. 3a, left panel 2). We performed western blot analysis to determine the active EGFR status in the SH3GL2-transfected cells. As shown in Fig. 3a (right panel), we observed marked reduction of active EGFR (Tyr845) expression (panel 2) in all the three SH3GL2-transfected cell lines compared to the controls. We also observed a concomitant increase in the expression of Tyr1045 phosphorylation of EGFR in the SH3GL2-transfected cells, which triggers EGFR degradation through the ubiquitin pathway by c-cbl [20]. Thus, forced overexpression of wt-SH3GL2 resulted in decreased expression of activated EGFR in the NSCLC cells.
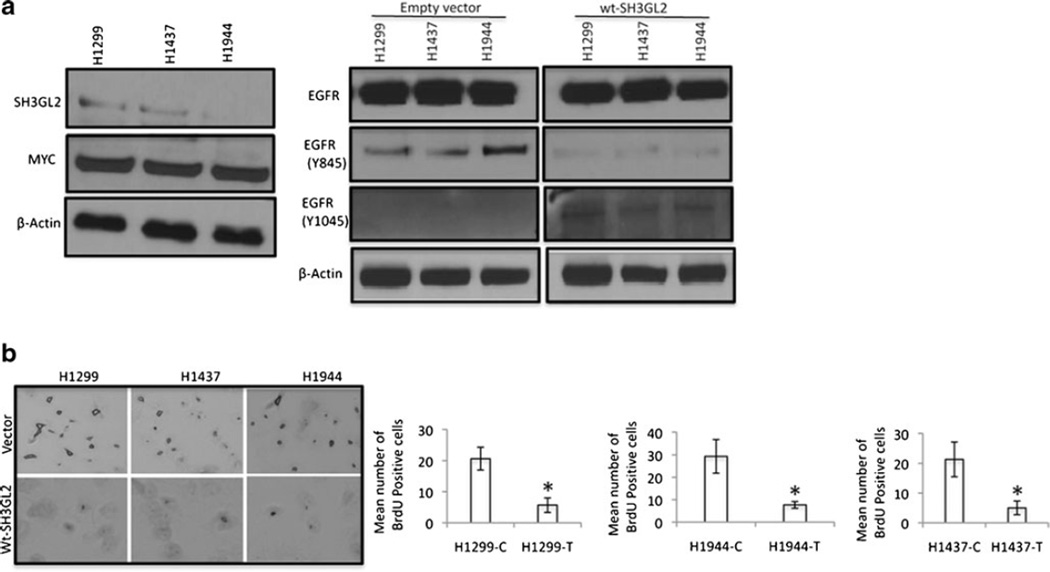
Fig. 3.
EGFR expression and proliferation of the transfected cells. a Expression of EGFR in the SH3GL2-transfected cells. Expression of endogenous SH3GL2 in H1299, H1437, and H1944 NSCLC cells (left panel). MYC-tagged SH3GL2 fusion protein in these cells was detectable by the anti-MYC antibody. β-actin was used a loading control. Right panel: Western blot analysis demonstrated a marked decrease in EGFR phosphorylation at Tyr845 (panel 2) and an increase in phosphorylation at Tyr1045 site (panel 3) in the wt-SH3GL2-transfected groups compared to the empty vector-transfected groups. β-actin was used as a loading control. b Compared to the empty vector-transfected group, BrdU incorporation was significantly lower (p=0.0015–0.030, right panel), indicated by asterisk in the wt-SH3GL2-transfected groups. Representative photomicrograph was shown in the left panel. Magnification ×200
SH3GL2 reduces NSCLC cell growth via modulation of downstream effectors
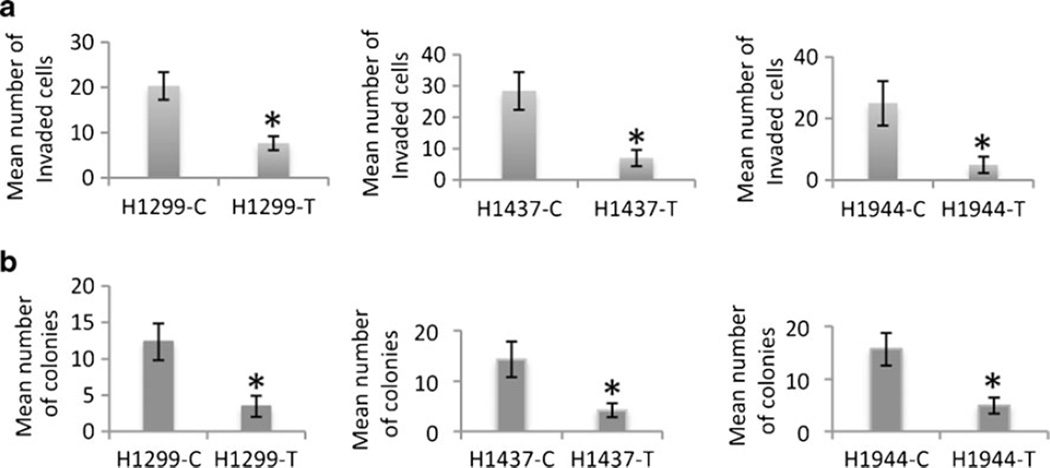
Since EGFR is a key molecule regulating cellular growth through continuous growth-promoting signaling in NSCLC [4], we examined proliferation, anchorage-independent growth, and invasion capabilities of the SH3GL2-transfected cells. We observed a significant decrease in the number of proliferating cells in the wt-SH3GL2-transfected groups (p=0.0015–0.030) compared to the control vector groups (Fig. 3b). Representative example was shown in the left panel. We also observed a significant decrease in the number of invading cells in the wt-SH3GL2-transfected groups (p=0.029–0.049) compared to the control groups (Fig. 4a). The number of soft agar colonies was also significantly decreased in the wt-SH3GL2-transfected group (p=0.023–0.039) compared to the control groups (Fig. 4b). Representative photomicrographs for invasion and colony formation were shown in Fig. S1a. In the in vivo setting, growth of SH3GL2-transfected H1299 cells was also significantly reduced (p=0.016) compared to the control vector-transfected group (79±18 vs. 273±54 mg; Fig. S1b).
Fig. 4.
In vitro invasion potential and anchorage-independent growth capabilities of the transfected cells. a Compared to the empty vector-transfected groups, the number of invaded cells significantly decreased (p=0.029–0.049, indicated by asterisk) in the wt-SH3GL2-transfected group. b Number of soft agar colonies was significantly decreased (p= 0.023–0.039, indicated by asterisk) in the wt-SH3GL2-transfected group
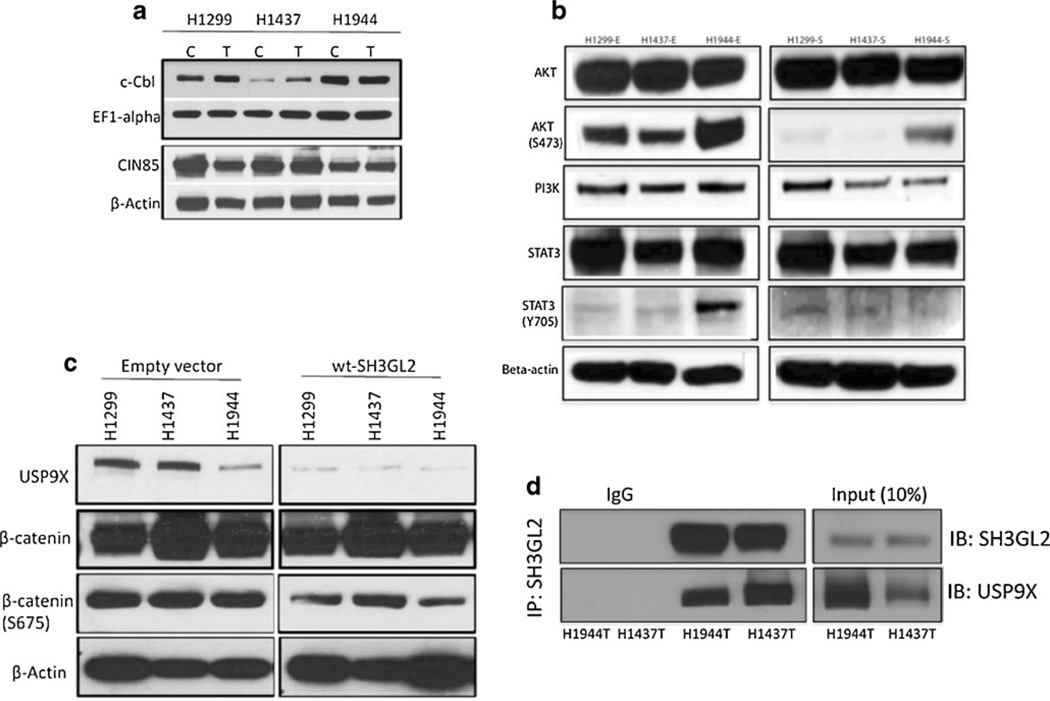
One earlier study has shown that SH3GL2 with its partner CIN85 and CbI mediates receptor endocytosis, and in this cascade, CIN85 remains constitutively bound to SH3GL2, whereas the binding of CIN85 to c-cbl depends on stimulation by the ligands [19]. To examine the expression status of CIN85 and c-Cbl in the transfected cells, we performed western blot analysis using specific antibodies. We did not detect any marked change in the expression of wild-type CIN85 and c-cbl in the transfected cells (Fig. 5a). Since we observed a marked decrease in active EGFR expression (Y845) and corresponding reduction in cell growth and invasion (Fig. 3 and Fig. 4), we examined the expression pattern of some of the key EGFR pathway molecules PI3K, AKT, and STAT3 in the transfected cells by western blot analysis. As shown in Fig. 5b, we observed a considerable reduction of PIK3 and phosphorylated AKT (Ser473) and STAT3 (Tyr705) expression level in the wt-SH3GL2-transfected groups compared to the empty vector-treated groups. Thus, overexpression of wt-SH3GL2 resulted in a considerable decrease in the expression level of some activated downstream EGFR signaling-associated molecules in the NSCLC cell lines.
Fig. 5.
Expression of SH3GL2 interacting CIN85, c-cbl, and EGFR signaling pathway molecules in the transfected cells. a Western blot analysis showing more or less similar expression pattern of CIN58 and c-cbl in the transfected cells. EF1-alpha and β-actin were used as loading control as indicated. C empty vector, T wt-SH3GL2 transfected. b Western blot analysis demonstrated marked reduction of AKT (Ser473), STAT3 (Tyr705), and PI3K expression in the wt-SH3GL2-transfected groups compared to the empty vector-transfected groups. Empty vector transfected; S wt-SH3GL2 transfected. c Western blot analysis showing marked reduction of USP9X and β-catenin (Ser675) in the wt-SH3GL2-transfected groups compared to the empty vector-transfected group. β-actin was used as loading control as indicated. d Co-immunoprecipitation analysis in SH3GL2-transfected H1437 and H1944 cells showed reciprocal interaction between SH3GL2 and USP9X
In recent studies, USP9X (ubiquitin-specific peptidase 9), a mammalian orthologue of Drosophila melanogaster FAM was found to be involved in enhancing survival of cancer cells through deubiquitination and stabilization of growth regulatory molecules including TGF-beta receptors, MCL-1 [21]. USP9X also stabilizes β-catenin (CTNNB1) expression [21] and predicted to interact with SH3GL2 (http://www.sigmaaldrich.com/life-science/your-favorite-gene-search.html). To examine the expression pattern of both USP9X and β-catenin in the SH3GL2-transfected lung cancer cells, we performed western blot analysis using antibodies specific to USP9X and (β-catenin. As shown in Fig. 5c, we observed a marked decrease in the expression level of both USP9X and phosphorylated (β-catenin (S675) in the SH3GL2-transfected NSCLC cell lines. Thus, forced overexpression of SH3GL2 also resulted in decreased expression level of USP9X and activated (β-catenin in the NSCLC cells. Since overexpression of SH3GL2 resulted in marked decrease in USP9X expression and their interaction was predicted as mentioned above, we performed co-immunoprecipitation analysis. Using total lysates from SH3GL2-transfected H1437 and H1944 cells, anti-SH3GL2 antibody effectively immunoprecipitates both SH3GL2 and USP9X (Fig. 5d).
Discussion
Lung cancer is a lethal disease with disappointing morbidity and development of new methods for early detection, monitoring, and therapeutic intervention are high priorities. Understanding the underlying molecular genetic changes involved in lung tumorigenesis is fundamental for developing improved diagnostic and treatment strategies. We performed a high-resolution genome wide scanning (Affymetrix 250K-NSpI array) of a small number of lung cancer patients. This array system has been shown to be a useful method for novel gene discovery in lung cancer [8]. We also procured multiple follow-up mucosal biopsies from these patients and scrutinized them in parallel. These biopsy specimens appeared abnormal under auto-fluorescence bronchoscopy but were histologically normal appearing [12]. In each case, tumor-free matched normal lymph nodes were used as control and all data were normalized as per Affymetrix guidelines and validated using standard data interpretation criteria to avoid false-positive results [8], Strikingly, one gene SH3GL2 that mapped to human chromosome 9p22 was found to be most frequently deleted in the tumors and corresponding all the histologically normal-appearing mucosal biopsies of the four patients. SH3GL2, also known as endophilin A1, is highly expressed in the brain and has an N-terminal BAR domain, a variable middle region, and a C-terminal SH3 domain [22]. It plays an important role in clathrin-dependent synaptic vesicle endo-cytosis. On the other hand, SH3GL2 also plays an important role in receptor tyrosine kinase (RTK) trafficking and signaling including EGFR [22]. Other than the RTKs, SH3GL2 was also implicated in the trafficking of other types of receptors including the beta-1 adrenergic receptor and the vesicular glutamate transporter [22]. Based on the frequent loss of SH3GL2 in various cancers, its emerging role in tumorigenesis has also been implicated [22].
In our pilot screening study, frequent deletion of SH3GL2 was observed in small subsets of patients with SCC histology [12]. These heavy smoker patients developed abnormal mucosal patches in the airway, which exhibited a number of clonal mitochondrial DNA mutations [12]. To identify nuclear genomic changes in these patients, we performed the genome wide scanning and identified frequent and clonal loss of SH3GL2 in the tumors and the corresponding normal-appearing mucosal biopsies. Remarkably, consistent SH3GL2 loss was evident in both SCC and ADC subtypes as documented in the GEO database (GSE #10245), indicating that SH3GL2 loss is not limited to specific subtype or among smokers. On the other hand, since ADC is the more frequently occurring subtype with both smoking and non-smoking history and showed SH3GL2 loss (GEO; GSE #10245), we examined a considerable number of primary NSCLC tumors with ADC histology. A cohort of never-smoker and smoker was chosen to determine any correlation with smoking. In some tested cases, LOH event appears to be accompanied with copy number loss as revealed by FISH analysis. Thus, copy number loss accompanied with frequent LOH at SH3GL2 locus appeared to be the major mechanism for inactivation of one allele in these samples. However, we did not detect any homozygous deletion of SH3GL2 in these cases. To our knowledge, this is the first report of frequent LOH and loss of protein expression of SH3GL2 in primary tumors of NSCLC patients. In our preliminary analysis, SH3GL2 loss was present and detectable in both the tumor and the corresponding histologically normal-appearing mucosa, thereby suggesting SH3GL2 as a potential tumor suppressor as proposed earlier [22]. The same tumors and corresponding normal-appearing mucosa also shared a number of mitochondrial DNA mutations [12]. Thus, the bron-choscopically abnormal but histologically normal-appearing mucosa harbored multiple nuclear and mitochondrial genetic changes, which may provide necessary drive for tumorigenic progression along with other acquired changes.
Recently, SH3GL2 loss was found to be associated with poor prognosis of the disease in a recent comprehensive analysis of 307 hepatocellular carcinoma patients [23]. Furthermore, LOH at SH3GL2 or loss of the locus containing SH3GL2 has been reported in breast, head and neck, and ovarian cancer [24–26]. Thus, it appears that SH3GL2 is a frequent target in different human cancers. Other than deletion analysis, we also performed promoter hypermethylation analysis of the SH3GL2 gene hypothesizing that methylation could be the mechanism for inactivation of the second allele of this gene. However, we did not find significantly higher methylation of SH3GL2 promoter in the 12 primary tumors analyzed. Notably, majority of these tumors harbored LOH of one allele. Since loss of gene expression by IHC analysis suggests complete inactivation of the SH3GL2 gene, mutation of the second copy of the SH3GL2 gene is possible, or other potential epigenetic mechanism(s) including micro-RNAs could be discovered.
In the present study, LOH and IHC analysis was performed in two different sets of specimens due to the lack of samples from the same cohort. However, we observed a similar frequency of SH3GL2 loss at both genomic and protein level, thereby indicating that SH3GL2 loss is a common event in these primary tumors.
In the first cohort of 60 cases, SH3GL2 loss was significantly higher among the never-smoker Asian patient population. Majority of these patients also harbored concomitant EGFR gene mutation. Overall, the never-smokers with EGFR gene mutation had a markedly higher (43 vs. 24 %) simultaneous SH3GL2 loss compared to the current smokers with KRAS gene mutation. However, it did not reflect any statistical significance likely due to a small sample size. Notably a subset of tumors, bearing simultaneous SH3GL2/ EGFR gene alteration(s) may have a selective growth advance for further progression. Identification of these changes in a substantial number of cases at various stages could be very useful for developing detection and monitoring strategies, which is our long-term goal. In the other cohort of 56 cases from Mayo Clinic, the patients with frequent loss of SH3GL2 were also never-smokers. However, we could not perform EGFR/KRAS mutation analysis in this cohort due to the lack of DNA samples, which is one of the drawbacks of this study.
In an earlier study conducted in HEK 293T and CHO cells, SH3GL2 with its binding partner CIN85 was shown to be involved in a ligand inducible interaction with EGFR, suggesting a role in controlling receptor endocytosis and receptor-mediated signaling [19]. It was further demonstrated in these cells that SH3GL2 with its constitutive binding partner CIN85 interacts with CbI and regulates EGFR internalization through ligand-specific stimulation [19]. By forced overexpression of SH3GL2 in three NSCLC cell lines, which minimally expressed endogenous SH3GL2 protein, we did not see any considerable changes in the level of expression of the wild-type CIN85 or cbl, the constitutive binding partner of SH3GL2 in the transfected cells. However, forced overexpression of SH3GL2 in these NSCLC cell lines significantly reduced active EGFR expression and possibly triggered its degradation via ubiquitin pathway as evident by increased phosphorylation of EGFR at Tyr1045 residue that creates a major docking site for c-Cbl that leads to EGFR ubiquitination and degradation following its activation [19, 20]. Notably, the complex responsible for EGFR degradation is composed of c-Cbl:SH3GL2:CIN85 [19]. Upon binding to EGF, EGFR is autophosphorylated and recruits the E3 ubiquitin ligase Cbl, which ubiquitylates EGFR and triggers its endocytosis (22). Besides serving as an ubiquitin ligase, Cbl also recruits SH3GL2 via the adapter protein CIN85, thereby allowing SH3GL2 to promote clathrin-mediated endocytosis of EGFR (19, 22). It is well-known that in never-smoker lung cancer patients, EGFR signaling pathway is specifically upregulated and transduces growth-promoting signal through the predominant STAT3 and AKT signaling pathways (4). In this context, loss of SH3GL2 could be beneficial for tumor progression through sustained EGFR signaling (wild type or mutant). Notably, we have seen simultaneous SH3GL2 loss and EGFR gene mutation in a subset of never-smoker lung cancer patients.
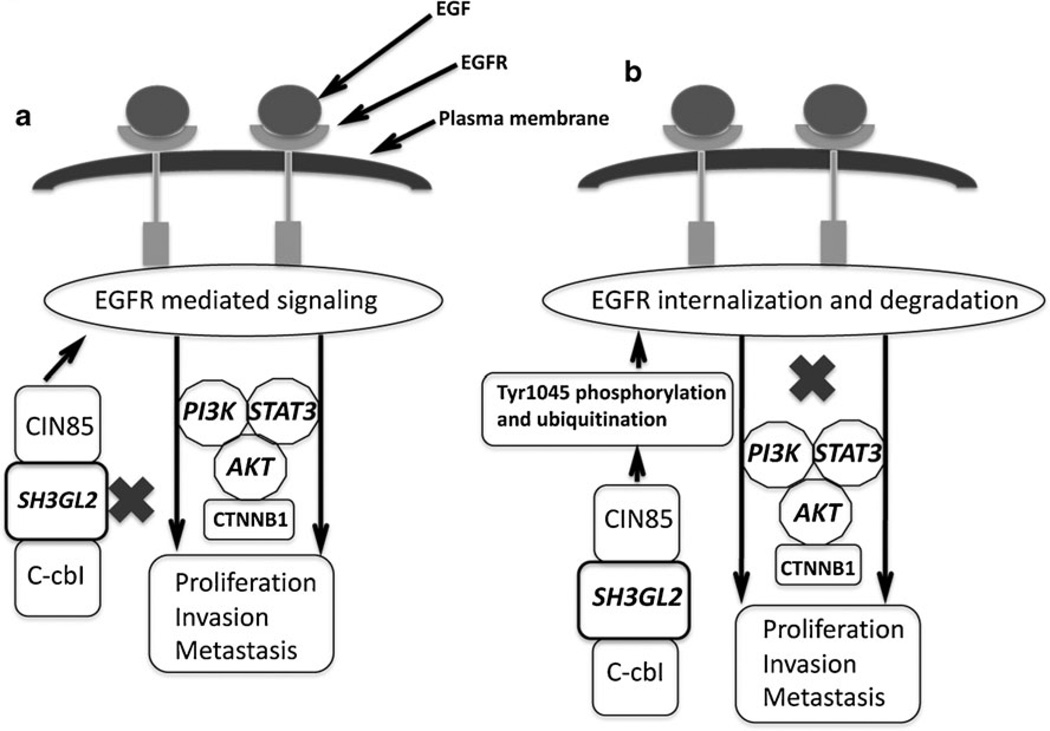
The canonical or non-canonical (β-catenin transactivation pathways play a major role in tumorigenesis keeping the interactor EGFR in the signaling loop as evident in numerous studies [27]. Some studies also demonstrated EGFR-PI3K-AKT-mediated transactivation of CTNNB1 pathway in prostate cancer [27]. Moreover, recent studies have shown that USP9X, a deubiquitinase can stabilize (β-catenin thereby inducing WNT signaling pathway [28]. Since SH3GL2 was predicted to interact with USP9X and we have observed down-regulation of PI3K and activated EGFR and AKT expression in the SH3GL2-transfected cells, we examined the status of USP9X and CTNNB1 in the SH3GL2-transfected cells. It is evident from our results that SH3GL2 downregulated the expression of both USP9X and activated CTNNB1 along with the concomitant low expression of activated EGFR and its downstream signaling molecules PI3K, activated AKT and STAT3. Moreover, a reciprocal interaction between SH3GL2 and USP9X was evident from the co-immunoprecipitation study indicating for a cross-talk between SH3GL2:USP9X in mediating tumor growth inhibition. Further studies are warranted to investigate the functional link between SH3GL2 and USP9X in lung tumorigenesis. Thus, it appears that these cascades of events blocking the EGFR signaling and associated CTNNB1-mediated transactivation pathway resulted in decreased tumor growth and invasion. Moreover, EGFR internalization and degradation markedly increased in the SH3GL2-overexpressing cells following EGF stimulation thereby restricting EGFR-mediated continuous signaling. As a result, our observed decreased tumor growth and invasion of the SH3GL2-overexpressing lung cancer cells were consistent with a perturbed EGFR signaling pathway. To our knowledge, this is also the first study to demonstrate a link between SH3GL2 and EGFR signaling pathway in NSCLC. Thus, loss of SH3GL2 from the CIN85–SH3GL2–Cbl complex that regulates EGFR endocytosis and signaling might allow the tumor cells to continuously receive growth-promoting signals through EGFR signaling (Fig. 6a). On the other hand, via the available wild-type SH3GL2, the normal functioning of the CIN85–SH3GL2–Cbl complex can be restored which in turn may inhibit EGFR signaling (Fig. 6b).
Fig. 6.
Schematic outline of the regulation of EGFR internalization and mediated signaling by the CIN85–SH3GL2–Cbl complex. a Deletion or inactivation of SH3GL2 (indicated by X) from this complex will allow tumor progression via continuous signaling through the EGFR pathway upon ligand (EGF) stimulation as depicted. b Presence of wild-type SH3GL2 and the appropriately assembled CIN85–SH3GL2– Cbl complex on the other hand will trigger EGFR internalization and subsequent degradation thereby resulting in decreased tumor growth via downregulation of the EGFR signaling pathway
In summary, we demonstrated in this study the frequent loss of SH3GL2 at both the DNA and protein level in primary NSCLC tumors and the impact of its forced expression on tumor growth, invasion, and active EGFR expression. Genomic loss of SH3GL2 and its expression appears to be an important and common inactivation event in lung cancer progression. Because its frequency of loss rivals that of any other discovered tumor suppressor gene, further work into its mechanism of action through EGFR signaling and other pathways is warranted.
Supplementary Material
Acknowledgments
This work was supported by EDRN-UO1 CA 084986 (DS); US–Egypt Joint Science and Technology fund (58-3148-169, SD), AD Williams (646299, SD) and Elsa U. Pardee Foundation (548424, SD), NIH-CA80127, and NIH-CA80354 (PY). PBF holds the Thelma Newmeyer Corman Chair in Cancer Research in the VCU Massey Cancer Center.
Footnotes
Conflict of interest No potential conflict of interests exists.
Electronic supplementary material The online version of this article (doi:10.1007/s00109-012-0955-3) contains supplementary material, which is available to authorized users.
Contributor Information
Santanu Dasgupta, Department of Human and Molecular Genetics, VCU Institute of Molecular Medicine, VCU Massey Cancer Center, Virginia Commonwealth University, School of Medicine, Richmond, VA, USA sdasgupta@vcu.edu.
Jin Sung Jang, Department of Medicine, Division of Pulmonary and Critical Care Medicine, Mayo Clinic, Rochester, MN, USA.
Chunbo Shao, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins University, Baltimore, MD, USA.
Nitai D. Mukhopadhyay, Department of Biostatistics, Virginia Commonwealth University, Richmond, VA, USA
Upneet K. Sokhi, Department of Human and Molecular Genetics, VCU Institute of Molecular Medicine, VCU Massey Cancer Center, Virginia Commonwealth University, School of Medicine, Richmond, VA, USA
Swadesh K. Das, Department of Human and Molecular Genetics, VCU Institute of Molecular Medicine, VCU Massey Cancer Center, Virginia Commonwealth University, School of Medicine, Richmond, VA, USA
Mariana Brait, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins University, Baltimore, MD, USA.
Conover Talbot, Institute for Basic Biomedical Sciences, Johns Hopkins University, Baltimore, MD, USA.
Rex C. Yung, Department of Pulmonary and Critical Care Medicine, Johns Hopkins University, Baltimore, MD, USA
Shahnaz Begum, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins University, Baltimore, MD, USA.
William H. Westra, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins University, Baltimore, MD, USA
Mohammad Obaidul Hoque, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins University, Baltimore, MD, USA.
Ping Yang, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA.
Joanne E. Yi, Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA
Stephan Lam, Department of Integrative Oncology, British Columbia Cancer Research Centre, Vancouver, BC, Canada.
Adi F. Gazdar, Hamon Center for Therapeutic Oncology Research, University of Texas Southwestern Medical Center, Dallas, TX, USA
Paul B. Fisher, Department of Human and Molecular Genetics, VCU Institute of Molecular Medicine, VCU Massey Cancer Center, Virginia Commonwealth University, School of Medicine, Richmond, VA, USA
Jin Jen, Department of Medicine, Division of Pulmonary and Critical Care Medicine, Mayo Clinic, Rochester, MN, USA.
David Sidransky, Department of Otolaryngology–Head and Neck Surgery, Johns Hopkins University, Baltimore, MD, USA dsidrans@jhmi.edu.
References
- 1.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thoracic Oncol. 2007;2:327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 2. http://www.cancer.gov/cancertopics/types/lung. Accessed 7 March 2012.
- 3.Andrews J, Yeh P, Pao W, Horn L. Molecular predictors of response to chemotherapy in non-small cell lung cancer. Cancer J. 2011;17:104–113. doi: 10.1097/PPO.0b013e318213f3cf. [DOI] [PubMed] [Google Scholar]
- 4.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nature Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 5.Jonsson S, Varella-Garcia M, Miller YE, Wolf HJ, Byers T, Braudrick S, Kiatsimkul P, Lewis M, Kennedy TC, Keith RL, et al. Chromosomal aneusomy in bronchial high-grade lesions is associated with invasive lung cancer. Am J Respir Crit Care Med. 2008;177:342–347. doi: 10.1164/rccm.200708-1142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varella-Garcia M, Chen L, Powell RL, Hirsch FR, Kennedy TC, Keith R, Miller YE, Mitchell JD, Franklin WA. Spectral karyotyping detects chromosome damage in bronchial cells of smokers and patients with cancer. Am J Respir Crit Care Med. 2007;6:505–512. doi: 10.1164/rccm.200609-1329OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massion PP, Zou Y, Uner H, Kiatsimkul P, Wolf HJ, Baron AE, Byers T, Jonsson S, Lam S, Hirsch FR, et al. Recurrent genomic gains in preinvasive lesions as a biomarker of risk for lung cancer. PLoS One. 2009;4:e5611. doi: 10.1371/journal.pone.0005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockwood WW, Chari R, Coe BP, Thu KL, Garnis C, Malloff CA, Campbell J, Williams AC, Hwang D, Zhu CQ, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med. 2010;7:e1000315. doi: 10.1371/journal.pmed.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas RK, Weir B, Meyerson M. Genomic approaches to lung cancer. Clin Cancer Res. 2006;12:4384s–4391s. doi: 10.1158/1078-0432.CCR-06-0098. [DOI] [PubMed] [Google Scholar]
- 12.Dasgupta S, Yung RC, Westra WH, Rini DA, Brandes J, Sidransky D. Following mitochondrial footprints through a long mucosal path to lung cancer. PLoS One. 2009;4:e6533. doi: 10.1371/journal.pone.0006533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta S, Koch R, Westra WH, Califano JA, Ha PK, Sidransky D, Koch WM. Mitochondrial DNA mutation in margins and tumors of recurrent HNSCC patients. Can Prev Res. 2010;3:1205–1211. doi: 10.1158/1940-6207.CAPR-10-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasgupta S, Soudry E, Mukhopadhyay N, Shao C, Yee J, Lam S, et al. Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol. 2011;227(6):2451–2460. doi: 10.1002/jcp.22980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta S, Bhattacharya-Chatterjee M, O’Malley BW, Jr., Chatterjee SK. Reversal of immune suppression following vaccination with recombinant vaccinia virus expressing IL-2 in an orthotopic murine model of head and neck squamous cell carcinoma. Cancer Therapy. 2004;2:375–388. [Google Scholar]
- 16.Dasgupta S, Hoque MO, Upadhyay S, Sidransky D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res. 2008;68:700–706. doi: 10.1158/0008-5472.CAN-07-5532. [DOI] [PubMed] [Google Scholar]
- 17.Szymkiewicz I, Kowanetz K, Soubeyran P, Dinarina A, Lipkowitz S, Dikic I. CIN85 participates in Cb-l-b down-regulation of receptor tyrosine kinases. J Biochem. 2002;277:39666–39672. doi: 10.1074/jbc.M205535200. [DOI] [PubMed] [Google Scholar]
- 18.Dasgupta S, Hoque MO, Upadhyay S, Sidransky D. Forced cytochrome B gene mutation expression induces mitochondrial proliferation and prevents apoptosis in human uroepithelial SV-HUC-1 cells. Int J Cancer. 2009;125:2829–2835. doi: 10.1002/ijc.24701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soubyran S, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. CbI-CIN85-endophlin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 20.Ettenberg SA, Keane MM, Nau MM, Frankel M, Wang LM, Pierce JH, Lipkowitz S. cbl-b inhibits epidermal growth factor receptor signaling. Oncogene. 2009;18:1855–1866. doi: 10.1038/sj.onc.1202499. [DOI] [PubMed] [Google Scholar]
- 21.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-CbI/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 22.Kjaerulff O, Brodin L, Jung A. The structure and function of endophilin proteins. Cell Biochem Biophys. 2011;60:137–154. doi: 10.1007/s12013-010-9137-5. [DOI] [PubMed] [Google Scholar]
- 23.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinha S, Chunder N, Mukherjee N, Alam N, Roy A, Roychoudhury S, Panda CK. Frequent deletion and methylation in SH3GL2 and CDKN2A loci are associated with early- and late-onset breast carcinoma. Ann Surg Oncol. 2008;5:1070–1080. doi: 10.1245/s10434-007-9790-0. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh A, Ghosh S, Maiti GP, Sabbir MG, Alam N, Sikdar N, Roy B, Roychoudhury S, Panda CK. SH3GL2 and CDKN2A/2B loci are independently altered in early dysplastic lesions of head and neck: correlation with HPV infection and tobacco habit. J Pathol. 2009;217:408–419. doi: 10.1002/path.2464. [DOI] [PubMed] [Google Scholar]
- 26.Osterberg L, Levan K, Partheen K, Delle U, Olsson B, Sundfeldt K, Horvath G. Potential predictive markers of chemotherapy resistance in stage III ovarian serous carcinomas. BMC Cancer. 2009;9:368. doi: 10.1186/1471-2407-9-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu T, Li C. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236. doi: 10.1186/1476-4598-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vucic D, Dixit VM, Wertz IE. Ubiquitination in apoptosis: a post translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.